Abstract
Particle tracking is an invaluable technique to extract quantitative and qualitative information regarding the transport of nanomaterials through complex biological environments. This technique can be used to probe the dynamic behavior of nanoparticles as they interact with and navigate through intra- and extra-cellular barriers. In this article, we focus on the recent developments in the application of particle-tracking technology to nanomedicine, including the study of synthetic and virus-based materials designed for gene and drug delivery. Specifically, we cover research where mean square displacements of nanomaterial transport were explicitly determined in order to quantitatively assess the transport of nanoparticles through biological environments. Particle-tracking experiments can provide important insights that may help guide the design of more intelligent and effective diagnostic and therapeutic nanoparticles.
Keywords: extracellular, intracellular, nanoparticle, particle tracking, virus
The ability to navigate intra- and extra-cellular barriers successfully is critical if nanoparticles (NPs) are to achieve diagnostic and/or therapeutic goals. This is no trivial task. The complex biological environments NPs must travel through are full of steric and adhesive obstacles that can severely limit effective targeting and delivery. NPs do not always behave as expected, owing to nanoscopic interactions that are not detected at the macroscopic level. Investigative tools to probe the behavior of NPs at the length scale of the particles may unveil important information that can be used to improve the design of engineered nanomaterials.
Quantitative particle tracking is one such technique. With particle tracking, transport properties of individual particles (or aggregates of particles depending on the colloidal status) can be extracted with high resolution. Quantitative parameters, such as diffusivity and velocity, and qualitative parameters, such as mode and directionality of transport, can be obtained for each NP. From such information, much insight can be attained regarding the biophysics characterizing the transport of nanomaterials through the complex intra- and extra-cellular environments. The dynamics of NP transport and their interaction with cellular or tissue components can be quantified in a simple, direct manner.
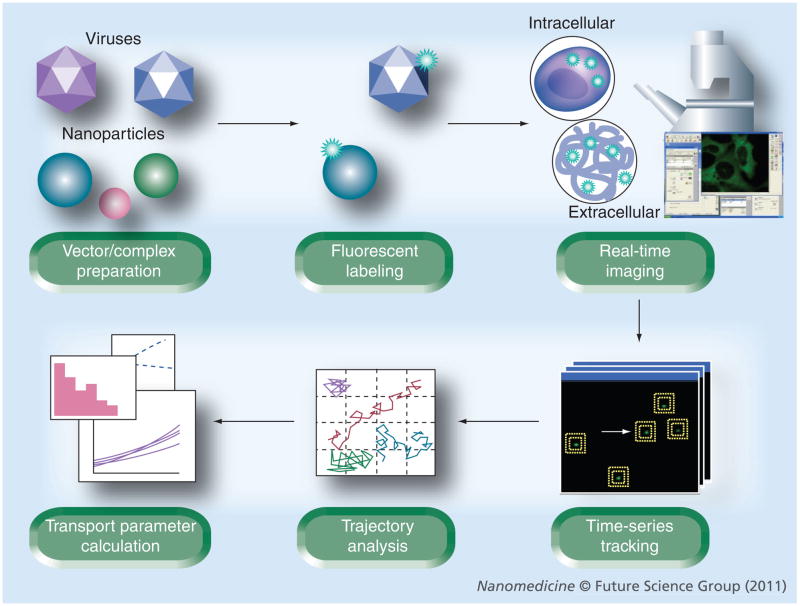
The experiment workflow for particle tracking is shown in Figure 1. Some nanomaterials, such as single-walled carbon nanotubes (SWNTs), exhibit intrinsic fluorescence that can be used for imaging. NPs lacking intrinsic fluorescence are first labeled and then added to either cells or extracellular models. A fluorescence/confocal microscope is used to capture high-resolution movies documenting the transport of NPs through the respective biological environments. Using commercially available, free or custom software, the movies are analyzed to extract x and y positional data over time. The obtained trajectories can be visually inspected for qualitative information regarding NP transport, and the positional data is used to calculate mean square displacement (MSD). In most cases, the optical slice of an image is much smaller than its x–y dimensions; therefore, the tracked particle movement can be simplified as 2D. MSD in two dimensions of a given trajectory of x,y coordinates is defined as [1]:
where xi and yi are the positions for each time point indicated by i, N is the total number of frames (or points of a trajectory), Δt is time between frames, and n is number of time intervals. Based on MSD versus t relationships, particle motion can be classified into four transport modes [2–4]:
Figure 1. Workflow for particle-tracking experiments.
Nanomaterials lacking intrinsic fluorescence are labeled with fluorophores. After adding nanoparticles to live cells or extracellular models, a confocal or epifluorescence microscope equipped with a charge-coupled device camera or photomultiplier tube is used to capture time series images at the desired temporal resolution. The movies are processed with commercial, free or custom software to generate x,y coordinate datasets to reconstruct trajectories of particles. Data are analyzed to calculate transport parameters, such as mean square displacement, diffusion coefficient and velocity.
- Normal diffusion (diffusion mode):
- Anomalous diffusion (hindered mode):
- Directed motion with diffusion (active transport mode):
-
Corralled diffusion (confined diffusion mode):
where D is the microscopic diffusion coefficient (α < 1), v is the velocity, C is the corral size, and A1 and A2 are constants determined by the corral geometry. Readers are directed to reviews elsewhere for a more in-depth description of particle-tracking technology [3–7].
In this article, we present contemporary research that applies particle-tracking technology to the study of synthetic and virus-based materials specifically designed for nanomedicine (Tables 1 & 2). We focus on primary reports that have quantified NP transport through the determination of MSD – the basic requirement for particle-tracking technology. The studies presented in this article help the nanomedicine community understand the behavior and transport mechanisms of engineered NPs, and provide a quantitative means for investigating the effects of NP physicochemical properties on transport through biological environments. Such insights can profoundly impact the way nanomaterials are designed for biomedical applications.
Table 1.
Intracellular nanoparticle transport studies using particle tracking.
| Author (year) | Vector | Vector type | Cargo | Diameter (nm) | Label | Cell | Microscope | Tracking software | Transport parameters | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Suh et al. (2003) | PEI | Nonviral | Salmon DNA | 156 | OG514-PEI | COS-7 | EF | MetaMorph | MSD, RC | [13] |
| Suh et al. (2004) | PEI | Nonviral | Salmon DNA | 138 | OG514-PEI | COS-7 | EF | MetaMorph | MSD, D | [14] |
| de Bruin et al. (2007) | LPEI, EGF-PEG-PEI | Nonviral | pDNA | 85–266 | Cy5-DNA Cy3-DNA |
HuH7 | WFF | MATLAB | MSD, D, v | [16] |
| Bausinger et al. (2006) | PEI | Nonviral | pCMVLuc | 85, 165 | Cy3-DNA | HuH7 | Confocal | Unspecified | MSD, D, v | [15] |
| Suh et al. (2007) | PEG-COOH-PS | Nonviral | None | 100 | Proprietary dye | HeLa | EF | MetaMorph | MSD, D, RC | [20] |
| Lai et al. (2007) | COOH-PS | Nonviral | None | 20, 40, 200 | Proprietary dye | HeLa, PHUVEC | Confocal | MetaMorph | MSD, D | [22] |
| Lai et al. (2008) | COOH-PS | Nonviral | None | 24, 43 | Proprietary dye | HeLa | Confocal | MetaMorph | MSD, D, RC | [21] |
| Sauer et al. (2009) | MgNP, lipoplex | Nonviral | GFP Nuc pDNA | 388 | TMR-MgNP, Cy5-DNA | HuH7 | WFF | MATLAB | MSD, D, v | [18] |
| Gopalakrishnan et al. (2006) | Lipid/QD | Nonviral | None | 100 | Intrinsic | HEK293 | Confocal | Unspecified | MSD | [19] |
| Jin et al. (2008) | SWNT | Nonviral | D(GT)15 | 2 × 20 | Intrinsic | NIH3T3 | Fluorescence | ImageJ | MSD | [2] |
| Neugart et al. (2007) | Nanodiamond | Nonviral | None | 50 | Intrinsic | HeLa | Confocal | Unspecified | MSD, D | [17] |
| Lakadamyali et al. (2003) | Influenza X31 | Viral | Unspecified | Unspecified | DiD, Cy3, CypHer5 | CHO HeLa |
DIC, EF | Unspecified | MSD, v | [24] |
| Babcock et al. (2004) | Influenza X31, vRNP | Viral | Unspecified | 30–100 | Cy3-vRNP | BS-C-1 | DIC, EF | Unspecified | MSD, v | [25] |
| Suk et al. (2007) | Ad, PEI | Viral | Salmon DNA | 143 | OG488-PEI GFP-Ad |
Rat primary neuron | Confocal | MetaMorph | MSD, D, RC | [26] |
| Akita et al. (2010) | R8-lip, Ad | Viral | pDNA | 189 | Rho-lip, TR-Ad | HeLa | WFF | G-track | MSD, D, v | [27] |
| Seisenberger et al. (2001) | AAV | Viral | Unspecified | 20 | Cy5-AAV | HeLa | EF | Unspecified | MSD | [23] |
AAV: Adeno-associated virus; Ad: Adenovirus; APS: Amine-modified polystyrene; CD: Cyclodextrin; D: Diffusion coefficient; DIC: Differential interference contrast; DiD: 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine; EF: Epifluorescence; FCS: Fluorescence correlation spectroscopy; GFP: Green fluorescent protein; HSNF: Human skin normal fibroblast; ICS: Spatio–temporal image correlation spectroscopy; LPEI: Linear polyethylenimine; MgNP: Magnetic nanoparticle; MSD: Mean square displacement; OG: Oregon Green; PEG: Poly(ethylene glycol); PEI: Polyethylenimine; PHUVEC: Primary human umbilical vein endothelial cells; PS: Polystyrene; QD: Quantum dot; R8-lip: Octa-arginine-modified liposome; RC: Relative change; Rho: Rhodamine; SWNT: Single-wall nanotube; TMR: Tetramethylrhodamine; TR: Texas Red; v: Velocity; vRNP: Ribonucleoprotein; WFF: Wide-field fluorescence.
Table 2.
Extracellular nanoparticle transport studies using particle tracking.
| Author (year) | Vector | Diameter (nm) | Label | ECM | Microscope | Tracking software | Transport parameters | Ref. |
|---|---|---|---|---|---|---|---|---|
| Lai et al. (2007) | PEG-COOH-PS | 100–500 | Proprietary dye | CVM | EF | MetaMorph | MSD, D | [28] |
| Wang et al. (2008) | PEG-COOH-PS | 200 | AF 647-PS | CVM | EF | MetaMorph | MSD, D | [30] |
| Tang et al. (2009) | PSA, PSA-PEG, PLGA | 100–200 | Dox/PSA, PLGA | CVM, CFS | EF | MetaMorph, MATLAB | MSD | [32] |
| Lai et al. (2009) | PEG-COOH-PS | 100, 200, 500, 1000 | Proprietary dye | CVM | EF | MetaMorph | MSD | [34] |
| Lai et al. (2010) | PEG-COOH-PS, HSV-1 | 100–1000, 180 | Proprietary dye-PS AF488-HSV GFP-HSV |
CVM | EF | MetaMorph | MSD, D | [35] |
| Dawson et al. (2003) | COOH-PS APS |
100–500 | Proprietary dye | CFS | EF, confocal | MetaMorph | MSD, D | [29] |
| Suk et al. (2009) | COOH-PS | 100, 200, 500 | Proprietary dye | CFS | EF | MetaMorph | MSD, D | [36] |
| Dawson et al. (2004) | PLGA-DDAB COOH-PS |
<200 | Proprietary dye | PGM | Confocal | MetaMorph | MSD, D | [33] |
| Cu and Saltzman (2009) | PEG-PLGA | 170 | Coumarin 6 | CM | Fluorescence | ImageJ | D | [31] |
| Kawai et al. (2009) | QD705, fluosphere | 20, 40, 100 | Intrinsic | Interstitium | EF | Unspecified | MSD, D, v | [1] |
APS: Amine-modified polystyrene; CFS: Cystic fibrosis sputum; CM: Cervical mucus; CVM: Cervicovaginal mucus; D: Diffusion coefficient; Dox: Doxorubicin; ECM: Extracellular matrix; EF: Epifluorescence; GFP: Green fluorescent protein; HSV: Herpes simplex virus; MSD: Mean square displacement; PEG: Poly(ethylene glycol); PGM: Pig gastric mucus; PLGA: Poly(D,L-lactic-coglycolic) acid; PS: Polystyrene; PSA: Poly(sebacic acid); QD: Quantum dot; v: Velocity.
Intracellular transport of synthetic NPs
Investigation of NP intracellular transport encompasses identifying endocytic pathways, quantifying transport rates and recognizing transport modes. This information can be used to pinpoint rate-limiting steps to intracellular NP delivery, which in turn will highlight ways to improve delivery efficiency. Towards this goal, the intracellular transport of a number of nanomaterials, including polyethylenimine (PEI), polystyrene (PS), carbon nanotubes, lipid/quantum dot polyplexes, magnetic NPs and nanodiamonds has been investigated with particle tracking.
Particle tracking has revealed important quantitative information regarding the cellular bottlenecks to gene delivery with synthetic vectors. The first nanomaterial to be studied with particle tracking was PEI, a cationic polymer used for its ability to condense negatively charged DNA [8] and mediate gene delivery [9–12]. In the study by Suh et al., the subcellular transport of PEI NPs was studied in live cells. The average size of the PEI NPs in this study was 156 nm [13,14]. Using real-time particle tracking, PEI/DNA nanocomplexes were shown to efficiently transport to the perinuclear region in a microtubule-dependent manner, suggesting that cytoplasmic transport of these NPs is not rate limiting. Therefore, efforts to improve cytoplasmic transport are not necessary for PEI/DNA complexes. Bausinger et al. investigated the transport of PEI NPs from surface binding to cell division [15]. They concluded that PEI/DNA complexes are engulfed by the cell membrane, and molecular motors then facilitate the active transport of PEI NP-containing vesicles along microtubules. During mitosis, polyplexes are associated with movement along microtublues of the spindle apparatus with a speed corresponding to motor proteins. de Bruin showed that targeting PEI NPs via attachment of EGF ligands speeds up their internalization compared with unmodified controls [16]. The uptake of NPs was classified into three phases, and phase 1 – involving particle internalization – was shortened, based on the fraction of particles that were able to move into the cell before all outside particles were quenched. Thus, conjugation of bioactive ligands may improve the rate of endocytosis in addition to benefits regarding target specificity.
Nanoparticles internalized by endocytosis can also undergo exocytosis as a mechanism by which a cell clears these exogenous materials. Particle tracking offers the capability of measuring rates of NP exocytosis in combination with endocytosis. A potential drug and gene delivery vector, SWNTs, have been investigated on this subject by Jin et al. [2]. SWNTs have a high aspect ratio and may potentially be able to deliver multiple genes to cells more efficiently. The average size of the SWNTs in this study was 1.6 nm in diameter. Over 10,000 trajectories of DNA-wrapped SWNTs were captured and endocytosis, intracellular trafficking and exocytosis of SWNTs were identified in NIH-3T3 cells. The authors concluded that the rates of endocytosis and exocytosis were regulated by the cell to keep the intracellular concentration of NPs below cytotoxic levels.
Not all types of NPs exhibit active transport behavior in cells; some are immobilized or slowed down in certain cellular compartments. Furthermore, endosomal escape of NPs has been observed to be a difficult barrier for intracellular delivery. Nanodiamonds with a doped-color center and with low cytotoxicity are being investigated as an alternative NP for live-cell imaging owing to their luminescent properties. Neugart et al. studied the dynamic behavior of 50-nm nanodiamonds in living cells [17]. When incubated with HeLa cells for 3 h, 60% of nanodiamonds interact with the cells, but only 10% are internalized. The nanodiamonds are seen distributed uniformly inside cells and are immobilized in the endosomal pathway. By attaching subcellular targeting ligands to the nanodiamonds, the intracellular distribution of the particles can potentially be altered allowing for the imaging of other subcellular structures. Other nanomaterials investigated with particle tracking include magnetic NPs [18] and quantum dots. Tetramethylrhodamine-labeled iron oxide magnetic particles complexed with Cy5-labeled DNA and cationic lipid were found to have similar internalization dynamics and intracellular behavior as PEI [16]. The endosomal release of complexes takes as long as 24 h, suggesting this barrier is rate-limiting. In Gopalakrishnan et al., researchers performed particle tracking on quantum dots that were embedded in the membranes of lipid vesicles and showed that the NPs are successfully immobilized within the lipid bilayers [19].
By comparing transport rates of different NP designs with particle tracking, the validity of design improvements can be determined quantitatively. For example, once NPs escape endosomes into the cytoplasm, they may need to move around to reach target organelles, proteins or biomolecules, for example, to carry out their function. Conjugation of polyethylene glycol (PEG) to NP surfaces, or PEGylation, can prevent nonspecific adhesion to intracellular structures and endow NPs with sufficient mobility. Suh et al. microinjected PEGylated PS or unmodified PS into live cells, and used particle tracking to quantify and compare their transport rates [20]. Notably, the diffusivities of NPs increase by 100% upon PEGylation. Analysis of transport modes indicates that 80% of the unmodified NPs experience hindered movement in the crowded cytoplasmic microenvironment. In comparison, only 50% of PEGylated NPs are hindered. Thus, PEGylation can substantially improve the cytoplasmic transport of PS NPs.
Effect of NP size on intracellular transport can also be assessed with particle tracking. Investigating different-sized carboxylated PS (COOH-PS) NPs has revealed a nondegradative trafficking pathway for nanomaterials, resulting in accumulation around the Golgi and endoplasmic reticulum (ER). Specifically, Lai et al. discovered a privileged nondegradative intracellular pathway for 24-nm NPs that is clathrin and caveolae independent [21,22]. By comparison, 43-nm NPs take the classic degradative pathway. The nonacidic vesicles in this pathway display hindered, diffusive and active transport modes, as well as the pearls-on-a-string motion on micro-tubules (possibly motor protein facilitated). The difference in dynamics is characterized by a lower amount of budding and fusion events for vesicles carrying 24-nm NPs than those carrying 43-nm NPs. The result of the alternative transport pathway is the accumulation of NPs around the Golgi and ER in nondegradative vesicles. This prolonged retention time of NPs near the nucleus can be advantageous when delivering therapeutic agents to this target organelle.
Intracellular transport of viral NPs
In contrast to synthetic gene-delivery vectors, viral vectors have been developed by nature over millions of years. Virus-based gene-delivery vectors, despite some intrinsic disadvantages, including immunogenicity, are currently the most popular platforms for gene therapy. Investigating their transport through intracellular barriers using high-resolution particle tracking can be highly useful in elucidating the infectious pathway quantitatively. This provides invaluable information for the improvement of viral vectors, as well as inspiring new ways to create non-viral vectors. One of the first viruses to be studied with particle tracking is the adeno-associated virus (AAV) [23]. AAV particles were labeled with a single Cy5 dye molecule and their transport within live cells was documented. MSD calculations were used to draw conclusions about the different modes of virus transport as the viruses moved through the cell cytoplasm and into the nucleus. This study yielded the first clues on the short-range dynamics of intracellular AAV transport. In Lakadamyali et al., the authors overcame previous difficulties in labeling the influenza virus to be able to perform quantitative single-virus particle tracking and investigate the transport and fusion of viruses [24]. The results indicate that there are three stages of virus transport dependent on actin filaments and microtubules. In Babcock et al., the authors studied influenza ribonucleoproteins and found that they diffuse through the cytoplasm and the nucleus [25]. Collectively, particle-tracking studies of AAV and influenza have further verified the important role of cytoskeletal elements in the effective infection process of viruses. Ability of synthetic NPs to harness these same intracellular highways may be an important design criterion.
Particle tracking is a useful technique to compare the quantitative transport of viruses and synthetic NPs side-by-side. Adenovirus is a popular viral gene-delivery vector exploited for its ability to efficiently deliver relatively large amounts of DNA to both dividing and nondividing cells. In Suk et al., the intracellular transport of adenoviruses was compared with that of nonviral PEI NPs using particle tracking [26]. Results indicate that adenoviruses escape endosomes more efficiently than PEI NPs, potentially explaining why these viral vectors are more effective at delivering genes compared with the nonviral vectors. In Akita et al., the authors compared the cytoplasmic transport of octaarginine-modified liposome (R8-lip) to that of adenovirus [27]. R8-lipo was directionally transported within vesicular compartments at a rate slower than that of adenoviruses that underwent fast directional transport on micro-tubules. By comparing the transport properties of efficient virus vectors to less efficient nonviral vectors, clues to how nonviral vector designs can be improved may be unveiled.
Extracellular transport of NPs
The effective transport of NPs through extracellular barriers, such as mucus and interstitium, may be critically important in order to achieve the desired therapeutic and/or diagnostic goals. Particle tracking can be an invaluable tool to probe the quantitative transport properties of engineered nanomaterials as they move through these complex barriers. Information gathered from particle-tracking experiments can feedback into the design process to improve NP physicochemical properties and reach design aims.
Mucus is composed of mucin fibers in a low viscosity interstitial fluid, and protects the body against toxic particles and pathogens by steric exclusion and adhesive trapping. Pathogens immobilized in mucus are then cleared rapidly. These same functions, however, also make mucus a potentially critical barrier to the transport of NPs if the target cells are underneath the mucus layer. NPs need to evade mucoadhesive trapping, navigate the lower viscosity pores of the gel and transport through to the other side rapidly before clearance can occur.
The transport behavior of NPs through mucus may be difficult to predict at the nanoscale. This is where particle-tracking technology can prove to be invaluable. In Lai et al., the authors tracked the transport of COOH-PS NPs of various physicochemical properties in mucus to determine which properties allowed the NPs to diffuse fastest through mucus. Surprisingly, they found that 200–500-nm particles are able to move faster in mucus than 100-nm particles [28]. This study exemplifies this nonintuitive behavior, so particles must be studied carefully using quantitative tools, such as particle tracking. In cystic fibrosis (CF) sputum, 100-, 200- and 500-nm COOH-PS NPs have large variant transport rates, with smaller NPs having few but fast-moving populations [29]. Furthermore, COOH-PS NPs may be more adhesive to CF sputum than amine-modified particles.
To improve the mucus transport of NPs, the effect of an antiadhesive coating provided by PEGylation was investigated with particle tracking. In Wang et al., the authors looked at different levels of PEGylation of COOH-PS NPs and their ability to speed-up travel through mucus [30]. Results indicate high coverage provided by the smaller 2-kDa molecular weight PEG is best for this purpose and produces a 1000-fold increase in MSD. In Cu et al., a diffusion model based on Fickian mass transport was used to solve the effective diffusion coefficients of various PEGylated and unmodified NPs in cervical mucus [31]. NPs of 170 nm and coated with PEG to neutralize surface charge move unhindered in the mucosal gel, suggesting PEGylated vectors can deliver greater drug doses to the underlying epithelium. PEGylation of a NP made from a different material, polysebacic acid, increases MSD two- to ten-fold through mucus [32].
Using a different material itself can impact transport rates as sub-200-nm poly (D,L-lactic-coglycolic) acid NP can travel through mucus ten-times faster than similarly sized PS NPs [33]. The authors suggest that differences in particle surface hydrophilicity and aggregate–mucus interaction may have been responsible for the difference in transport between the two particles. Lai et al. went on to develop different-sized nonadhesive PEGylated COOH-PS NPs, and quantified their transport through cervicovaginal mucus using particle tracking [34]. Results indicate mucus becomes highly permeable to NPs smaller than 500 nm, which makes small non-adhesive delivery systems ideal for mucosal delivery. In fact, the engineered NPs display greater transport rates than herpes simplex viruses that are even slightly smaller in size [35]. Remarkably, effective diffusivities of the NPs in cervicovaginal mucus are just slightly lower than in water. In Suk et al., the authors were interested in developing a delivery vector that would be able to penetrate CF mucus [36]. They discovered that PEGylated COOH-PS NPs up to 200 nm were able to travel through this mucus without sticking.
Delivering NPs to the tumor interstitium is an important goal for many NP-based technologies. Studying this process in a quantitative fashion will unveil ways to improve NP design. Notably, Kawai et al. quantitatively characterized the transport of model drug delivery vectors in vivo through tumor vessels and the interstitium after intravascular injection [1]. This study uncovered the behavior of different-sized NPs within different interstitial spaces. NP trajectories were constructed from confocal images taken at the perivascular, interstitia and intercellular areas of the xenograft tumor. Experiments show MSD, velocity and diffusion coefficients of the NPs are inversely related to their size. The findings provide guidelines for designing therapeutic NPs that can effectively transport through interstitium to reach tumor cells.
Future perspective
Quantitative particle tracking has proven to be invaluable for studying the transport of biomedical nanomaterials within live cells and through extracellular environments. A number of quantitative and qualitative transport parameters can be determined to provide a comprehensive picture of NP transport. Continued investigation of NP transport in a quantitative fashion should reveal more ways to improve the design of current vectors. Correlations between NP physiochemical properties and transport behavior obtained with particle-tracking experiments can be powerful design paradigms for creating the next generation of sophisticated materials for nanomedicine. For example, by choosing the right combination of NP size and surface properties, transport through extracellular environments can be controlled. All of the examples presented in this article are focused on developing NPs with faster transport through the extracellular space. Future work in this area may involve generation of NPs with more sophisticated transport behaviors, such as those that can switch between different modes of transport depending on the local conditions. Furthermore, by quantitatively characterizing how viruses transport within live cells, ideas of how to improve the efficiency of nonviral vectors can be obtained. Using nature as inspiration may yield the next generation of biomimetic artificial viruses that can effectively and intelligently transport to target subcellular sites and mediate desired diagnostic and/or therapeutic functions.
Executive summary.
Particle tracking is a useful technique to investigate nanoparticle transport, both quantitatively and qualitatively.
Nanoparticle transport through intra- and extra-cellular barriers can be studied with particle tracking.
Experimental workflow for particle tracking involves labeling nanoparticles, capturing movies with a fluorescence/confocal microscope, analyzing the movies and calculating transport parameters, such as diffusivity and velocity.
Intra- and extra-cellular transport of a variety of synthetic nanoparticles has been studied with particle tracking.
Physicochemical properties affect intra- and extra-cellular transport behavior of nanoparticles.
Transport of a handful of viruses has been studied with particle tracking.
Further correlations between nanoparticle properties and biological transport behavior can serve as design paradigms for creating the next generation of sophisticated nanomaterials with novel functions.
Footnotes
Financial & competing interests disclosure
This work was supported by the NIH (1RC2GM092599–01), National Science Foundation Center for Biological and Environmental Nanotechnology (EEC-0647452), NIH Nanobiology Interdisciplinary Graduate Training Program (T32EB009379) to C Dempsey, and National Science Foundation Research Experiences for Undergraduates (DBI-1004476) summer fellowship to D Chona. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
For reprint orders, please contact: reprints@futuremedicine.com
Bibliography
- 1.Kawai M, Higuchi H, Takeda M, Kobayashi Y, Ohuchi N. Dynamics of different-sized solid-state nanocrystals as tracers for a drug-delivery system in the interstitium of a human tumor xenograft. Breast Cancer Res. 2009;11(4):R43. doi: 10.1186/bcr2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin H, Heller DA, Strano MS. Single-particle tracking of endocytosis and exocytosis of single-walled carbon nanotubes in NIH-3T3 cells. Nano Lett. 2008;8(6):1577–1585. doi: 10.1021/nl072969s. [DOI] [PubMed] [Google Scholar]
- 3.Saxton MJ, Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 4.Suh J, Dawson M, Hanes J. Real-time multiple-particle tracking: applications to drug and gene delivery. Adv Drug Deliv Rev. 2005;57(1):63–78. doi: 10.1016/j.addr.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Kusumi A, Sako Y, Yamamoto M. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys J. 1993;65(5):2021–2040. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai SK, Hanes J. Real-time multiple particle tracking of gene nanocarriers in complex biological environments. Methods Mol Biol. 2008;434:81–97. doi: 10.1007/978-1-60327-248-3_6. [DOI] [PubMed] [Google Scholar]
- 7.Qian H, Sheetz MP, Elson EL. Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys J. 1991;60(4):910–921. doi: 10.1016/S0006-3495(91)82125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Utsuno K, Uludag H. Thermodynamics of polyethylenimine-DNA binding and DNA condensation. Biophys J. 2010;99(1):201–207. doi: 10.1016/j.bpj.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boussif O, Lezoualc’h F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Aneed A. An overview of current delivery systems in cancer gene therapy. J Control Release. 2004;94(1):1–14. doi: 10.1016/j.jconrel.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. J Control Release. 1999;60(2–3):149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- 12.Lungwitz U, Breunig M, Blunk T, Gopferich A. Polyethylenimine-based non-viral gene delivery systems. Eur J Pharm Biopharm. 2005;60(2):247–266. doi: 10.1016/j.ejpb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Suh J, Wirtz D, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc Natl Acad Sci USA. 2003;100(7):3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suh J, Wirtz D, Hanes J. Real-time intracellular transport of gene nanocarriers studied by multiple particle tracking. Biotechnol Prog. 2004;20(2):598–602. doi: 10.1021/bp034251y. [DOI] [PubMed] [Google Scholar]
- 15.Bausinger R, Von Gersdorff K, Braeckmans K, et al. The transport of nanosized gene carriers unraveled by live-cell imaging. Angew Chem Int Ed Engl. 2006;45(10):1568–1572. doi: 10.1002/anie.200503021. [DOI] [PubMed] [Google Scholar]
- 16.de Bruin K, Ruthardt N, Von Gersdorff K, et al. Cellular dynamics of EGF receptor-targeted synthetic viruses. Mol Ther. 2007;15(7):1297–1305. doi: 10.1038/sj.mt.6300176. [DOI] [PubMed] [Google Scholar]
- 17.Neugart F, Zappe A, Jelezko F, et al. Dynamics of diamond nanoparticles in solution and cells. Nano Lett. 2007;7(12):3588–3591. doi: 10.1021/nl0716303. [DOI] [PubMed] [Google Scholar]
- 18.Sauer AM, De Bruin KG, Ruthardt N, Mykhaylyk O, Plank C, Brauchle C. Dynamics of magnetic lipoplexes studied by single particle tracking in living cells. J Control Release. 2009;137(2):136–145. doi: 10.1016/j.jconrel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Gopalakrishnan G, Danelon C, Izewska P, et al. Multifunctional lipid/quantum dot hybrid nanocontainers for controlled targeting of live cells. Angew Chem Int Ed Engl. 2006;45(33):5478–5483. doi: 10.1002/anie.200600545. [DOI] [PubMed] [Google Scholar]
- 20.Suh J, Choy KL, Lai SK, et al. PEGylation of nanoparticles improves their cytoplasmic transport. Int J Nanomedicine. 2007;2(4):735–741. [PMC free article] [PubMed] [Google Scholar]
- 21.Lai SK, Hida K, Chen C, Hanes J. Characterization of the intracellular dynamics of a non-degradative pathway accessed by polymer nanoparticles. J Control Release. 2008;125(2):107–111. doi: 10.1016/j.jconrel.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai SK, Hida K, Man ST, et al. Privileged delivery of polymer nanoparticles to the perinuclear region of live cells via a non-clathrin, non-degradative pathway. Biomaterials. 2007;28(18):2876–2884. doi: 10.1016/j.biomaterials.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 23.Seisenberger G, Ried MU, Endress T, Buning H, Hallek M, Brauchle C. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science. 2001;294(5548):1929–1932. doi: 10.1126/science.1064103. [DOI] [PubMed] [Google Scholar]
- 24.Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. Visualizing infection of individual influenza viruses. Proc Natl Acad Sci USA. 2003;100(16):9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babcock HP, Chen C, Zhuang X. Using single-particle tracking to study nuclear trafficking of viral genes. Biophys J. 2004;87(4):2749–2758. doi: 10.1529/biophysj.104.042234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suk JS, Suh J, Lai SK, Hanes J. Quantifying the intracellular transport of viral and nonviral gene vectors in primary neurons. Exp Biol Med (Maywood) 2007;232(3):461–469. [PubMed] [Google Scholar]
- 27.Akita H, Enoto K, Masuda T, Mizuguchi H, Tani T, Harashima H. Particle tracking of intracellular trafficking of octaarginine-modified liposomes: a comparative study with adenovirus. Mol Ther. 2010;18(5):955–964. doi: 10.1038/mt.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai SK, O’hanlon DE, Harrold S, et al. Rapid transport of large polymeric nanoparticles in fresh undiluted human mucus. Proc Natl Acad Sci USA. 2007;104(5):1482–1487. doi: 10.1073/pnas.0608611104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dawson M, Wirtz D, Hanes J. Enhanced viscoelasticity of human cystic fibrotic sputum correlates with increasing microheterogeneity in particle transport. J Biol Chem. 2003;278(50):50393–50401. doi: 10.1074/jbc.M309026200. [DOI] [PubMed] [Google Scholar]
- 30.Wang YY, Lai SK, Suk JS, Pace A, Cone R, Hanes J. Addressing the PEG mucoadhesivity paradox to engineer nanoparticles that “slip” through the human mucus barrier. Angew Chem Int Ed Engl. 2008;47(50):9726–9729. doi: 10.1002/anie.200803526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cu Y, Saltzman WM. Controlled surface modification with poly(ethylene)glycol enhances diffusion of PLGA nanoparticles in human cervical mucus. Mol Pharm. 2009;6(1):173–181. doi: 10.1021/mp8001254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang BC, Dawson M, Lai SK, et al. Biodegradable polymer nanoparticles that rapidly penetrate the human mucus barrier. Proc Natl Acad Sci USA. 2009;106(46):19268–19273. doi: 10.1073/pnas.0905998106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dawson M, Krauland E, Wirtz D, Hanes J. Transport of polymeric nanoparticle gene carriers in gastric mucus. Biotechnol Prog. 2004;20(3):851–857. doi: 10.1021/bp0342553. [DOI] [PubMed] [Google Scholar]
- 34.Lai SK, Wang YY, Cone R, Wirtz D, Hanes J. Altering mucus rheology to “solidify” human mucus at the nanoscale. PLoS ONE. 2009;4(1):E4294. doi: 10.1371/journal.pone.0004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai SK, Wang YY, Hida K, Cone R, Hanes J. Nanoparticles reveal that human cervicovaginal mucus is riddled with pores larger than viruses. Proc Natl Acad Sci USA. 2010;107(2):598–603. doi: 10.1073/pnas.0911748107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suk JS, Lai SK, Wang YY, et al. The penetration of fresh undiluted sputum expectorated by cystic fibrosis patients by non-adhesive polymer nanoparticles. Biomaterials. 2009;30(13):2591–2597. doi: 10.1016/j.biomaterials.2008.12.076. [DOI] [PMC free article] [PubMed] [Google Scholar]