Abstract
Background & Aims
Several small molecule inhibitors of the hepatitis C virus (HCV) NS3/4A protease have advanced successfully to clinical trials. However, the selection of drug-resistant mutants is a significant issue with protease inhibitors (PIs). A variety of amino acid substitutions in the protease domain of NS3 can lead to PI resistance. Many of these significantly impair the replication fitness of HCV RNA replicons. However, it is not known whether these mutations also adversely affect infectious virus assembly and release, processes in which NS3 also participates.
Methods
We studied the impact of 25 previously identified PI-resistance mutations on the capacity of genotype 1a H77S RNA to replicate in cell culture and produce infectious virus.
Results
Most PI-resistance mutations resulted in moderate loss of replication competence, although several (V36A/L/M, R109K, and D168E) demonstrated fitness comparable to wild type, whereas others (S138T and A156V) were severely impaired both in RNA replication and infectious virus production. Although reductions in RNA replication capacity correlated well with decreased yields of infectious virus for most mutations, a subset of mutants (Q41R, F43S, R155T, A156S, and I170A/T) reproducibly demonstrated greater impairment in their ability to produce virus than predicted from reductions in RNA replication capacity. More detailed examination of the I170A mutant demonstrated no defect in release of virus from cells, and no significant difference in specific infectivity of extracellular virus particles.
Conclusions
Replicon-based assays might underestimate the loss of fitness caused by PI-resistance mutations, as some mutations in the NS3 protease domain specifically impair late steps in the viral lifecycle that involve intracellular assembly of infectious virus.
Keywords: Antiviral drug, NS3/4A protease, viral assembly, viral replication
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis, frequently culminating in liver cirrhosis and hepatocellular carcinoma. The current standard-of-care therapy for patients with chronic hepatitis C is a combination of pegylated interferon-α and ribavirin. However, it is only partially effective, as only about 50% of patients with genotype 1 HCV infection achieve a sustained virological response1. Accordingly, there is intense interest in developing novel, small-molecule, direct-acting antiviral compounds.
The HCV NS3/4A protease is a particularly promising target for direct-acting antiviral therapies. Several chemical classes of NS3/4A protease inhibitors (PI) have been developed that potently inhibit HCV replication. Two linear peptidomimetic ketoamides (telaprevir and boceprevir) have entered phase 3 studies2–4, and several macrocyclic inhibitors (e.g., danoprevir, vaniprevir and TMC435) are in phase 2 development5–7. Despite this progress, the selection, emergence, and persistence of drug-resistant viruses are significant concerns with these antiviral compounds8, 9. Drug-resistant variants exist at varying frequencies in untreated patients as part of the viral quasispecies10, 11. This reflects the highly replicative nature of HCV infections as well as the error-prone nature of the HCV RNA-dependent RNA polymerase12. Resistant viruese are rapidly selected and may become dominant among the quasispecies under the pressure of antiviral exposure. Both in vitro and in vivo studies have shown that a variety of amino acid substitutions within the NS3 protease domain can lead to PI resistance, with the specific mutations dependent on both chemical class and the individual compound (Supplementary Table 1).
While the impact of these mutations on resistance to NS3/4A inhibitors has been well characterized, their impact on viral fitness, both in cell culture and in infected patients, is less well understood. Viral fitness can be defined as the ‘relative ability of a virus to produce infectious progeny’13. However, previous efforts to characterize the ‘fitness’ of PI-resistant mutants in vitro have been limited to RNA replicons that are incapable of producing infectious virus. Thus ‘fitness’ has generally been characterized in terms of ‘relative replication capacity’. PI-resistant mutants often have inferior replication capacity in cell culture. For example, a common PI-resistance mutation (A156T) in the NS3 protease drastically reduces the capacity of replicons to replicate in cell culture14, 15. However, replicons containing drug-resistance mutations are not universally impaired in their replication capacity: replicons with V36A, Q41R, T54A, A156S, or I170A mutations have been reported to replicate at least as well as replicons with wild-type NS3 sequence16–19. The replication capacity of R155K was also similar to wild type in some reports17, 20, but significantly reduced in others16.
A better understanding of the impact of PI-resistance mutations on the fitness of HCV is important, since fitness is a critical factor in determining the probability with which mutants will emerge, persist, and potentially cause disease in patients treated with PIs. Our aim here was to determine whether these mutations adversely affect the production of infectious virus – a more definitive measure of viral fitness than used in prior studies. The NS3 helicase domain is involved in an early step in the assembly of infectious virus particles21. While the extent to which the protease domain of NS3 also contributes to assembly is unknown, it is possible that PI-resistance mutations could adversely impact virus assembly as well as polyprotein processing. To assess this possibility, we analyzed an extensive panel of PI resistance mutations placed in the background of an infectious molecular clone of a genotype 1a virus, pH77S.2, that when transfected as RNA into permissive cells replicates efficiently and produces infectious virus. Our results confirm that most PI-resistance mutations adversely impact the replication of HCV RNA, and in some cases also specifically impair the production of infectious virus.
Materials and Methods
Cells and reagents
Details of the cells used in these studies are provided on-line in the Supplementary Materials and Methods. Stock solutions of ciluprevir (BILN2061), boceprevir (SCH 503024), danoprevir (ITMN-191), and vaniprevir (MK7009) were prepared in DMSO.
Plasmids
pH77S.3 is derived from pH77S22, an infectious molecular clone of a genotype 1a HCV, and contains an additional cell culture-adaptive mutation in E2. When transfected into permissive cells, genome-length RNA transcribed from pH77S.3 replicates efficiently and produces virus that is infectious for naïve cells. To monitor replication, the Gaussia luciferase (GLuc) sequence, fused at its C terminus to the FMDV 2A autoprotease, was inserted between p7 and NS2 of pH77S.2. Detailed descriptions of these plasmids, cloning procedures, and construction of PI resistant mutants are provided on-line in the Supplementary Methods.
Viral Fitness
Fitness of the mutated HCV RNAs was determined by measuring the capacity of each to replicate in transfected cells and also to produce infectious virus. Genome-length RNA was synthesized in vitro and transfected into cells as described in detail in the Supplementary Methods. Replication of the RNA was assessed by monitoring GLuc activity secreted into the medium of transfected cells, or by northern analysis of extracted RNA. Yields of infectious virus released into cell culture supernatant fluids were determined by a quantal fluorescent focus assay. Details of these methods are provided on-line in the Supplementary Methods. RNA replication capacity and infectious virus yields were normalized to those produced by wild-type RNA in each experiment, and compared across the panel of NS3 mutants.
Antiviral activity assays
Antiviral activities were determined based on the ability of compounds to inhibit the production of GLuc by HCV RNA-transfected cells. Further details are provided on-line in the Supplementary Methods.
Statistical analysis
Student’s t test was used to compare the magnitude of the reductions in RNA replication capacity and/or infectious virus yield imposed by PI mutations, after normalization to wild-type controls.
Results
PI resistance profile of genotype 1a H77 virus
Since NS3 participates in the assembly of virus particles21, we set out to determine the impact of PI-resistance mutations in NS3 on fitness of a molecular clone of genotype 1a HCV, pH77S.3, a derivative of pH77S that contains cell culture-adaptive mutations and produces fully infectious virus when transfected as RNA into Huh7 cells22. To facilitate monitoring viral RNA replication, we inserted the Gaussia luciferase (GLuc) sequence isn frame between the p7 and NS2 sequences of pH77S.3. Details concerning this construct and validation of GLuc expression as a surrogate marker of the replication of RNA transcribed from it are described in detail on-line in the Supplementary Methods and Results. To profile resistance to PIs in this genotype 1a virus, we used site-directed mutagenesis to construct 25 different H77S.3/GLuc2A mutants, each with a specific amino acid substitution in NS3 reported previously to cause resistance in genotype 1b virus against at least one of 7 candidate PIs: ciluprevir (BILN2061), telaprevir (VX-950), boceprevir (SCH503034), SCH446211 (Sch6), danoprevir (ITMN191), TMC435, or vaniprevir (MK7009) (Supplementary Table 1). These mutations include those identified in vitro in replicon-based studies in vitro and also in vivo, in clinical trials, and involve 11 different residues in the protease domain of NS3. For the most part, these mutations have been studied previously only in the context of genotype 1b replicons. To confirm that they also confer PI resistance in a genotype 1a background, we assessed the ability of selected PIs (ciluprevir, boceprevir, danoprevir and vaniprevir) to inhibit replication of a subset of the H77S.3/GLuc2A mutants. Antiviral EC50 values were determined from the concentration of the compound required to cause a 50% reduction in secretion of GLuc by RNA-transfected cells (see Supplementary Methods). With the exception of R155Q, which did not lead to resistance against boceprevir as expected, the mutations caused significant increases in the EC50 of one or more PI (Table 1). Because the EC50 of boceprevir against the wild-type H77S.3/GLuc2A construct (490 nM) was over 100-fold higher than that of the other PIs tested, the fold-change in the EC50 was generally less dramatic with this compound (Table 1). Notably, in two cases, S138T and A156V, the RNA replication fitness of the genotype 1a mutant was so impaired as to prevent a reliable measurement of the EC50 (see below).
Table 1.
EC50 of PI’s against mutant H77S.3/GLuc2A RNAs
| Boceprevir |
MK7009 |
ITMN191 |
BILN2061 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutant | Mean [nM] |
S.D. [nM] |
FC* | Mean [nM] |
S.D. [nM] |
FC | Mean [nM] |
S.D. [nM] |
FC | Mean [nM] |
S.D. [nM] |
FC |
| wt | 490 | 80 | - | 0.8 | 0.3 | - | 0.5 | 0.1 | - | 1.5 | 0.5 | - |
| V36A | 1780 | 240 | 3.6 | |||||||||
| V36G | 3860 | 360 | 7.9 | |||||||||
| V36L | 1500 | 94 | 3 | |||||||||
| V36M | 1500 | 130 | 3 | |||||||||
| Q41R | 750 | 100 | 1.5 | 1.9 | 0.1 | 2.4 | 1.5 | 0.4 | 3 | 9.1 | 2.1 | 6.2 |
| F43S | 3360 | 620 | 6.9 | 17.8 | 3 | 44 | ||||||
| T54A | 2710 | 300 | 5.5 | |||||||||
| Q80R | 14 | 1 | 9.3 | |||||||||
| R109K | 610 | 30 | 1.2 | |||||||||
| S138T | ||||||||||||
| R155K | 1830 | 240 | 3.7 | 430 | 80 | 538 | 130 | 6 | 316 | 750 | 71 | 510 |
| R155T | 2540 | 610 | 5.2 | 250 | 15 | 312 | 18.3 | 9.2 | 45 | 680 | 42 | 460 |
| R155Q | 210 | 70 | 0.4 | 400 | 8 | 267 | ||||||
| R155G | ||||||||||||
| A156S | 3420 | 320 | 7 | 14 | 0.8 | 17.5 | 7.4 | 1.7 | 18 | |||
| A156T | 6670 | 820 | 13.7 | 200 | 64 | 250 | 7.2 | 1.9 | 18 | 1060 | 96 | 706 |
| A156V | ||||||||||||
| D168A | 540 | 10 | 1.1 | 720 | 54 | 900 | 135 | 21 | 330 | 680 | 75 | 453 |
| D168E | 23 | 5.8 | 56 | |||||||||
| D168H | 66 | 1 | 160 | |||||||||
| D168V | 510 | 40 | 1 | 1360 | 210 | 1700 | 68 | 1 | 166 | 1100 | 270 | 733 |
| D168G | 12.8 | 2 | 31 | |||||||||
| D168N | 8.1 | 0.4 | 20 | |||||||||
| I170A | 1060 | 80 | 2.2 | |||||||||
| I170T | 1020 | 90 | 2.1 | |||||||||
FC = fold change in EC50 from wild-type.
Impact of PI-resistance mutations on RNA replication capacity
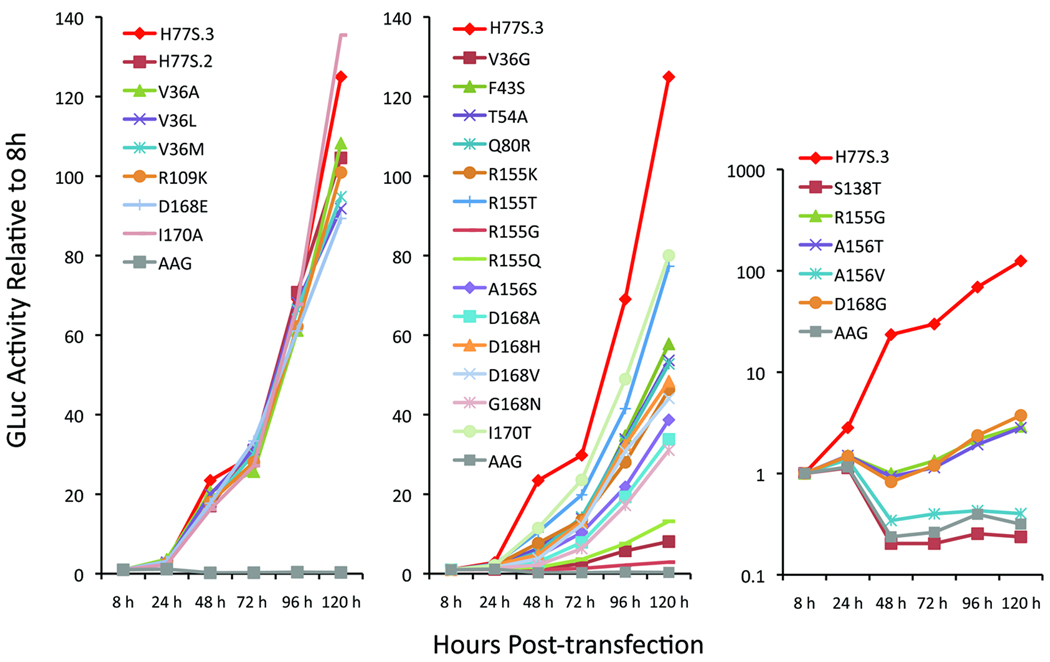
To determine the impact of the resistance mutations on the replication capacity of H77S.3/GLuc2A RNA, we measured GLuc activity in media collected at 24h intervals following transfection. The results, normalized to the activity present 8 hrs after transfection, allowed classification of the 25 mutants into 4 groups based on RNA replication kinetics (Fig. 1). The first of these groups, comprising the V36A/L/M, Q41R (H77S.2/GLuc2A), R109K, D168E, and I170A mutants, demonstrated little loss of replication capacity, with fold-increases in GLuc activity ≥85% of that observed with H77S.3/GLuc2A (Fig. 1, left panel). A second group, comprising the majority of the mutants, demonstrated moderately impaired replication capacity (GLuc activity <85% H77S.3/GLuc2A) but still generated GLuc activities that increased continuously after transfection (Fig. 1, middle panel). A third group, comprised of R155G, A156T, and D168G, demonstrated more severe impairments in replication, with the GLuc activity at 48h consistently less than at 8h, but generally increasing after 72h (Fig. 1, right panel, note the log scale for GLuc in this panel). A fourth group, comprised only of S138T and A156V, was defined by the absence of any increase in GLuc activity after 8–24h, and thus demonstrated GLuc expression similar to that observed with the lethal AAG mutant (Fig. 1, right panel). These results show that the replication competence of genotype 1a RNA is variably affected by PI-resistance mutations, with the impact on replication ranging from none to very severe. Although some patterns were evident (see Discussion), loss of replication competence did not correlate strictly with the specific NS3 residue involved or the magnitude of PI resistance (fold-change in EC50).
Figure 1. Impact of drug-resistance mutations on genotype 1a HCV RNA replication.
Medium was collected and replaced at 8, 24, 48, 72, 96, 120h after transfection of H77S.3/GLuc2A RNAs carrying the indicated mutations, and GLuc activity determined at each point in time. Results were normalized to the 8h GLuc activity, and represent the mean of triplicate samples and are representative of multiple experiments. The mutants are grouped in 3 panels, with increasingly negative impact on replication capacity from left to right.
Impact of PI-resistance mutations on infectious virus production
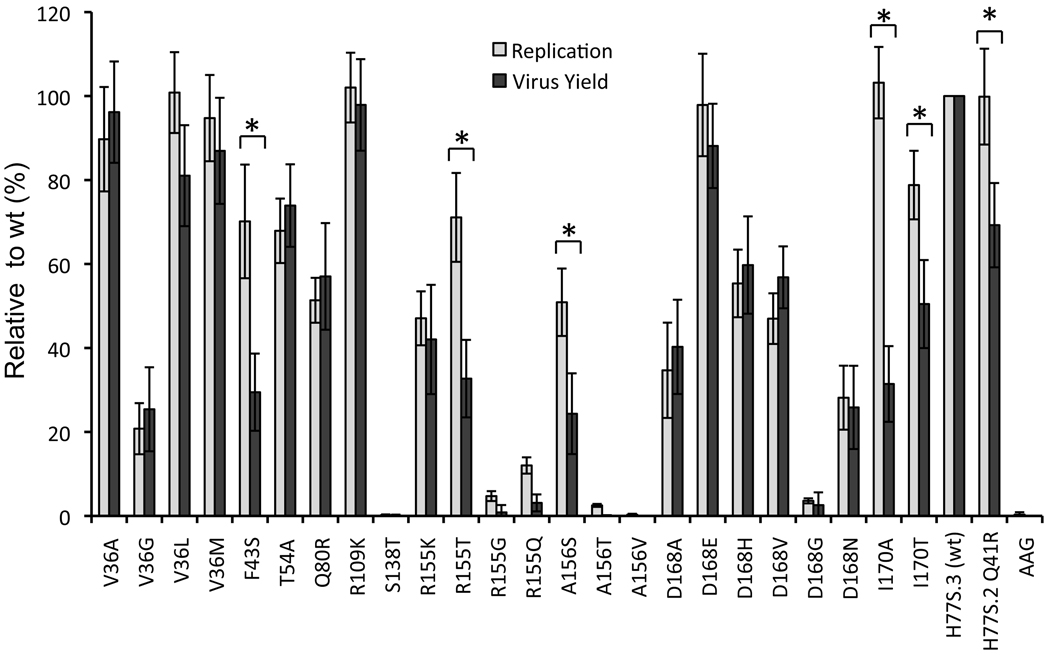
We next assessed the impact of each of the PI-resistance mutations on production of infectious virus by H77S.3 RNA. Cell culture supernatant fluids were collected 72h and 96h after transfection were inoculated onto naïve cells, and foci of infected cells detected by immunofluorescence 96h later. As shown in Figure 2, infectious virus yields varied significantly among the different mutant RNAs, most often correlating closely with the relative RNA replication capacity of the related H77S.3/GLuc2A mutant. This is not surprising, as RNA replication is essential for production of infectious virus. However, 6 mutants (Q41R, F43S, R155T, A156S, I170A and I170T), demonstrated a reproducible discordance between replication capacity (GLuc expression relative to wild-type) and infectious virus yield (FFU/ml relative to wild-type) (Fig. 2). In replicate experiments, the yields of infectious virus from these mutants were significantly less than expected from the RNA replication assay results (p<0.001 by Student’s t test). These results suggest that this subset of resistance mutations specifically impedes some aspect of infectious virus assembly and/or release, above and beyond any negative effect of the mutation on genome amplification. R155G and R155Q also demonstrated very substantial defects in production of infectious virus that were greater than the observed defect in replication (Fig. 2). Thus like the Thr substitution at Arg155 in R155T, Gly and Gln substitutions at residue 155 may also negatively modulate the production of infectious virus. However, the replication of these RNAs was so severely reduced that it was difficult to document an additional, statistically significant defect in infectious virus yield.
Figure 2. Impact of drug-resistance mutations on HCV RNA replication and infectious virus production.
GLuc activity secreted by RNA-transfected cells at 96 vs. 8 h, as shown in Fig. 1, was normalized to that of the wild-type H77S.3 RNA (100% = 66 ± 19 fold-increase, lightly shaded bars) and plotted adjacent to infectious virus yield (combined data from 72 and 96 hr endpoints) from each mutant, similarly normalized to H77S.3 RNA (100% = 3.4 ± 1.3×103 FFU/ml, solid bars). The data shown represent the mean ± S.D. from at least three (replication) or four (virus yield) independent experiments. An asterix [*] designates mutants for which the difference between the relative capacities to replicate as RNA and to produce infectious virus is significant by student’s t-test (p<0.001). “wt” refers to wild type.
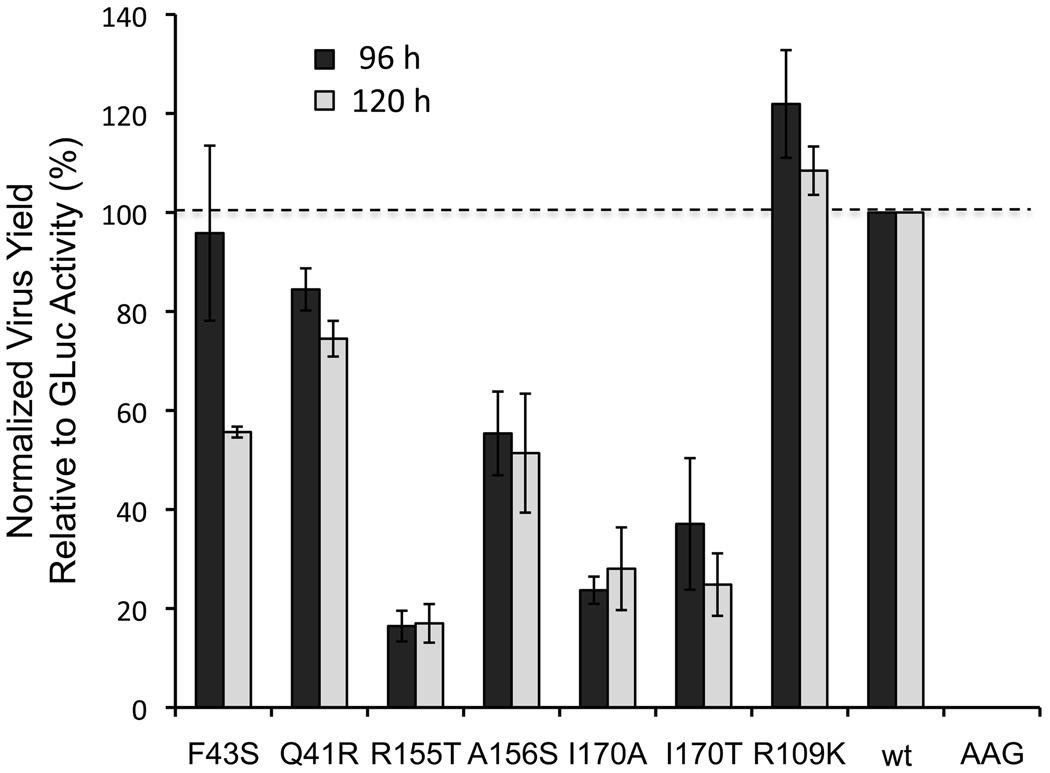
To confirm that the discordance we observed between the impact of the Q41R, F43S, R155T, A156S and I170A/T mutations on infectious virus yields from H77S.3 RNA and the capacity of the mutated H77S.3/GLuc2A RNAs to replicate was not in some way related to the GLuc2A insertion, we carried out two additional sets of experiments. First, we directly assessed the production of infectious virus from the mutated H77S.3/GLuc2A RNAs, comparing GLuc activity and infectious virus titer present in supernatant culture fluids obtained from cells transfected with the mutated H77S.3/GLuc2A RNAs. We determined the FFU/GLuc activity ratio of each mutant, and normalized this to that observed with the wild-type H773.3/GLuc2A RNA that has no mutation in the NS3 protease domain. R109K mutant, that has no defect in either RNA replication or infectious virus yield (Figs. 1 and 2), was included as an additional control. These results (Fig. 3) confirmed that the R155T, A156S and I170A/T mutations, placed in the H77S.3/GLuc2A background, result in reduced production of infectious virus in addition to their impact on viral RNA replication. The F43S and Q41R mutants appeared to have lesser specific defects in infectious virus yield, while the R109K mutant produced infectious virus titers equivalent to wild-type, as anticipated.
Figure 3. Infectious virus yield versus GLuc expression from H77S.3/GLuc2A mutants.
Supernatant culture fluids were assayed for GLuc activity as well as infectious virus 96h and 120h following transfection of H77S.3/GLuc2A and related mutant RNAs. Results are shown as the ratio of infectious virus to GLuc activity, normalized to wild-type H77S.3/GLuc2A RNA (100% = 90 ± 12 FFU/ml/103 light units (L.U.) at 96h, and 88 ± 11 FFU/ml/103 L.U. at 120h). Results are means ± S.D. from 3 experiments.
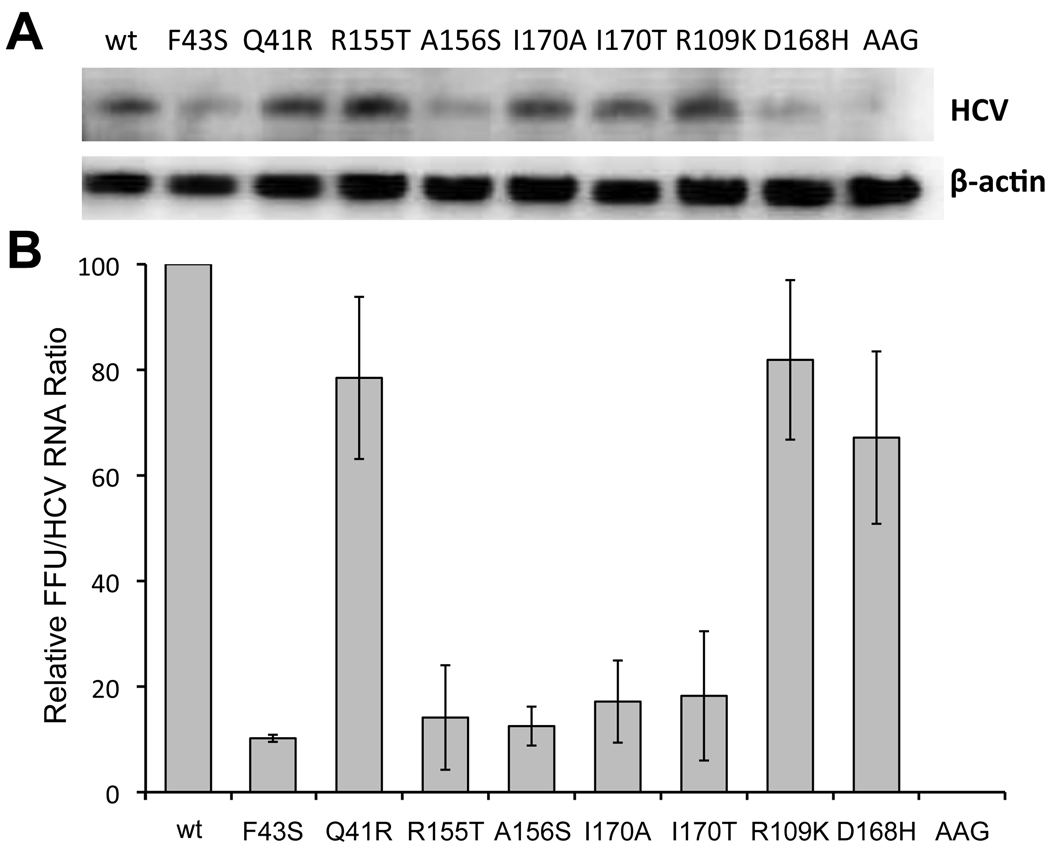
We also compared infectious virus yields produced by the mutated parental H77S.3 RNAs with viral RNA replication assessed directly by quantitative northern analysis. Virus yields were determined in supernatant fluids collected 96h after transfection, at which time total RNA was extracted from the cells for northern analysis. Northern blots (Fig. 4A) were quantified by phosphor imaging, and the abundance of HCV RNA normalized to that of β-actin included as a loading control. This provided an FFU/HCV RNA ratio for each mutant, which was then normalized to that observed with wild-type H77S.3 RNA. The results of these experiments (Fig. 4B) were remarkably similar to those shown in Fig. 2 and 3, demonstrating that the yield of infectious virus, normalized to HCV RNA replication, was substantially less than wild-type in the F43S, R155T, A156S, and I170A/T mutants. Q41R demonstrated only a minor defect in the production of infectious virus (~20% reduced), while R109K and D168H were similar to the wild-type H77S.3 RNA (Fig. 4B).
Figure 4. Infectious virus yield vs. RNA abundance of H77S.3 mutants.
H77S.3 and related RNAs containing the indicated mutations were transfected into Huh-7.5 cells. (A) Cells were lysed and total RNA extracted at 96h, then subjected to northern blotting with HCV-and β-actin-specific probes. (B) Ratio of extracellular infectious virus at 96h to intracellular RNA abundance quantified by phosphor image analysis of northern blots (HCV/β-actin RNA), normalized to yields from wild-type H77S.3 RNA (1.2 ± 0.52×103 FFU/ml). Results shown are means ± range from duplicate experiments.
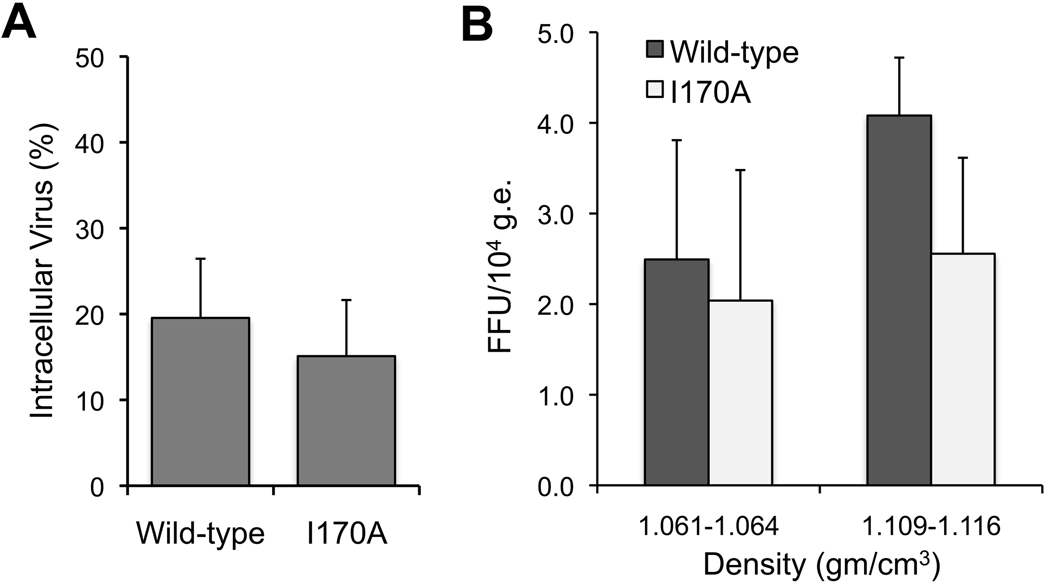
To further assess the nature of the defect in infectious virus production, we selected the I170A mutant that has significantly reduced infectious virus yield but no impairment in RNA replication (Figs. 2–4). When compared with wild type, we found no difference in the proportion of infectious I170A virus present within cell lysates and culture supernatant fluids (Fig. 5A). Thus, the reduction in yield is not due to impaired release of infectious virus from cells. We also compared the buoyant densities of the extracellular viruses by equilibrium gradient centrifugation (see Supplemental Methods). Both the wild-type and I170A viruses showed two major peaks of infectivity at ~1.062 and ~1.112 gm/cm3 (data not shown), with the latter predominant. No significant differences were apparent in the specific infectivity of the viruses present in these peak fractions (Fig. 5B). Collectively, these data indicate that the I170A mutation results in a defect in intracellular infectious virus assembly.
Figure 5.
A PI resistance mutation causes a defect in assembly of infectious virus. (A) Intracellular infectious virus titer as the percent of extracellular wild-type (H77S.3) and I170A virus 96h after RNA transfection. Results are means ± S.D. (B) Specific infectivity of the major species of extracellular wild-type (H77S.3) and I170A virus particles isolated by equilibrium gradient centrifugation. Results are mean FFU/104 HCV genome equivalents (g.e.) ± S.E.M. in 3 consecutive gradient fractions centered on each peak of infectivity.
Discussion
Despite progress in the clinical development of direct acting antiviral inhibitors of HCV replication, the selection and persistence of drug resistant mutants remains a major concern and will mandate the use of combinations of therapeutic agents in future treatment regimes12. Given the highly replicative nature of HCV infection, with as many as 1012 new genomes produced each day in the typical patient23, and the highly error-prone nature of the HCV polymerase, viral RNAs harboring specific resistance mutations are likely to be present within the RNA quasispecies prior to treatment in virtually all patients. Our data (Fig. 1) and data from others14–20 suggest that most of these resistance mutations negatively impact fitness; thus, viruses that harbor them represent a very small proportion of the quasispecies pre-treatment. Nonetheless, these mutants may gain a significant growth advantage over more prevalent drug-susceptible quasispecies upon the initiation of PI therapy, leading to their emergence as dominant quasispecies. This is a major clinical concern as direct acting antivirals enter the clinical armamentarium, making it important to gain a better understanding of the impact of specific NS3 mutations on antiviral susceptibility and replication fitness. Here, we have studied this in the context of a genotype 1a viral RNA (H77S.3) that replicates efficiently in cultured cells and produces virus that is infectious in cell culture22 and in the chimpanzee model (see below). Our results show that NS3 mutations conferring resistance against a variety of chemically diverse PIs have variable impact on genotype 1a RNA replication, and provide novel evidence that a subset of these mutations also specifically impair a later step in the virus life cycle involved in the assembly of infectious virus.
The resistance mutations that we constructed in the genotype 1a H77S.3 background were identified by a review of published data obtained from both cell culture-based and clinical studies of PIs (Supplementary Table 2). Previous studies of the impact of PI-resistance mutations on viral fitness generally have used subgenomic replicons derived from genotype 1b viruses, and have typically focused on small numbers of mutations selected by single PIs. Our data are unique in that they are derived from studies of a full-length genotype 1a RNA, and include analysis of a comprehensive panel of mutants selected by a variety of linear ketoamide and macrocyclic inhibitors of NS3/4A in previous studies. As expected, we confirmed that these mutations, introduced into the genotype 1a background, generally cause resistance against PIs in the patterns expected (Table 1). Thus, we found a significant increase in the EC50 of boceprevir, a linear ketoamide, for the V36G mutant, while the D168A and D168V mutations markedly increased the EC50 for the macrocyclic compounds, danoprevir, vaniprevir and ciluprevir, but not boceprevir (Table 1). Importantly, the R155T and A156T mutations substantially increased the EC50 for each of the compounds tested.
Although some PI-resistance mutations, including V36A, Q41R, T54A, A156S and V170A, have been suggested in prior studies to replicate more efficiently than related wild-type RNAs16–19, we did not observe this. H77S RNA containing Q41R (H77S.2) and I170A mutations replicated as efficiently as wild-type virus (Figs. 1 and 4), but none of the mutants we studied replicated significantly more efficiently than the wild type. This difference might be due to the use of RNAs derived from different HCV strains, or possibly our use of genome-length versus subgenomic RNA. In general, we found most resistance mutations have a negative impact on replication of H77S.3 RNA (Fig. 1). There was little correlation between loss of fitness, however, and the degree of antiviral resistance caused by specific mutations. While D168A demonstrated greater resistance against danoprevir than D168G (Table 1), D168G was severely compromised for replication while the loss of replication competence was only moderate for D168A (Fig. 2). Similarly, R155T demonstrated robust replication, yet caused very substantial increases in the EC50 for each of the PIs tested. This likely reflects differences in how various PIs interact with the substrate-binding pocket in the protease, and how this fit is impacted by the resistance mutation compared with binding of the native polyprotein substrate. It is likely that the loss of RNA replication fitness stems from reduced catalytic activity of the mutant protease, and therefore reduced competence in processing of the viral polyprotein. Enzymatic activity is known to be sharply decreased with the A156T mutation, and both subgenomic replicons14, 15 and genome-length H77S RNA (Fig. 1) with this mutation show reduced replication capacity.
The most novel aspect of this study has been our ability to test the impact of PI-resistance mutations on the production of infectious virus as well as RNA replication capacity. For most mutants these measures of fitness correlated well, but in a subset of six mutants, i.e., Q41R, F43S, R155T, A156S, I170A and I170T, we observed a significantly greater negative impact on the ability to produce infectious virus than on replication capacity Q41R and I170A RNAs replicated as efficiently as wild-type H77S.3 RNA, but produced infectious virus at rates that were ~20% and ~80% reduced. Although modest in magnitude, such defects in infectious virus production are likely to be exponentially magnified during the multiple cycles of cell infection occurring an infected patient. Importantly, of the subset of resistance mutations causing such defects, all but Q41R have been identified in patients enrolled in clinical trials of PIs (Table 1), making these results relevant to the setting in vivo. Q41R is a particularly interesting mutation. We identified this early in the development of the H77S clone as a cell-culture adaptive mutation24, and it is present in the H77S and H77S.2 constructs (see Supplementary Fig. S1A). In a chimpanzee that was persistently infected with virus produced by cells transfected with H77S.2 RNA, we observed a complete reversion of this mutation, suggesting that the wild-type Gln41 residue is preferred at this position in vivo. Although we found that H77S RNA encoding Arg41 replicated just as efficiently as Gln41 (Figs. 1, 2 and 4), it had a small but reproducible impairment in infectious virus production (Figs. 2, 3, and 4). This provides a potential explanation for the reversion of the Q41R mutation during persistent infection, and an interesting in vivo correlate of the findings in cell culture.
A comparison of intracellular and extracellular infectious virus titers indicated that the reduction in yield of the I170A mutant is not due to impaired release of infectious virus from the cell (Fig. 5A). Furthermore, within the range of experimental error, we could not demonstrate any consistent difference in the specific infectivity of wild-type and I170A viruses isolated on equilibrium density gradients (Fig. 5B). This suggests that the reduced infectivity of extracellular fluids is not due to impaired virus entry.
The impaired catalytic activity observed with some resistance mutations reduces the ability of the protease to cleave both TRIF and MAVS (unpublished data), cellular substrates of NS3/4A that are essential adaptor proteins in signaling pathways that induce interferon responses25. This could limit the ability of the virus to evade innate immune responses, and possibly further impair viral RNA replication. However, the fact that wild-type and I170A viruses have similar specific infectivity (Fig. 5A) and identical RNA replication profiles in transfected cells (Fig. 1, left panel) indicates that this is not the cause of the reduction observed in production of infectious I170A virus (Fig. 2). Moreover, the disparities in RNA replication and infectious virus production shown in Fig. 4 were observed in Huh-7.5 cells that lack both TLR3 and RIG-I signaling. Collectively, the data point to a defect in virus assembly.
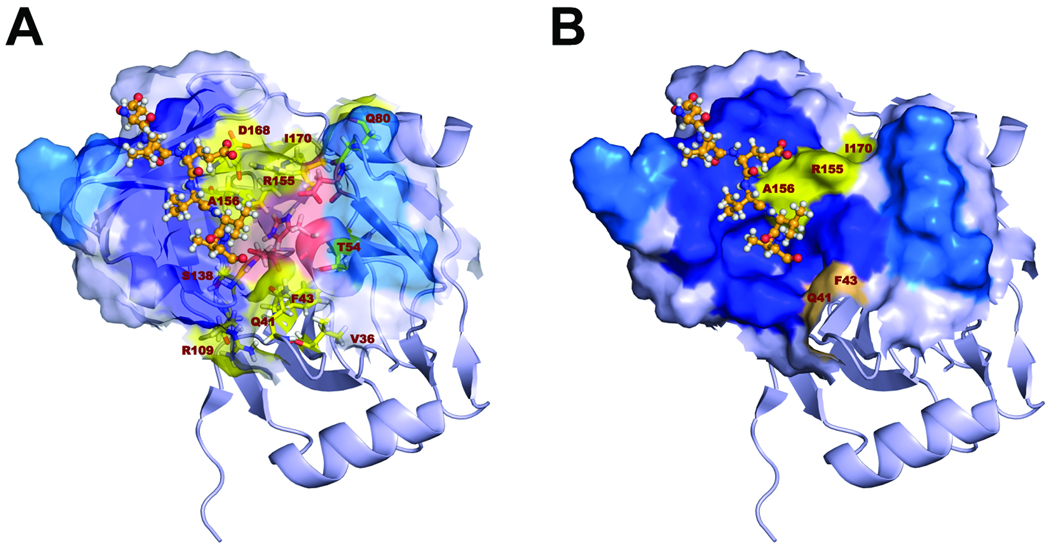
It is likely that this defect in assembly of infectious virus is due to negative effects of the mutations on essential protein-protein interactions, rather than reduced catalytic activity of the protease as discussed above. NS3 interacts with multiple nonstructural proteins, including NS2, and these interactions are important for production of infectious virus (Ma et al., submitted for publication). Resistance mutations might also influence the structure or function of the NS3 helicase domain, for which genetic evidence strongly supports a role in particle assembly21. Consistent with this hypothesis, the NS3 residues at which we found resistance mutations to have the greatest specific effect on production of infectious virus, Arg155, Ala156, and Ile170 (Figs. 3 and 4), are nestled together on the surface of the protease, at one edge of the substrate-binding site (Figs. 6A and B). This surface of the protease abuts the helicase domain in the crystallographic structure of the full-length NS3 molecule26. Although it is not known if NS3 actually adopts this conformation in vivo, prior studies do suggest a modulating influence of the upstream protease domain on NS3 helicase activity27. The two other residues within the protease domain that we found to influence production of infectious virus, Gln41 and Phe43, are also surface-exposed, but on the opposing side of the substrate-binding domain (Fig. 6B).
Figure 6.
Structural model of the substrate-binding site in the genotype 1a NS3 protease domain (PDB2OC0). (A) The surface depicted faces onto the NS3 helicase domain. The C-terminal NS3 product of proteolysis is shown in orange ball-and-stick mode (created by superimposing the structure from 1CU1). Residues where resistance mutations were introduced are labeled and shown as yellow sticks. Catalytic residues are shown as red sticks. The surface of the substrate-binding site is partially transparent. Surface coloring in red and yellow highlights the surface involvement of catalytic residues and residues involved in PI-resistance mutations, respectively. (B) Similar to panel A, but with the surface now non-transparent, and with the surface residues that interact with the helicase domain shown in dark blue. The C-terminal NS3 cleavage product is shown in ball-and-stick mode (orange). Surface-exposed residues at which PI resistance mutations were introduced in H77S.3 are colored in salmon and yellow. Those that specifically impact infectious virus yield cluster in two areas on the surface of the protease adjacent to the substrate-binding pocket.
The data presented here represent an advance over previous studies of the fitness of PI-resistant mutants in that they assess the impact of resistance mutations on steps in the viral life cycle beyond RNA replication. They demonstrate that the use of replicon-based assays, which assess only viral RNA replication, may significantly underestimate the loss of fitness caused by some PI-resistance mutations. Nonetheless, caution is warranted in extrapolating even from these data to the situation in vivo. The transient transfection assay we used here did not allow for the emergence of compensatory mutations capable of rescuing the impaired replication capacity of resistant viruses. In longer-term experiments, we have documented such compensatory mutations in replicons containing the A156T mutation15. Antiviral drug resistance will inevitably be an issue as PIs enter clinical practice, and continuing efforts will be needed to monitor resistance and to relate data emerging from ongoing clinical studies to results obtained using available in vitro systems.
Supplementary Material
Acknowledgements
We thank Dr. Arvind Patel for AP33 antibody and Dr. Charles Rice for Huh-7.5 cells.
Grant support: This work was supported in part by a contract and grants from the National Institutes of Health (NO1-AI25488, U19-AI040035, and R01-AI075090) and the Deutsche Forschungsgemeinschaft (WE4388/3-1).
Abbreviations
- FMDV
foot-and-mouth disease virus
- GLuc
Gaussia luciferase
- HCV
hepatitis C virus
- NS
nonstructural protein
- PI
protease inhibitor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: SML has served as a consultant for Abbott, Hoffman-LaRoche, Juvaris Biotherapeutics, GSK, Merck, Novartis, and Pfizer; SZ has served as a consultant for Abbott, BMS, Gilead, Merck, Novartis, Pfizer, Roche, Tibotec, and Vertex.
Writing Assistance: The authors thank Mardelle Susman who provided editorial assistance under support from the NIH.
Author Contributions:
TS: study concept and design, acquisition, analysis and interpretation of data, drafting of manuscript;
DY, DMcG: acquisition, analysis and interpretation of data;
CW: analysis and interpretation of data, critical revision of manuscript;
MY: technical and material support, critical review of the manuscript;
SZ: critical review of the manuscript for important intellectual content.
SML: obtained funding, study concept and design, analysis and interpretation of data, critical review of the manuscript.
References
- 1.Fried M, Shiffman M, Reddy K, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 2.McHutchison J, Everson G, Gordon S, et al. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360:1827–1838. doi: 10.1056/NEJMoa0806104. [DOI] [PubMed] [Google Scholar]
- 3.Hézode C, Forestier N, Dusheiko G, et al. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N Engl J Med. 2009;360:1839–1850. doi: 10.1056/NEJMoa0807650. [DOI] [PubMed] [Google Scholar]
- 4.Sarrazin C, Rouzier R, Wagner F, et al. SCH 503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alpha-2b for genotype 1 nonresponders. Gastroenterology. 2007;132:1270–1278. doi: 10.1053/j.gastro.2007.01.041. [DOI] [PubMed] [Google Scholar]
- 5.Reesink H, Fanning G, Farha K, et al. Rapid HCV-RNA decline with once daily TMC435: a phase I study in healthy volunteers and hepatitis C patients. Gastroenterology. 2010;138:913–921. doi: 10.1053/j.gastro.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 6.Seiwert S, Andrews S, Jiang Y, et al. Preclinical characteristics of the hepatitis C virus NS3/4A protease inhibitor ITMN-191 (R7227) Antimicrob Agents Chemother. 2008;52:4432–4441. doi: 10.1128/AAC.00699-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liverton N, Carroll S, Dimuzio J, et al. MK-7009, a potent and selective inhibitor of hepatitis C virus NS3/4A protease. Antimicrob Agents Chemother. 2010;54:305–311. doi: 10.1128/AAC.00677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarrazin C, Kieffer T, Bartels D, et al. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology. 2007;132:1767–1777. doi: 10.1053/j.gastro.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 9.Susser S, Welsch C, Wang Y, et al. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology. 2009;50:1709–1718. doi: 10.1002/hep.23192. [DOI] [PubMed] [Google Scholar]
- 10.Bartels D, Zhou Y, Zhang E, et al. Natural prevalence of hepatitis C virus variants with decreased sensitivity to NS3.4A protease inhibitors in treatment-naive subjects. J Infect Dis. 2008;198:800–807. doi: 10.1086/591141. [DOI] [PubMed] [Google Scholar]
- 11.Kuntzen T, Timm J, Berical A, et al. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naïve patients. Hepatology. 2008;48:1769–1778. doi: 10.1002/hep.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimakami T, Lanford RE, Lemon SM. Hepatitis C: recent successes and continuing challenges in the development of improved treatment modalities. Curr Opin Pharmacol. 2009;9:537–544. doi: 10.1016/j.coph.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domingo E, Menéndez-Arias L, Holland J. RNA virus fitness. Rev Med Virol. 1997;7:87–96. doi: 10.1002/(sici)1099-1654(199707)7:2<87::aid-rmv188>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 14.Lin C, Gates C, Rao B, et al. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J Biol Chem. 2005;280:36784–36791. doi: 10.1074/jbc.M506462200. [DOI] [PubMed] [Google Scholar]
- 15.Yi M, Tong X, Skelton A, et al. Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor. Reduced RNA replication fitness and partial rescue by second-site mutations. J Biol Chem. 2006;281:8205–8215. doi: 10.1074/jbc.M510246200. [DOI] [PubMed] [Google Scholar]
- 16.Tong X, Bogen S, Chase R, et al. Characterization of resistance mutations against HCV ketoamide protease inhibitors. Antiviral Res. 2008;77:177–185. doi: 10.1016/j.antiviral.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 17.He Y, King M, Kempf D, et al. Relative replication capacity and selective advantage profiles of protease inhibitor-resistant hepatitis C virus (HCV) NS3 protease mutants in the HCV genotype 1b replicon system. Antimicrob Agents Chemother. 2008;52:1101–1110. doi: 10.1128/AAC.01149-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Bartels D, Hanzelka B, et al. Phenotypic characterization of resistant Val36 variants of hepatitis C virus NS3-4A serine protease. Antimicrob Agents Chemother. 2008;52:110–120. doi: 10.1128/AAC.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong X, Chase R, Skelton A, et al. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antiviral Res. 2006;70:28–38. doi: 10.1016/j.antiviral.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, Müh U, Hanzelka B, et al. Phenotypic and structural analyses of hepatitis C virus NS3 protease Arg155 variants: sensitivity to telaprevir (VX-950) and interferon alpha. J Biol Chem. 2007;282:22619–22628. doi: 10.1074/jbc.M610207200. [DOI] [PubMed] [Google Scholar]
- 21.Ma Y, Yates J, Liang Y, et al. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J Virol. 2008;82:7624–7639. doi: 10.1128/JVI.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi M, Villanueva R, Thomas D, et al. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci U S A. 2006;103:2310–2315. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann AU, Lam NP, Dahari H, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 24.Yi M, Lemon SM. Adaptive mutations producing efficient replication of genotype 1a hepatitis C virus RNA in normal Huh7 cells. J Virol. 2004;78:7904–7915. doi: 10.1128/JVI.78.15.7904-7915.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemon SM. Induction and evasion of innate antiviral responses by hepatitis C virus. J Biol Chem. 2010;285:22741–22747. doi: 10.1074/jbc.R109.099556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao N, Reichert P, Taremi SS, et al. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure.Fold.Des. 1999;7:1353–1363. doi: 10.1016/s0969-2126(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 27.Howe AY, Chase R, Taremi SS, et al. A novel recombinant single-chain hepatitis C virus NS3-NS4A protein with improved helicase activity. Protein Science. 1999;8:1332–1341. doi: 10.1110/ps.8.6.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.