Abstract
The discovery and development of new biomarkers continues to be an exciting and promising field. Improvement of prediction of risk of developing disease is one of the key motivations in these pursuits. Appropriate statistical measures are necessary for drawing meaningful conclusions about the clinical usefulness of these new markers. In this review, we present several novel metrics proposed to serve this purpose. We use reclassification tables constructed based on clinically meaningful disease risk categories to discuss the concepts of calibration, risk separation, risk discrimination, and risk classification accuracy. We discuss the notion that the net reclassification improvement is a simple yet informative way to summarize information contained in risk reclassification tables. In the absence of meaningful risk categories, we suggest a ‘category-less’ version of the net reclassification improvement and integrated discrimination improvement as metrics to summarize the incremental value of new biomarkers. We also suggest that predictiveness curves be preferred to receiver-operating-characteristic curves as visual descriptors of a statistical model’s ability to separate predicted probabilities of disease events. Reporting of standard metrics, including measures of relative risk and the c statistic is still recommended. These concepts are illustrated with a risk prediction example using data from the Framingham Heart Study.
Keywords: reclassification, risk prediction, NRI, IDI, calibration, discrimination
Introduction
The discovery and study of new biomarkers are among the most extensively developing areas of clinical chemistry and clinical medicine in the last two decades. A few examples (from a very long and constantly expanding list) include the role of biomarkers such as plasminogen-activator inhibitor type 1, gamma glutamyl transferase, C-reactive protein (CRP), B-type natriuretic peptide, urinary albumin-to-creatinine ratio, or fibrinogen (1–5) in the pathogenesis of cardiovascular disease (CVD). One of the most useful potential applications of newer biomarkers might be in the field of prediction of the risk of developing disease. It is hoped that addition of markers that lie along the causal pathway of the disease outcome of interest to the prediction models containing standard risk factors might lead to improved quantification of true future risk of developing disease in individuals and of the overall disease burden at the population level. Sometimes biomarkers may not be causally related to disease but serve as ‘risk markers’ (as opposed to risk factors that are causally related) because they aid risk stratification or identify subgroups of individuals who are more responsive to select therapeutic agents. Furthermore, in some cases controlled clinical trials have been conducted to determine if altering levels of new biomarkers might lead to improved prognosis of patients. For example, in the cardiovascular field, the inflammatory marker CRP has been extensively evaluated for its potential ability to quantify the risk of developing CVD (4–6). In a recent development, the FDA advisory panel voted in favor of recommending rosuvastatin for patients with no history of heart disease as long as they have elevated circulating levels of CRP (>2mg/dl), based on the results of the JUPITER trial (7).
Paralleling the growth of biomarker research, risk prediction algorithms have emerged as among the most popular tools in preventive medicine. They have been developed for a wide range of disease outcomes, including CVD and its components (coronary heart disease [CHD], stroke, heart failure), different forms of cancer, atrial fibrillation, hypertension, diabetes and many other adverse health outcomes (8–16). Such algorithms are usually based on a set of standard risk factors that are combined to form a prediction rule using statistical modeling methods.
The inevitable question that arises is how best to evaluate the incremental contribution of a new biomarker to a conventional risk prediction model. Statistical significance and magnitude of the effect size for the association of a biomarker with a disease outcome (typically expressed with measures of relative or absolute risk) are an important starting point and must be reported, but these metrics may not offer sufficient information regarding the incremental clinical contribution or usefulness of a putative new biomarker. Since risk prediction models use binary or survival outcomes, metrics such as the linear regression R-square (for assessing incremental gain) are not directly applicable here. The c statistic (17), which originated in diagnostic studies and quantifies discriminatory ability of risk classification algorithms, has been the measure of choice for evaluation of biomarkers, but in recent years researchers have emphasized several limitations of the c statistic in the context of risk prediction (18, 19). This situation arises due to the broader focus of risk prediction as compared to diagnostic studies. For diagnostic purposes, it is assumed some people do and others do not have the disease condition of interest, and the task at hand is to distinguish between the two groups by means of ‘separation’. Optimal cut-offs are selected based on pre-defined criteria, and they can be readily re-calibrated if these criteria change. Relative risks play a dominant role as key metrics in this circumstance. However, in the context of risk prediction, the disease event has not happened yet, and we are attempting to predict it in the future time horizon. Distinguishing disease events from ‘nonevents’ remains very important, but correct calibration is also critical because we want to assign absolute disease risks that correspond to reality (actual experience of individuals, or observed risk). Furthermore, for survival data, it might be desirable to distinguish between disease events that happen at different time points (time-to-event analyses, (20)). Risk thresholds that correspond to disease incidence are established, and often not easily modified. Accordingly, clinical decisions are often made based on these risk thresholds (9). This suggests that evaluating categories of disease risk corresponding to these established risk thresholds may offer a very meaningful clinical perspective when evaluating a new biomarker.
In this review, we examine several contemporary methods of quantifying the additional contribution of a new biomarker to a standard risk prediction model. First, we introduce a data set from a Framingham Heart Study sample that will serve to illustrate the concepts being discussed. Next, we discuss standard measures applied to new biomarkers including p-values, measures of relative risk and increase in the c statistic with their properties and limitations. The main focus of our review is on the concept of risk reclassification tables, and the continuous metrics that parallel these tables in the case of no risk thresholds. We discuss the importance of calibration, separation (spread), discrimination and risk classification accuracy. Net reclassification improvement (NRI), discrimination slopes, integrated discrimination improvement (IDI) and predictiveness curves are presented in a detailed example (19, 21). In the discussion concluding this review we outline other topics in risk prediction-oriented biomarker studies.
Materials and Methods
Here we introduce a practical example that will serve as an illustration for the concepts developed further. For ease of reference, we use the same data set and analysis as the one presented by Pencina et al. (19). We note that this example is intended to illustrate concepts discussed in this review rather than serve as a substantive analysis.
The Framingham Heart Study originated in 1948 (22), enrolling the ‘original’ cohort of 5209 individuals. In 1971 children and spouses of the ‘original’ cohort were invited to join the study, and 5124 attended the first examination of the Framingham Offspring cohort. Of these, 3951 Offspring cohort participants aged 30 to 74 attended the fourth Framingham Offspring examination between 1987 and 1992. We excluded participants with prevalent CVD and missing standard risk factors including lipid levels, leaving 3264 individuals eligible for this analysis. During 10 years of follow-up, 183 individuals experienced their first CHD event (myocardial infarction, angina pectoris, coronary insufficiency, or coronary heart disease death). All participants gave written informed consent and the study protocol was approved by the Institutional Review Board of the Boston Medical Center.
We applied the SAS software, version 9.1 (Copyright 2002–2003 SAS Institute Inc., Cary, NC, USA) to fit two proportional hazards regression models: the first one (which we will call “old”) included sex, diabetes and smoking as dichotomous and age, systolic blood pressure, total cholesterol as continuous predictors; the second model (called “new”) used the same predictors plus HDL cholesterol. The metrics employed to determine the usefulness of HDL cholesterol when added to the “old” model are summarized in table 1 and described in the Results.
Table 1.
Statistical methods used to determine usefulness of new biomarkers
| Name | Symbol | Purpose | Description | Advantages | Limitations |
|---|---|---|---|---|---|
| p-value | p-value | Assessment of statistical significance | Probability of occurrence of observed effect if null hypothesis true | Well known Easy to interpret |
Gives no sense of strength of association or effect size |
| Hazard ratio (Odds ratio, risk ratio) |
HR (OR, RR) |
Quantifying relative risk | Ratio of risks between to exposure groups expressed as hazards (odds, event rates) | Known Easy to interpret |
Describes only individual variables with focus on relative and not absolute risk Not possible compare variables with different distributions |
| Increase in c statistic (increase in AUC) | IAUC | Quantifying improvement in discrimination (model’s ability to distinguish events from nonevents) | Difference of probability that model assigns higher risk for those who develop events vs. those who do not between new and old models | Known Compares models and not individual variables Biomarkers differently distributed can be compared Based on ranks (not affected by calibration) |
Does not change materially even for powerful predictors Directly affected by goodness of baseline model Does not have well accepted interpretation |
| Reclassification table | Describing change in categories between models with and without new biomarker | Cross-tabulation of risk categories between model with and without new biomarker | Compares models and not individual variables Biomarkers differently distributed can be compared Presents full data Allows multiple simple checks |
Dependent on choice and number of categories No simple summary measure Can be interpreted differently by different readers |
|
| Difference between predictiveness curves | Depiction of difference in risk spread between models with and without new biomarker | Plots of model-based predicted probabilities as function of risk quantiles for models with and without new biomarker | Compares models and not individual variables Biomarkers differently distributed can be compared Intuitive and easy to interpret Allows depiction of entire distributions |
Measures spread and not necessarily correctness Does not change materially even for powerful predictors |
|
| Net Reclassification Improvement with categories | NRI | Quantifying amount of correct reclassification introduced by using model with new biomarker | Net proportion of events reclassified correctly plus net proportion of nonevents reclassified correctly | Compares models and not individual variables Biomarkers differently distributed can be compared Summary measure for reclassification tables Intuitive and related to clinical practice |
Dependent on choice and number of categories Ranges of meaningful improvement not established |
| Net Reclassification Improvement without categories | NRI(>0) | Quantifying amount of correct change in model-based probabilities introduced by using model with new biomarker | Net proportion of events with increased model-based probability plus net proportion of nonevents with decreased model-based probability | Compares models and not individual variables Biomarkers differently distributed can be compared Summary measure Not affected by calibration Does not directly depend on goodness of baseline model |
Ranges of meaningful improvement not established |
| Integrated Discrimination Improvement | IDI | Quantifying increase in separation of events and nonevents | Difference of differences between means of model-based probabilities for events and nonevents for model with and without new biomarker | Compares models and not individual variables Biomarkers differently distributed can be compared Summary measure Intuitive and meaningful |
Sensitive to differences in event rates Ranges of meaningful improvement not established |
| Relative Integrated Discrimination Improvement | rIDI | Quantifying increase in separation of events and nonevents on a relative scale | Ratio of differences between means of model-based probabilities for events and nonevents for model with and without new biomarker minus 1 | Compares models and not individual variables Biomarkers differently distributed can be compared Summary measure Relative scale may improve interpretability |
Ranges of meaningful improvement not established |
Results
Standard Metrics
The most basic requirement put on a new marker under consideration is that it is statistically significantly associated with the outcome of the study. The purpose behind this is simple: we need to rule out the possibility that the observed association might be due to chance. Statistical significance is determined using a p-value and the level of “less than 0.05” is universally accepted. However, what is frequently overlooked is that p-value is a function of both effect size (how strong the association is) as well as sample size (how large is our study or how many events we observed). Thus, large studies are very likely to declare statistical significance, even though the magnitude of the effect might be miniscule. Hence, the concept of clinical significance has been put forth to reduce the impact of sample size. It is usually associated with the observed relative risk expressed using risk, odds or hazard ratios, or as an absolute risk expressed as risk difference, or as its inverse known as the number-needed-to-treat (23). In the context of risk prediction and adding new biomarkers to existing models, these risk metrics are estimated using models that already adjust for standard risk factors. The phrase independent contribution is used in clinical literature to emphasize that the analysis took into account existing risk factors. In simple terms, a new marker that is associated with the risk of outcome on its own may not be of much use in risk prediction, if its correlation with standard factors diminishes its contribution to the overall model. In our example, the hazard ratio per one standard deviation increase in HDL cholesterol was 0.65 (95% confidence interval: 0.53, 0.80), and it was significant with p-value < 0.001.
While these standard risk metrics are commonly used and well understood and provide useful information about the magnitude of association, they may not give a complete assessment of the added contribution of the new marker in the context of risk prediction. It is generally true that the higher the magnitude of absolute or relative risk for a new biomarker in a model adjusted for standard risk factors, the more the ‘gain’ in model performance that can be expected. However, the questions of “how much ‘gain’ is good enough” and “how to compare contributions of markers evaluated on different scales (continuous, ordinal, binary)” have not been answered. The latter question has become even more relevant with flexible modeling techniques (splines etc.) increasing in popularity and application.
The area under the receiver-operating-characteristics (ROC) curve (AUC), often called the c statistic, has become the standard metric for assessing performance of models for binary outcomes. Hanley and McNeil (17) have shown that AUC is equal to the probability that given two subjects, one with and one without an event of interest, the one with event will have a higher model-based predicted probability of an event. In simpler terms this means that the model is more likely to assign higher risks to people with events, which is obviously a desirable property. On the other hand, the relationship between the AUC and the plot of ‘Sensitivity’ versus ‘1 – Specificity’ (the ROC curve) is appealing for the purpose of risk classification. Any paper attempting to propose a new risk prediction model is expected to report the value of the c statistic.
Given the above, it seems natural to use the increment in the c statistic as a method of quantifying the added value offered by new biomarkers, in a manner similar to the increment in R-square used in linear regression. However, recently researchers have observed (based on empirical applications) that the IAUC is very small when the baseline model’s c is large (24). On one hand, this observation should not be surprising: good models are harder to improve upon. However, the extent to which IAUC depends on baseline c (rather than the effect size of association for a new biomarker) seems undesirably large.
The above empirical finding can be illustrated with the help of the following simple simulation. A baseline model for a binary outcome with the c statistics ranging from 0.50 to 0.99 was constructed using a single predictor with necessary effect size, and then a new marker uncorrelated with the first one was added to that model. The effect sizes (ES) of the new marker of 0.18, 0.41 and 0.69 were selected to correspond to odds ratios of 1.2, 1.5 and 2.0, respectively. Then, the IAUCs between the baseline and new models were calculated and plotted against the baseline c and are displayed in Figure 1.
Figure 1.
Increase in area under the curve (c statistic) as a function of baseline c statistic for three different effect sizes
We observe how quickly the IAUCs decline as a function of the baseline c. Baseline models with c above 0.75 cannot be improved by more than 0.05, even if the effect size is as large as 0.69 (OR per one standard deviation of 2.0). A marker with odds ratio of 1.2 per standard deviation can add only a miniscule amount to the c statistic. Note that these results were obtained in the optimistic scenario of no correlation between the new marker and baseline model predictors. In case of correlation greater than zero between a new biomarker and standard risk factors, odds ratios of the magnitudes presented here will be even harder to attain. In our HDL example, the c statistics for the old and new models were 0.762 and 0.774, yielding a small IAUC = 0.012 (p-value = 0.092).
Another criticism of the c statistic and the IAUC came from the CVD risk prediction domain, where treatment guidelines are based on generally accepted risk categories based on absolute 10-year predicted event rates. Since the c statistic is not influenced by absolute risk levels, it was argued it cannot adequately capture the clinical usefulness of new markers in situations where treatment decisions are based on established categories (18). Reclassifications tables were proposed instead (18), and appropriate ways of interpreting them were put forth (19).
Reclassification Table Metrics and their Continuous Counterparts
As mentioned above, some clinical decisions based on risk prediction algorithms are based on established categories of absolute risk. Primary prevention of CVD or CHD serves as the key example, where people with predicted 10-year risk above 20% are recommended for treatment, whereas those with risk below 10% (or more recently 6%) are considered ‘low risk’ (9,10,25). Adherence to such categories suggests that a lot can be learned about the usefulness of a new marker from simple cross-tabulation of risk categories (for example, ‘low risk’ 0–6%, ‘medium risk’ 6–20%, ‘high risk’ > 20%) based on predicted probabilities obtained using models without and with the new marker. Janes et al. (26) observe that key information is contained in the margins of the reclassification table and suggest looking at the following three characteristics (“checks”) that can be derived from such a table:
Calibration;
Risk stratification capacity;
Classification accuracy.
We illustrate their meaning and interpretation using data published in (19) and described in the Materials and Methods section looking at the added usefulness of HDL cholesterol in a 10-year risk prediction model of CHD. The reclassification table is given as Table 2 and consists of 3 parts, first for individuals with CVD events, second for individuals who do not experience CVD events (nonevents) and third for all individuals combined. The first two have been presented in (19), the third one is added here to illustrate additional concepts.
Table 2.
Reclassification table for models without and with HDL cholesterol
| Old Model |
New Model | ||||
|---|---|---|---|---|---|
| 0–6% | 6–20% | >20% | Total | ||
| Event | 0–6% | 39 | 15 | 0 | 54 |
| 6–20% | 4 | 87 | 14 | 105 | |
| >20% | 0 | 3 | 21 | 24 | |
| Total | 43 | 105 | 35 | 183 | |
| Nonevent | 0–6% | 1959 | 142 | 0 | 2101 |
| 6–20% | 148 | 703 | 31 | 882 | |
| >20% | 1 | 25 | 72 | 98 | |
| Total | 2108 | 870 | 103 | 3081 | |
| All | 0–6% | 1998 | 157 | 0 | 2155 (66%) %event =2.5% |
| 6–20% | 152 | 790 | 45 | 987 (30%) %event=10.6% |
|
| >20% | 1 | 28 | 93 | 122 (4%) %event=19.7% |
|
| Total (%) | 2151 (66%) %event=2.0% |
975 (30%) %event=10.8% |
138 (4%) %event=25.4% |
3264 | |
Good calibration means that model-based predicted event rates closely match those observed in practice. In the context of a reclassification table, the simplest calibration check looks at the observed event rates presented in the margins of the third part of the table, which show data on all individuals combined and determines if they fall into the risk categories to which they correspond. In our example, event rates for the “old model” (i.e., the one without HDL) are given in the row margins as 2.5%, 10.6% and 19.7% for risk categories of 0–6%, 6–20% and >20%, respectively. For simplicity, we used crude event rates, but Kaplan-Meier rates could be presented instead, and would be even more appropriate. It is evident that that the first two rates fall into the respective categories, while the third one is on the border (19.7% is very close to but technically outside the >20% category). The model with HDL does a little better with respective rates of 2.0%, 10.8% and 25.4% well within their respective categories.
The above ‘check’ is descriptive. A formal test would follow the Hosmer – Lemeshow (27) chi-square approach (or Nam and D’Agostino’s analog for survival data (28)), and one could argue that the more traditional, decile-based presentation is equally if not more meaningful. Another approach would look at the plot of predicted vs. observed risk – an illustration of this approach has been presented in a recent paper on reporting guidelines for biomarker studies in CVD risk prediction by Hlatky et al. (29).
It is important to note that using standard analytic methods, which include logistic and proportional hazards regressions, one is almost guaranteed to obtain a reasonable degree of model calibration. Largest violations are observed when the mean of model-based predictions is different than the incidence rate in the cohort being analyzed (it is referred to as bias (D’Agostino et al. (30))). However, the two regressions mentioned above introduce a minimal bias in the sample in which the model was developed, and thus should lead to a reasonably good calibration if the number of predictors is sufficient.
The second ‘check’ suggested by Janes et al. (26) relates to discrimination understood as the model’s ability to spread predicted risk across individuals. A simple check coming from the reclassification table looks at the sizes of the three risk categories. Better models will tend to have more people in the lowest and highest risk groups. In our example, again looking at the third part of the reclassification table 1, these numbers are approximately identical for the old and new models, with 66%, 30% and 4% across the three risk groups. In particular, we observe very little shrinkage of the middle risk group.
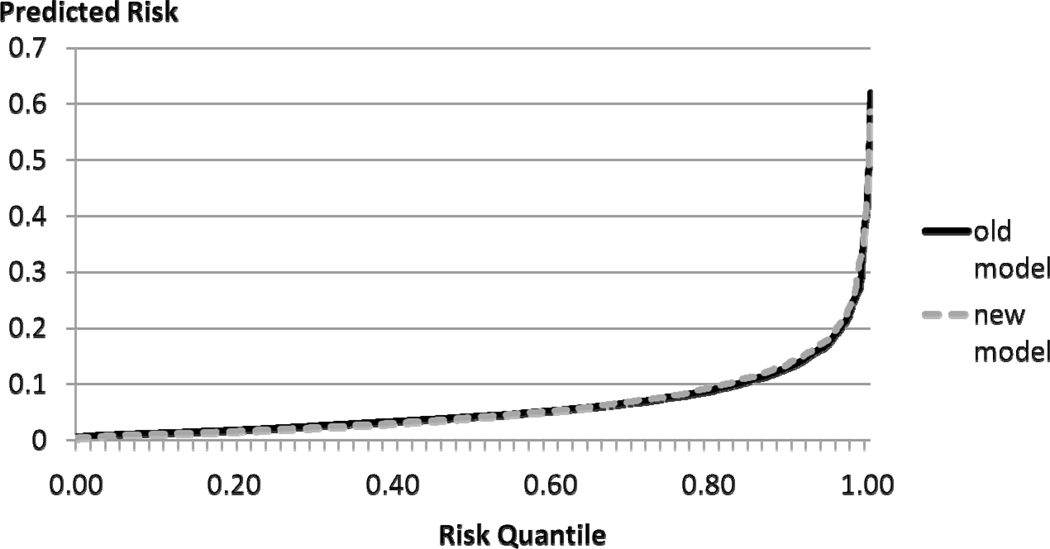
A more general way of inspecting the amount of spread offered by a given model can be depicted using the predictiveness curve introduced by Pepe et al. (21). It is constructed by ranking all predicted probabilities and plotting them against the quantile of risk. Ideally, the predictiveness curve remains very close to the horizontal axis and then increases very rapidly. If meaningful categories exist, they can be marked on the vertical axis and then corresponding quantiles of risk determined on the horizontal axis to determine the size of the middle risk groups, similarly to the reclassification table. Other useful properties of this curve have been described by Pepe et al. (21). Figure 2 shows two predictiveness curves, one for the model without and one for the model with HDL.
Figure 2.
Predictiveness curves for coronary heart disease risk prediction model without (old model) and with HDL cholesterol (new model)
The curves are virtually overlapping. The plot leads to the same conclusion as the one derived from looking at the margins of the reclassification table: there is very little difference between the ‘spread-ability’ of the two models. In our experience, this conclusion is very common when examining the usefulness of new biomarkers. It is a question for future research to determine if this is caused by the inadequacy of the markers under consideration, or if it is an inherent property of the predictiveness curve itself.
While it is desirable ‘to spread’ the predicted risks as much as possible, the fact of adequate separation does not have to guarantee that events get higher predicted probabilities and non-events get low predicted probabilities, a condition necessary for a good prediction model. When significant predictors are used, it is very unlikely that a large separation would not imply good classification accuracy, but direct measures of the latter are clearly desirable.
In the context of reclassification tables, the net reclassification improvement (NRI) offers a simple yet meaningful check of classification accuracy (19). In its simplest form for binary outcomes, NRI is calculated by examining events and non-events separately (first two parts of reclassification Table 1). Only people who change risk categories contribute to the NRI, since for those who do not, the models perform the same. It is desirable to increase the predicted probabilities of event for those who experience events, and hence any upward reclassification among events is considered beneficial. It is quantified by (bold-face) numbers above the diagonal in the first (event) part of Table 1: we get (15+0+14) = 29. This gain is offset by people with events who move down in categories using the new model. Their number is below the diagonal, and can be calculated as (4+0+3) = 7. Hence the net improvement is (29−7) = 22 out of 183 people with events, which gives a percentage improvement of (22/183) = 12.0%. For nonevents, the reasoning is reversed: downward movement in categories is considered beneficial. Looking at the second part of reclassification Table 1, we see (148+1+25) = 174 individuals without events moving down (bold-face numbers below the diagonal) and (142+0+31) = 173 moving upwards for a net gain of (174−173) = 1 person out of 3081, or 0.0%. The total NRI is calculated as the sum of the two, which in this case is 12%. The total NRI assumed implicit weighting of importance proportional to the odds of nonevents, in this case (incidence rate = 6%) it is 0.94/0.06 = 15.7:1. This means that each misclassified event is 15.7 more important than each misclassified ‘nonevent.’ Other weightings are also possible (cf. Vickers (31)). But at least as informative as the total NRI are its two components – the one calculated for events and one for nonevents. There is a 12% improvement in reclassification of events, and no change in reclassification of nonevents. Thus, the improvement in reclassification of events comes only at the cost of measuring HDL cholesterol.
Another useful look at the NRI focuses on a prospective interpretation and allows extension to survival data. We calculate the Kaplan-Meier event rates for individuals reclassified upwards and downwards, and compare them to the overall Kaplan-Meier rate. Ideally, the rate for those going up will be much higher than the overall rate, while the rate for those going down will be lower. In our example, the rate for those reclassified upwards is 15.1% and for those reclassified downwards it is 4.1% as compared to the rate of 6.0% for everyone combined, which fulfills the desired relationship: 15.1% > 6.0% > 4.1%. We conclude that people reclassified upwards are at higher risk than the average, while people reclassified downwards are at a lower risk.
Seeing the simple and informative nature of NRI and its components one wants to ask whether this approach can be generalized to situations where there are no categories. There are two ways in which NRI can be extended to situations where no meaningful categories exist. The first one defines any change in predicted probability as either upward or downward movement, depending on the direction. Since predicted probabilities are continuous if at least one of the predictors is, this implies that every person will be reclassified. In our example, this category-less NRI(>0) is 30.2%, with the corresponding event and nonevent components of 24.6% and 5.6%, respectively. These numbers are much larger than the ones we observed when categories were present – this is to be expected as presence of categories substantially reduces the amount of reclassification estimated. This example also illustrates the phenomenon of NRI increasing with the number of categories with the category-less NRI(>0) usually serving as the upper limit. However, the conclusions about reclassification of events and nonevents remain similar – much more improvement is seen for events than for nonevents.
Another way to extend NRI to the case of no categories is accomplished by assigning a weight to each movement that is equal to difference of probabilities from the old and new models. This leads to another measure of accuracy of prediction model which Pencina et al. (19) called the integrated discrimination improvement (IDI). They have also shown that IDI is equal to the difference in discrimination slopes as proposed by Yates (32). The discrimination slope has a nice intuitive definition: it is the difference in means of predicted probabilities for events and nonevents (in other words, it is a measure of separation in predicted probabilities for event and nonevents). In our example, these slopes for models with and without HDL are 0.0715 and 0.0630, yielding an IDI of 0.0085. Discrimination slopes and IDI depend on incidence of the outcome of interest, and further research is needed to gain some intuition of what an IDI of 0.0085 really means beyond its statistical significance at p-value = 0.016. One potential direction is to look at the relative IDI (rIDI) defined as the increase in discrimination slopes divided by the slope of the old model (33). In our case it is 13.5% (0.0085/0.0630). The following, heuristic argument might help assess the magnitude of rIDI. If every variable was to contribute equally to discrimination slope, and the old model had 6 predictors, the average contribution would be 16.6%. This is the incremental contribution that would be expected from the new variable. HDL with 13.5% rIDI comes close to the expectation. Alternatively, treating age as a time-scale adjustment rather than a risk factor, one looks at the relative increase beyond age. The contribution of the five risk factors other than age to the old model’s slope is 0.0400, resulting in non-age rIDI of 21%. The expected number based on 5 risk factors would be 20%, indicating that HDL offers a similar magnitude of improvement as the average of risk factors used in the old model.
Discussion
The purpose of this review was to summarize several contemporary methods of measuring the contribution of new markers to risk prediction models. We concluded that if meaningful risk categories exist that relate to clinical decisions, reclassification tables can provide very useful summaries of added benefit of new markers. Looking at the margins of reclassification table that combines all study individuals, we can get a sense of calibration and separation improvements offered by a putative new biomarker. However, the most important check looks at the improvement in classification accuracy, and can be accomplished by calculating the NRI, separately for events and nonevents. Various weights can then be applied to obtain a full sample summary measure. In cases where meaningful risk categories are not available or categorical analysis is not desired due to concerns of information loss, continuous equivalents to the reclassification table summaries can be calculated. In general continuous analysis of risk reclassification is preferable; however, if risk categories are used in practical applications, it is intuitive to employ them. Continuous equivalents of NRI include its category-less version, and difference in discrimination slopes called the IDI. For ease of interpretation, relative IDI might be preferred.
The appearance of new metrics for measuring the added usefulness of new biomarkers does not mean that the more standard and widely used statistics should not be calculated. On the contrary, we recommend reporting of measures of relative risk with their 95% confidence intervals and/or p-values as well as c statistic for the baseline and new model. C statistics have been in common use for the last 30 years and the familiarity aspect is important here. Before evaluating the new biomarker, it is essential to ascertain that the baseline model obtained before the addition of the new biomarker is as good as possible. One of the easiest ways to ascertain it is by looking at the c statistic.
Our focus here was limited to quantifying the improvement in risk prediction offered by new biomarker. As such, we did not address numerous key conditions necessary for successful research intended to assess the usefulness of biomarkers in risk prediction. We outline some of them here. First, as postulated for genetic markers, there are many components necessary to establish clinical utility (34). At its core are clinical and analytic validity. The former was partially addressed in this paper. The latter pertains to the quality and reproducibility of the biomarker assay. A biomarker with great potential for improvement in risk prediction in one study can prove practically useless if it cannot be reliably measured elsewhere. Second, it is necessary that the sample on which we evaluate the usefulness of new biomarkers is representative of the population of interest, and the definition of outcome in the study must agree with that to be used for its general clinical applications. In this context external validation of risk prediction models in different samples from the same population is essential to establish generalizability and avoid over-optimistic performance. For the same reasons we also suggest that new biomarkers be evaluated after standard variables are entered into the risk prediction model individually rather than in single fixed-weight score coming from existing sources. Third, in some settings measurement of standard risk factors may be impossible or inaccurate. Then biomarkers might be use as an alternative rather than addition to existing models. In these settings we would compare two models, one based on standard risk factors and one based on biomarkers. In this context all methods presented in table 1 expect for first two (p-values and relative risks) remain applicable. Fourth, our example and descriptions focused on a follow-up study; however, the methods presented in table 1 can be extended to case-control studies.
Finally, we need to ask a question how much improvement is really possible with new biomarkers. Hand (35) in his informative review makes two important observations. First, the level of complexity of the risk prediction model put forth is not directly related to the amount of improvement in performance. Generally, simple models are able to capture the majority of variability that remains to be explained. Second, new variables to be considered for risk prediction are likely to be correlated with those already present in the model and hence offer limited gains in performance. Furthermore, risk prediction models are based on baseline level of risk factors. It is unrealistic to assume that all risk factor levels will remain constant during the follow-up. Their trajectories are not known and not measurable at baseline when risk prediction is made. Thus it is possible that lack of perfection is an inherent feature of risk prediction models. However, this should not discourage further research as in all fields there remain ample opportunities for improvement in existing models.
Non-Standard Abbreviations
- CVD
cardiovascular disease
- CHD
coronary heart disease
- NRI
net reclassification improvement
- IDI
integrated discrimination improvement
- rIDI
relative integrated discrimination improvement
- ROC
receiver operating characteristics
- AUC
area under (the ROC) curve
- IAUC
increase in area under curve
Footnotes
Conflict of Interest
Have you accepted any funding or support from an organization that may in any way gain or lose financially from the results of your study or the conclusions of your review? No
Have you been employed by an organization that may in any way gain or lose financially from the results of your study or the conclusions of your review? No
Do you have any other conflicting interests? No
References
- 1.Wang TJ, Gona P, Larson MG, Tofler G, Levy D, Newton-Cheh C, et al. Multiple biomarkers for the prediction of first major cardiovascular events and death. N Engl J Med. 2006;355:2631–2639. doi: 10.1056/NEJMoa055373. [DOI] [PubMed] [Google Scholar]
- 2.Lee DS, Evans JC, Robins SJ, Wilson PWF, Albano I, Fox CS, et al. Gamma glutamyl transferase and metabolic syndrome, cardiovascular disease, and mortality risk: the Framingham Heart Study. Arterioscler Thromb Vasc Biol. 2007;27:127–133. doi: 10.1161/01.ATV.0000251993.20372.40. [DOI] [PubMed] [Google Scholar]
- 3.Schnabel RB, Larson MG, Yamamoto J, Sullivan LM, Pencina MJ, Meigs JB, et al. Relations of biomarkers of distinct pathophysiological pathways and atrial fibrillation incidence in the community. Circulation. 2010;121:200–207. doi: 10.1161/CIRCULATIONAHA.109.882241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ridker PM, Buring JE, Rifai N, Cook N. Development and validation of improved algorithms for the assessment of global cardiovascular risk in women. JAMA. 2007;297:611–619. doi: 10.1001/jama.297.6.611. [DOI] [PubMed] [Google Scholar]
- 5.Ridker PM, Paynter NP, Rifai N. C-reactive protein and parental history improve global cardiovascular risk prediction: the Reynolds Risk Score for men. Circulation. 2008;118:2243–2251. doi: 10.1161/CIRCULATIONAHA.108.814251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson PWF, Pencina MJ, Jacques P, Selhub J, D’Agostino RB, O’Donnell CJ. C-reactive protein and reclassification of cardiovascular risk in the Framingham Heart Study. Circ Cardiovasc Qual Outcomes. 2008;1:92–97. doi: 10.1161/CIRCOUTCOMES.108.831198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridker PM, Danielson E, Fonseca FAH, Genest J, Gotto AM, Kastelein JJP, et al. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. NEJM. 2008;359:2195–2207. doi: 10.1056/NEJMoa0807646. [DOI] [PubMed] [Google Scholar]
- 8.Wilson PWF, D’Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–1847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 9.Executive summary of the third report of the National Cholesterol Education Program (NCEP) Expert Panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III) JAMA. 2001;285:2486–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 10.D'Agostino RB, Vasan RS, Pencina MJ, Wolf PA, Cobain M, Massaro JM, Kannel WB. General cardiovascular risk profile for use in primary care. Circulation. 2008;117:743–753. doi: 10.1161/CIRCULATIONAHA.107.699579. [DOI] [PubMed] [Google Scholar]
- 11.Conroy RM, Pyorala K, Fitzgerald AP, Sans S, Menotti A, DeBacker G, et al. Estimation of ten-year risk of fatal cardiovascular disease in Europe: the SCORE project. Eur Heart J. 2003;24:987–1003. doi: 10.1016/s0195-668x(03)00114-3. [DOI] [PubMed] [Google Scholar]
- 12.Parikh NI, Pencina MJ, Wang TJ, Benjamin EJ, Lanier KJ, Levy D, et al. A risk score for predicting near-term incidence of hypertension: the Framingham Heart Study. Ann Intern Med. 2008;148:102–110. doi: 10.7326/0003-4819-148-2-200801150-00005. [DOI] [PubMed] [Google Scholar]
- 13.Meigs JB, Shrader P, Sullivan L, McAteer JB, Fox CS, Dupuis J, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. NEJM. 2008;359:2208–2219. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gail MH, Brinton LA, Byar DP, Corle DK, Green SB, Shairer C, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81:1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 15.Kannel WB, D’Agostino RB, Silbershatz H, Belander AJ, Wilson PWF, Levy D. Profile for estimating risk of heart failure. Arch Intern Med. 1999;159:1197–1204. doi: 10.1001/archinte.159.11.1197. [DOI] [PubMed] [Google Scholar]
- 16.Schnabel RB, Sullivan LM, Levy D, Pencina MJ, Massaro JM, D’Agostino RB, et al. Development of a risk score for atrial fibrillation (Framingham Heart Study): a community-based cohort study. Lancet. 2009;373:739–745. doi: 10.1016/S0140-6736(09)60443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143(1):29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 18.Cook NR. Use and misuse of the receiver operating characteristics curve in risk prediction. Circulation. 2007;115:928–935. doi: 10.1161/CIRCULATIONAHA.106.672402. [DOI] [PubMed] [Google Scholar]
- 19.Pencina MJ, D'Agostino RB, Sr, D'Agostino RB, Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Statist Med. 2008;27:157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 20.Pencina MJ, D’Agostino RB. Overall c as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Statist Med. 2004;23:2109–2123. doi: 10.1002/sim.1802. [DOI] [PubMed] [Google Scholar]
- 21.Pepe MS, Feng Z, Huang Y, Longton G, Prentice R, Thompson I, et al. Integrating the predictiveness of a marker with its performance as a clasifier. Am J Epidemiol. 2008;167:362–368. doi: 10.1093/aje/kwm305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kannel WB, McGee D. A general cardiovascular risk profile: the Framingham Study. Am J Cardiol. 1976;38:46–51. doi: 10.1016/0002-9149(76)90061-8. [DOI] [PubMed] [Google Scholar]
- 23.Laupacis A, Sackett DL, Roberts RS. An assessment of clinically useful measures of the consequences of treatment. NEJM. 1988;318:1728–1733. doi: 10.1056/NEJM198806303182605. [DOI] [PubMed] [Google Scholar]
- 24.Tzoulaki I, Liberopoulos G, Ioannidis JPA. Assessment of claims of improved prediction beyond the Framingham risk score. JAMA. 2009;302:2345–2352. doi: 10.1001/jama.2009.1757. [DOI] [PubMed] [Google Scholar]
- 25.Pencina MJ, D’Agostino RB, Larson MG, Massaro JM, Vasan RS. Predicting the 30-year risk of cardiovascular disease. the Framingham Heart Study. Circulation. 2009;119:3078–3084. doi: 10.1161/CIRCULATIONAHA.108.816694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janes H, Pepe M, Gu W. Assessing the value of risk predictions by using risk stratification tables. Ann Intern Med. 2008;149:751–760. doi: 10.7326/0003-4819-149-10-200811180-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosmer DW, Jr, Lemeshow S. Applied Logistic Regression. New York: John Wiley and Sons, Inc.; 1989. [Google Scholar]
- 28.D’Agostino RB, Nam BH. Handbook of Statistics. Vol. 23. Amsterdam: Elsevier; 2004. Evaluation of the performance of survival analysis models: discrimination and calibration measures. [Google Scholar]
- 29.Hlatky MA, Greenland P, Arnett DK, Ballantyne CM, Criqui MH, Elkind MSV, et al. Criteria for evaluation of novel cardiovascular risk: a scientific statement from the American Heart Association. Circulation. 2009;119:2408–2416. doi: 10.1161/CIRCULATIONAHA.109.192278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D'Agostino RB, Griffith JL, Schmidt CH, Terrin N. Proceedings of the biometrics section. Alexandria VA: American Statistical Association, Biometrics Section; 1997. Measures for evaluating model performance; pp. 253–258. [Google Scholar]
- 31.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yates JF. External correspondence: decomposition of the mean probability score. Organ Behav and Hum Per. 1982;30:132–156. [Google Scholar]
- 33.Pencina MJ, D'Agostino RB, Sr, D'Agostino RB, Jr, Vasan RS. Comments on integrated discrimination and net reclassification improvements – practical advice. Statist Med. 2008;27:207–212. [Google Scholar]
- 34.Haddow JE, Palomaki GE. ACCE: A model process for evaluating data on emerging genetic tests. In: Khoury M, Little J, Burke W, editors. Human genome epidemiology: a scientific foundation for using genetic information to improve health and prevent disease. Oxford University Press; 2003. pp. 217–233. [Google Scholar]
- 35.Hand DJ. Classifier technology and the illusion of progress. Stat Sci. 2006;21:1–14. [Google Scholar]