Abstract
Studies of Drosophila and mammals have documented circadian changes in the morphology and biochemistry of glial cells. In addition, it is known that astrocytes of flies and mammals contain evolutionarily conserved circadian molecular oscillators that are similar to neuronal oscillators. In several sections of this review, I summarize the morphological and biochemical rhythms of glia that may contribute to circadian control. I also discuss the evidence suggesting that glia-neuron interactions may be critical for circadian timing in both flies and mammals. Throughout the review, I attempt to compare and contrast findings from these invertebrate and vertebrate models so as to provide a synthesis of current knowledge and indicate potential research avenues that may be useful for better understanding the roles of glial cells in the circadian system.
Keywords: Drosophila, circadian, glia, astrocyte
INTRODUCTION
Comparisons of vertebrate and insect glial cells have suggested that certain classes have common morphological attributes and molecular signatures (see review of M. Freeman, this issue). Much is known about the glial cell classes populating the mammalian brain, and recent studies have described the development, morphology and functions of the glial cell classes in the larval and adult Drosophila nervous systems (Awasaki et al., 2008; Doherty et al., 2009; Edwards and Meinertzhagen, 2010; Parker and Auld, 2006; Pereanu et al., 2005; Silies et al., 2007; Spindler et al., 2009). Other studies have documented parallels between Drosophila and mammals with regard to the functions of glial cells in development, physiology, and neural responses to injury (Bainton et al., 2005; Cafferty and Auld, 2007; Crews, 2010; Doherty et al., 2009;Freeman and Doherty, 2006; Jackson and Haydon, 2008; Silies et al., 2007). These same topics are covered in other reviews present in this special issue of Glia. I note that more general reviews have described roles for glial cells in regulating neurotransmission and complex behaviors of Drosophila and mammals (Haydon et al., 2009; Jackson and Haydon, 2008). Therefore, this review will focus specifically on the functions of glial cells in the regulation of circadian behavior, with an emphasis on a comparison of findings from studies of flies and mammals.
THE MOLECULAR CLOCKS OF FLIES AND HUMANS ARE SIMILAR
There are several previous reviews comparing the clock systems of Drosophila, mammals and other species (Helfrich-Forster, 2004; Reppert and Weaver, 2000; Young and Kay, 2001). Therefore, I will not provide a detailed description of the molecular and cellular biology of circadian clocks in this review. Rather, it is sufficient to note that the molecular oscillators driving behavioral rhythmicity in flies and mammals, including humans, are quite similar. In all animal systems, including Drosophila (Hardin, 2005), they are composed of multiple, interlocked transcriptional/translational feedback loops in which clock proteins (PERIOD and others) translocate from the cytoplasm to the nucleus to regulate the activities of transcription factors (CLOCK and others) that are required for clock gene expression (Fig. 1A shows a schematic model of the fly molecular clock). The subsequent degradation of clock proteins results in 24-h rhythms of clock gene transcription and protein abundance which allow animals to keep time (see Fig. 1B). Layered upon this transcriptional rhythm are post-transcriptional events such as translational regulation, protein phosphorylation, and ubiquitination which regulate clock protein abundance and activities (not shown in Fig. 1), providing time lags and adding to the stability of the timekeeping mechanism (Zheng and Sehgal, 2008). Understanding the cellular and molecular basis of timekeeping has great biomedical significance because of the many neurological and psychiatric disorders associated with altered circadian function (Wulff et al., 2010). Indeed, single gene mutations in clock loci have been associated with human sleep disorders and polymorphisms have been linked to sleep and affective disorders including seasonal affective disorder and other types of depression.
Fig. 1.

Molecular clock and temporal expression of clock products. A: Schematic diagram of transcriptional interactions amount clock components. activation; repression. CWO may also mediate repression of output genes (not shown). B: Diurnal expression profiles for clock RNAs and proteins. Black lines represent RNA abundance profiles for per, tim and Clk. Colored lines indicate protein abundance profiles for clock proteins and immunofluorescence signals for Pigment Dispersing Factor (PDF), a clock output peptide. pCLK represents phosphorylated, activated CLK which has a circadian phase hours later than the Clk transcription rhythm. The PDF curve illustrates peptide immunoreactivity in the s-LNv dorsal projections; reduced amounts during the day are presumed to reflect release of the peptide.
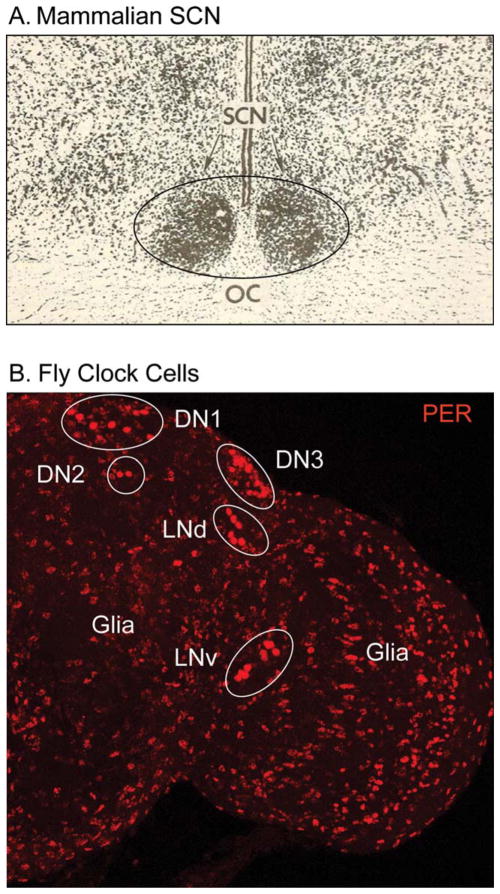
Both Drosophila and mammals possess intracellular circadian molecular oscillators. In Drosophila, each of ~150 communicating clock neurons contains a circadian oscillator and these neurons are distributed within several regions of the fly brain (Nitabach and Taghert, 2008). This neuronal population includes the Pigment Dispersing Factor (PDF)-containing lateral ventral neurons (LNv) that are known to be critical for behavioral rhythmicity. In contrast, the mammalian pacemaker contains approximately 100 times as many neurons (~20,000) that are localized to a discrete region of brain: the Suprachiasmatic Nuclei (SCN) within the anterior hypothalamus (Moore and Eichler, 1972; Silver and Schwartz, 2005; Stephan and Zucker, 1972) (see Fig. 2). In both flies and mammals, light stimuli—which reset the clock—are delivered to clock neurons via multiple photoreceptors, emphasizing the importance of environmental input to the timing system. Glia of the fly brain and astrocytes of the SCN also contain PERIOD (PER)-based molecular oscillators, consistent with the idea that these cells may be important components of the circadian circuitry. That concept is developed in more detail in the remainder of this review.
Fig. 2.
Locations of circadian clock cells in (A) the rat suprachiasmatic nuclei (SCN) and (B) the Drosophila brain. Only one “hemisphere” of the fly brain is pictured in this image, but clock cells are bilaterally localized in the brain. The white ovals encompass clock neurons of the fly brain; PER-expressing glial cells are distributed throughout the brain. The black oval in A delineates the clock neurons and astrocyctes of the rat brain. The image of the SCN was a gift from Dr. J. Levine (U. Toronto). OC, optic chiasm; LNv, ventral lateral clock neurons; LNd, dorsal lateral clock neurons; DN1, 2 and 3, the three groups of dorsal clock neurons.
GLIAL MOLECULAR CLOCKS
Glial clocks of flies
Early studies of the canonical clock protein PERIOD (PER) demonstrated that it was localized to neurons and glial cells of the fly brain, and that it showed robust circadian rhythms in abundance in both cell types (Zerr et al., 1990). This was the first work to suggest that glia may contain PER-based molecular oscillators. A later study explored the requirement for PER in different regions and cell types of the fly brain, using genetic mosaic analysis coupled with the characterization of circadian locomotor activity rhythms (Ewer et al., 1992). The observation that certain weakly rhythmic mosaic flies contained detectable PER only in glia of the brain was interpreted as evidence for a role of glial oscillators in the pacemaker driving rhythmic behavior. Indeed, both PER and TIMELESS (TIM), which function together in the fly clock (Hardin, 2005), are localized to a subset of adult glial cells, indicative of oscillator function (Suh and Jackson, 2007). This begs the question: do glial circadian oscillators have a direct role in the regulation of behavioral rhythmicity? Such oscillators might be important for free-running behavior (in constant conditions) or they might modulate behavior in other environmental conditions; e.g., they might regulate pacemaker light sensitivity by modulating dopaminergic functions (see discussion of Ebony in “Clock-controlled glial molecular rhythms”). Although clock proteins are localized to Drosophila glial cells, it is also not known whether the fly glial oscillators continue to run autonomously in the absence of neuronal clocks.
Astroglial clocks of mammals
Two studies have utilized reporter gene constructs to demonstrate rhythmic expression of clock genes in rat astrocytes (Prolo et al., 2005; Yagita et al., 2010). The earlier extensive analysis of Prolo et al. employed rat and mouse astroglia obtained from transgenic animals expressing a per-luciferase (per-luc) reporter. Those studies indicate that astrocytes contain a PER-based molecular oscillator that damps in the absence of neuronal signals. Primary cultures of cortical glia expressing the per-luc transgene exhibited circadian bioluminescence rhythms that damped after several cycles. Rhythms could be reinitiated by culture medium replacement or treatment with the calcium ionophore Calcimycin or the adenylate cyclase agonist Forskolin. Interestingly, culture medium replacement had a phase-dependent effect on re-initiation of rhythmicity, indicating the presence of a cellular oscillator. Finally, a fraction (30%) of astrogial cultures showed sustained rhythmicity (seven days or longer) when co-cultured with SCN explants, whereas cortical explants did not influence rhythmicity. This suggests that a secreted neuronal factor expressed in the SCN may be required for sustained rhythms. Thus, astroglia cultures behave as damped circadian oscillators that require neuronal signaling for either the maintenance of individual cell oscillations or the synchronization of glial cell clock populations.
RHYTHMIC CHANGES IN THE MORPHOLOGIES OF GLIA AND NEURONS
Morphology rhythms in flies
It is known that glial support functions or signaling (gliotransmission) can influence neuronal excitability, with both molecular and morphological changes perhaps contributing to altered glia-neuron communication. In both the housefly (Musca domestica) and the blowfly (Calliphora vicina), the L1 and L2 monopolar inter-neurons of the visual system (secondary neurons within the optic lamina) exhibit circadian changes in axon caliber that are postulated to regulate visual processing (Pyza and Meinertzhagen, 1995; Pyza and Cymborowski, 2001). Interestingly, the axonal size rhythm is apparently driven by alterations of glial cell morphology: epithelial glia of the lamina that are intimately associated with L1 and L2 display rhythmic changes in size with a phase opposite that of the neurons; i.e., axonal caliber is largest when glial cells shrink to minimum size (Pyza and Gorska-Andrzejak, 2004). Injections of glial cell metabolic inhibitors (fluorocitrate, iodoacetate) into live flies disrupt the glial circadian rhythm, leaving glia at a small or intermediate size, but such inhibitors enhance the amplitude of the neuronal rhythm, indicating that increases in glial size may normally counteract the neuronal size change (Pyza and Gorska-Andrzejak, 2004). In contrast, Pyza and Gorska-Andrzejak showed that octanol injections eliminate both the glial and L1/L2 rhythms of houseflies, indicating that gap junctional coupling, which is present among all glia of the lamina, may be important for the morphology rhythms.
Given the results with houseflies, it is not surprising that there are circadian changes in the visual system monopolar neurons of Drosophila. Visualization of a GFP fluorescence reporter in the L2 neurons of Drosophila indicates that there are diurnal changes in axonal caliber, nuclear morphology and dendritic spine morphology when animals are maintained in a light-dark (LD) cycle (Gorska-Andrzejak et al., 2005). This does not, however, provide evidence that the L2 morphology changes are driven by an endogenous clock. However, other studies describe a modest circadian rhythm in the size of the L2 dendritic tree which persists in constant darkness (DD) and is eliminated in the Drosophila per01 mutant (Weber et al., 2009), indicative of circadian control. In unpublished work, the lab of Elżbieta Pyza has documented epithelial glial size rhythms in Drosophila (E. Pyza, pers. comm.), similar to those observed in other flies. In addition to regulating axon caliber, it is known that epithelial glia of the Drosophila lamina and Muller radial glial cells of the mammalian retina—participate in neuronal neurotransmitter recycling and other functions (Bringmann et al., 2006; Edwards and Meinertzhagen, 2010). Furthermore, one rhythmic glial factor—the Drosophila Ebony enzyme (Suh and Jackson, 2007)—participates in histamine recycling in the lamina (Stuart et al., 2007). Thus, glia may have multiple functions in the circadian regulation of visual processing and other processes in vertebrate and invertebrate species.
Astrocyte morphology rhythms in the mammalian brain
Similar to the fly studies, a previous analysis documented effects of glial inhibitors on the circadian rhythm of SCN neuronal firing observed in mammalian brain slice preparations: fluorocitrate induced ultradian rhythmicity (multiple peaks of firing during a single cycle) whereas octanol disrupted the rhythm (Prosser et al., 1994). This study and others (Shinohara et al., 2000; Van den Pol et al., 1992) suggested that glia and glia-neuron interactions might be important in the mammalian clock system as well.
Astrocytes and neurons in many regions of the mammalian brain exhibit activity-dependent morphological remodeling (Theodosis et al., 2008), but, in most cases, the circadian clock does not contribute to such changes. However, within the mammalian suprachiasmatic nuclei (SCN, the anatomical locus of the pacemaker), there are rhythmic changes in the abundance of Glial Fibrillary Acidic Protein (GFAP)—a molecular marker of astrocytes—and in the apparent morphology of astrocytes. In the hamster SCN, GFAP immunoreactivity is most anatomically restricted at CT14 (during the subjective night), a time corresponding with a pattern of staining that emphasizes individual, stellate-shaped astrocytes (Lavialle and Servière, 1993). At other times of the cycle, GFAP staining has the appearance of a network in which immunoreactivity is seen throughout the SCN. Interestingly, there is a rhythm of SCN glucose consumption in both rats and hamsters (Lavialle and Servière, 1993; Schwartz and Gainer, 1977), reflecting energy utilization, and consumption is lowest in the night. As astrocytes are a major source of glycogen in the brain (Brown and Ransom, 2007), Lavialle and Servière postulate that the changes in GFAP immunoreactivity may reflect the participation of SCN astrocytes in the circadian regulation of energy metabolism.
There are also rhythms in the morphology of rat SCN synapses and astrocytes in LD but these rhythms do not persist in DD, suggesting that the changes are a response to lighting conditions rather than endogenous mechanisms (Becquet et al., 2008; Girardet et al., 2010). Using a marker of presynaptic endings, along with Vasoactive Intestinal Polypeptide (VIP) staining, Girardet et al. showed that the number of synapses on VIP-containing SCN neurons is increased by 36% during the day (Zeitgeber Time 2 or ZT2), relative to night (ZT18). Both glutamatergic (from the retina) and non-glutamatergic innervation contributes to this change (Girardet et al., 2010), and the authors suggest that such changes may facilitate entrainment to light. There are also obvious daily changes in the glial coverage of VIP and AVP neuronal dendrites (Becquet et al., 2008). Glial coverage of soma and dendrites is higher at night than during the day for VIP neurons of the SCN “core” region which receives photic input from the retina. In contrast, AVP neurons in the SCN “shell” region have increased glial coverage during the day. Similar to the studies of Lavialle and Servière, Becquet et al. also report a robust diurnal rhythm in GFAP within the ventrolateral portion of the rat SCN—a region containing many VIP neurons—but this rhythm does not persist in DD, in contrast to findings in the hamster. Becquet et al. postulate that changes in astrogial coverage of VIP neurons may influence pacemaker resetting, but such a rhythm may also indicate the participation of astrocytes in neuronal energy metabolism. However, adrenalectomized rats—reported to have decreased corticosterone and altered photic entrainment (Sage et al., 2004)—exhibit a less robust diurnal GFAP rhythm (Becquet et al., 2008), and this has been interpreted as evidence for modulation of astrocytes and photic sensitivity by the steroid hormone. Consistent with such an interpretation, other studies indicate that neurons of the SCN core express androgen receptors and that androgen hormone modulates photic input to the pacemaker (Karatsoreos and Silver, 2007).
In addition to SCN astrocyte rhythms, it has been reported that astrocytes in other regions of the brain exhibit diurnal changes in morphology. For example, astrocytes surrounding Gonadotropin-Releasing Hormone (GnRH) neuronal cell bodies of the Organum Vasculosum of the Lamina Terminalis (OVLT) and the Medial Preoptic Nucleus (rMPN) have increased surface area during the early morning relative to other times of day (Gerhold and Wise, 2006). Changes in astrocyte surface area are postulated to contribute to reproductive behavior, with a reduction thought to stimulate neuronal inputs to the GnRH neurons to induce preovulatory synthesis and release of the neuropeptide. The diurnal change in surface area is apparently driven by VIP neuronal output from the pacemaker, because antisense oligos targeting the neuropeptide gene, when delivered to the SCN, eliminate rhythmicity. This may be a direct effect of VIP on glial cells, since astrocytes of the OVLT and rMPN regions express VPAC2, a receptor for the neuropeptide. Such a result suggests that VIP participates in neuron-to-glia signaling (see “Neuron-glia communication in the mammalian SCN” below).
CLOCK-CONTROLLED GLIAL MOLECULAR RHYTHMS
Ebony, a rhythmic Drosophila glial-specific protein
Null mutants of a Drosophila gene known as ebony exhibit arrhythmic patterns of locomotor activity (Newby and Jackson, 1991) because of elimination of a rhythmic glia-specific enzyme (Suh and Jackson, 2007). Ebony is localized to a subset of adult brain glial cells and it exhibits circadian rhythms in abundance in these cells (see Fig. 3). Genetic rescue analysis demonstrates that Ebony is required within glia for the manifestation of normal locomotor activity rhythms (Suh and Jackson, 2007). Interestingly, we and others have verified that the Ebony-containing glia are predominantly astrocyte-like cells of the fly adult brain (T. Awasaki, pers. comm.; F. Ng and F.R. Jackson, unpublished results) providing a further parallel between the glial modulation of rhythmicity in Drosophila and mammals. Similar to mammalian astrocytes, many Ebony-positive astrocytes contain PER-based molecular oscillators (Suh and Jackson, 2007). In addition, certain Ebony glial cells are close to projections of clock neurons including the PDF-containing neurons that are critical for behavioral rhythms.
Fig. 3.

Ebony-containing glial cells of the fly’s optic lobe (OL) and central brain. Ebony is predominantly cytoplasmic but is not well detected in most astrocytic processes. Thus, the typical fly astrocytic cell architecture cannot be seen in this image. All Ebony-containing cells of the fly brain are glia, as indicated by a glial marker protein(not shown in this image). Note that the optic lamina, which also contains Ebony-expressing glia, is not included in this particular specimen.
Mutants of the ebony gene were originally described by Bridges and Morgan (Bridges and Morgan, 1923), two of the founders of Drosophila genetics, on the basis of a dark body color phenotype (due to a role of the gene in cuticle pigmentation). Decades later, alleles were isolated in a screen for visual mutants by the lab of Seymour Benzer (Hotta and Benzer, 1969). Through the work of many investigators, it was discovered that Ebony protein has enzymatic activity (N-β-alanyl amine synthetase) that is responsible for conjugation of amines (histamine, dopamine, serotonin and others) to β-alanine (Hovemann et al., 1998; Richardt et al., 2003; Wright, 1987). Such an enzymatic function explains the cuticle and visual phenotypes of ebony mutants as dopamine is a substrate for melanin in the Drosophila cuticle pigmentation pathway (Wright, 1987) and histamine is the neurotransmitter of fly photoreceptor neurons (Burg et al., 1993)(the neurons that innervate the L1 and L2 monopolar cells described above).
A clue about which cell types require Ebony within the nervous system comes from the work of Hovemann, Meinertzhagen and colleagues. Immunostaining studies by those investigators have demonstrated that Ebony is localized to at least two types of optic lobe glia, including epithelial glia of the lamina that are adjacent to projections of the histaminergic photoreceptor neurons (Richardt et al., 2002). These and other studies (Suh and Jackson, 2007) indicate that Ebony has a completely glia-specific pattern of localization within the adult optic lobes and central nervous system of Drosophila. A role for Ebony in terminating the action of amine neurotransmitters, including the visual neurotransmitter histamine, is supported by its presence within laminar epithelial glia and other evidence demonstrating the importance of optic lobe glial cells for fly vision (reviewed in Edwards and Meinertzhagen, 2010). A role for glia in recycling histamine neurotransmitter (i.e., neurotransmitter retrieval)—by returning N-β-alanyl histamine to neurons—is also suggested by the localization of Tan within the photoreceptor neurons; Tan encodes a fungal-like acyltransferase that can cleave N-β-alanyl histamine, liberating histamine for neurotransmitter recycling. Not surprisingly, tan mutants, like ebony flies, have altered electroretinograms, indicative of abnormal vision (Hotta and Benzer, 1969; Pak et al., 1969). The roles of Ebony and Tan in the fly histamine recycling pathway are discussed in greater detail in recent reviews (Stuart et al., 2007; Edwards and Meinertzhagen, 2010). Recycling of histamine in the Drosophila optic lobe also involves the activity of a vesicular monoamine transporter (VMAT) variant that is exclusively expressed in fenestrated (non-Ebony) glia of the distal optic lamina, adjacent to photoreceptor neurons (Romero-Calderon et al., 2008). This raises the question: does glial VMAT or the neuronal isoform of the protein show circadian changes in abundance that might be important for the diurnal modulation of vision?
The proximity of Ebony-containing glial cells to aminergic neurons of several types (histamine, serotonin, dopamine) within the fly optic lobe and central nervous system suggests that the rhythm phenotype of mutants may be due to defective amine recycling. Consistent with a role for dopamine in the phenotype, the ebony1 mutation suppresses the hyperactivity associated with a Drosophila Dopamine Transporter (DAT) allele (Suh and Jackson, 2007). Thus, Ebony-containing glia may collaborate with dopaminergic neurons or other types of fly aminergic neurons known to regulate locomotor activity, sleep or arousal (Andretic et al., 2005; Crocker et al., 2010; Hardie et al., 2007;Kume et al., 2005; Lebestky et al., 2009).
Serotonin and dopamine have also both been implicated in circadian photoreception; neural dopamine is required for entrainment of activity rhythms to low ambient light (Hirsh et al., 2010) and serotonin receptor 1B is expressed in clock neurons and thought to contribute to photic sensitivity (Yuan et al., 2005). In addition, serotonergic and dopaminergic neuronal processes are in close proximity to dendrites or axonal projections of certain clock neurons in the larval and adult brains (Hamasaka and Nassel, 2006). Thus, glia may participate in modulating pacemaker light sensitivity by regulating amine recycling. Alternatively—or perhaps in addition—glial cells of the fly brain may release N-β-alanyl amine compounds that serve as gliotransmitters which regulate clock neurons. With regard to a role for glia in the modulation of circadian photic sensitivity, it is of interest that light stimulation which resets the mammalian pacemaker results in upregulation of the c-fos immediate early gene in both neurons and astrocytes of the SCN (Castel et al., 1997).
Rhythms in a fly Na+/K+-ATPase
Similar to vertebrates, a sodium pump (a Na+/K+-ATPase), composed of α catalytic and β subunits, is expressed in the nervous system and other tissues of Drosophila; it is sensitive to the sodium pump inhibitor ouabain and it regulates ionic homeostasis (Lebovitz et al., 1989; Sun and Salvaterra, 1995). Previous studies indicate that both α and β subunits have broad expression patterns in the adult fly brain, with localization to neurons and glial cells. Mutation of the α subunit leads to a sensitivity to mechanical stress (banging) and paralysis (Schubiger et al., 1994; Sun et al., 2001).
Interestingly, the sodium pump α subunit protein shows a circadian rhythm in abundance within epithelial glia of the optic lamina and in glia of the optic medulla (Gorska-Andrzejak et al., 2009), with a peak near the beginning of subjective night (in DD). The RNA encoding the ATPase also exhibits corresponding changes in abundance, suggesting a transcriptional regulation of the ATPase rhythm. Similarly, GFP expressed from a UAS transgene driven by Nrv2-Gal4 (reflecting Na+/K+-ATPase β subunit expression) shows day/night differences in fluorescence intensity, although rhythmicity is not incredibly robust, perhaps because both GAL4 and GFP are long lived and thus contribute to a high baseline for the signal.
Whereas the α subunit is localized to most glial cells of the optic lobe (with highest signal in the lamina), β subunit expression, based on Nrv2-Gal4-driven GFP fluorescence, is primarily in medulla neuropil glia, some of which also contain Ebony protein (Gorska-Andrzejak et al., 2009). There is no expression of the Nrv2-Gal4 transgene in the lamina epithelial glia cells, most of which express Ebony. The expression patterns of the fly ATPase subunits suggest that the ATPase oscillates in abundance within Ebony-positive and Ebony-negative glial cells of the optic lobe. Of note, there is co-localization of Nrv2-Gal4 and PER in certain glia of the medulla neuropil. An obvious question is: do glial oscillators contribute to the ATPase rhythm in those cells?
Regulation of astrocyte physiology by clock genes
Recent studies have explored the role of the mammalian PER-based oscillator in regulating glial physiology. It has been reported that there is a diurnal rhythm in Glast (EAAT1) glutamate transporter gene expression and protein amount within the SCN with peak protein occurring at the beginning of the photoperiod in an LD 12:12 light:dark cycle, although it was not determined whether this rhythm persists in constant conditions (Spanagel et al., 2005). However, the observation that EAAT1 protein levels do not show obvious rhythmicity in the Per2 mutant (Spanagel et al., 2005) suggests circadian control. The same study showed that Per2 mutant mice have reduced EAAT1 protein at most times of day and that extracellular glutamate levels are elevated in vivo. Spanagel et al. demonstrated that cultured astrocytes from the mutant show reduced glutamate uptake, consistent with the in vivo phenotype. The study also documented changes in alcohol consumption for mutants, implicating glutamatergic function in alcoholism.
A more recent study used cultured cortical astrocytes from clock mutant animals to replicate the effects of the Per2 mutation on EAAT1; it also showed that EAAT1 mRNA and protein levels are reduced in astrocytes obtained from Clock and NPAS2 mutant mice (Beaule et al., 2009). Although glutamate uptake is reduced in mutant animals, surprisingly, neither EAAT1 mRNA expression nor glutamate uptake, as assayed in cultured astrocytes and tissue slices of somatosensory cortex, were found to be circadian in nature (Beaule et al., 2009). Thus, the authors concluded that a PER-based oscillator does not control EAAT1 expression even though earlier studies (Spanagel et al., 2005) demonstrated a rhythm in EAAT1 mRNA abundance within the SCN, in vivo, that was eliminated in the Per2 mutant. However, I note that it might be the case that EAAT1 only oscillates in the SCN and/or that neuronal communication with astrocytes is required for EAAT1 rhythmicity (the latter is lacking for cultured astrocytes; see discussion of Beaule et al. in “Neuron-glia communication and circadian behavior”). If EAAT1 is actually not under circadian control, then the results of Beaule et al. suggest that clock proteins have non-circadian roles in the regulation of EAAT1 gene expression. For example, mouse Clock (McClung et al., 2005) and the fly per, clock, and cycle genes (Andretic et al., 1999) are apparently required, in a noncircadian manner, for normal cocaine sensitivity and/or sensitization.
Astrocyte ATP rhythms
It has been reported that there are diurnal rhythms of ATP content in certain brain regions of the adult rat (Dworak et al., 2010), but the changes seem to be driven in most regions by sleep rather than the circadian clock; i.e. levels are high during the sleep period and increases are prevented by sleep deprivation. In contrast, there are robust rhythms of ATP release from explants of the rat SCN and in cultured astrocytes (Womac et al., 2009), with a peak during the dark period of a light/dark cycle (the active period). Importantly, rhythmicity persists in constant darkness, indicative of circadian control and suggesting that a timed release of ATP from astrocytes might be important for cell-cell communication in the mammalian pacemaker.
Upon release from neurons or glia, ATP can be converted to ADP, AMP and adenosine through the action of extracellular ectonucleotidases, and, with the exception of AMP, all bind to A1 or A2 purinergic receptors on neurons, glia and muscles to regulate Ca2+, cAMP, IP3 and other second messenger pathways (Fields and Burnstock, 2006). Purinergic and other forms of intercellular signaling mechanisms permit astrocytes to communicate with neurons or other glial cells, and glia-to-neuron signaling is known to be critical for regulation of neuronal water/ion homeostasis (Simard and Nedergaard, 2004), neurovascular coupling (Gordon et al., 2007), neuronal energy metabolism (Brown and Ransom, 2007), synaptic transmission (Halassa and Haydon, 2010), and other biological processes (Fields and Burnstock, 2006). Similarly, is circadian release of ATP from glial cells critical for temporally synchronizing glial cell populations or modulating the activities of clock neurons? Interestingly, there are Drosophila genes encoding enzymes homologous to the extracellular ectonucleotidases; it will thus be possible to evaluate the importance of ATP-mediated gliotransmission in circadian and other forms of behavior in this genetic model.
NEURON-GLIA COMMUNICATION AND CIRCADIAN BEHAVIOR
Fly glial cells can modulate synaptic transmission
Results from the analysis of Drosophila Ebony suggest that glial cells are important for Drosophila circadian behavior (Suh and Jackson, 2007), but they do not indicate whether glia actively modulate pacemaker neurons in the adult brain. While studies are underway in my lab to evaluate the importance of glia-to-neuron communication, in vivo, for the modulation of the Drosophila circadian circuitry and behavior, it is premature to discuss results in this review. Nor are there in vivo studies in mammalian models that demonstrate a physiological regulation of circadian pacemaker neurons by astrocytes, although it has been shown that astrocytes communicate with neurons, via adenosine signaling, to modulate sleep homeostasis and cognitive processes affected by sleep deprivation (Halassa et al., 2009). A similar mechanism may contribute to the glial regulation of circadian behavior in mammals and Drosophila.
Regarding the capacity for regulation of neurotransmission by fly glia, previous studies and reviews highlight evidence for the presence of aminergic and glutamatergic tripartite synapses in the Drosophila nervous system (Edwards and Meinertzhagen, 2010; Jackson and Haydon, 2008), similar to the glutamatergic tripartite synapse of mammals. The work on Ebony, discussed in an earlier section, strongly suggests that glial cells modulate aminergic neuronal function. In addition, the review of D. Featherstone in this issue of Glia describes the glial regulation of Drosophila glutamatergic synapses by the EAAT1 transporter and an SLC7-like cystine/glutamate transporter called Genderblind (also, see comments about dEAAT1 in the next section).
Is neuron-glia communication needed for clock gene regulation of astrocyte eaat1 activity?
The previously mentioned work of Beaule et al. (2009) was interpreted as indicating a role for astrocyte clock genes in the regulation of EAAT1 activity. However, the study does not definitively indicate which cell types require clock gene function for normal EAAT1 activity. The mutant animals employed in the study have altered clock protein function in both neurons and glia, so it is possible that neuronal rather than autonomous glial factors regulate EAAT1 activity. Indeed, Yang et al. have shown that presynaptic neurons regulate the astroglial GLT-1 transporter (Yang et al., 2009). Similarly, both GLT1 and EAAT1 are upregulated when astrocytes are treated with dibutyryl-cAMP or co-cultured with neurons (Swanson et al., 1997). Thus, both transporters can be regulated by neuronal factors, and this may occur in the SCN or other brain regions.
Although the expression pattern and function of the Drosophila dEAAT1 homolog has been well characterized by the elegant studies of Serge Birman and colleagues (Rival et al., 2004, 2006), it is unknown whether it is regulated in a clock gene-dependent manner, similar to mammalian EAAT1. However, those studies demonstrate a broad expression pattern for dEAAT1 throughout the neuropil (synaptic regions) of the adult brain (Rival et al., 2004) in glial cell projections now known to be astrogial in nature (Doherty et al., 2009)(also see Fig. 4). Similar to mammalian EAAT1, the fly homolog is essential for viability and a lack of it results in neuropil degeneration (Rival et al., 2004). Interestingly, dEAAT1 is present in glial extensions adjacent to motor axons of the neuromuscular junction (NMJ), but not until adulthood, suggesting a physiological role for the glial transporter. Consistent with that idea, electrophysiological recordings from adult NMJs deficient for glial dEAAT1 demonstrate enhanced responses of muscles to motor nerve stimulation (Rival et al., 2006).
Fig. 4.

Cellular localization dEAAT1 in the fly brain as reflected by a dEAAT1-Gal4 driver regulating expression of a UAS-CD8-GFP transgene. The CD8 moiety directs GFP to membranes of glial cell bodies and processes in the synaptic neuropil.
As insect motor neurons utilize glutamate as a transmitter, the studies of Rival et al. (2006) provide strong evidence that Drosophila glial cells regulate glutamatergic neurotransmission. Thus, it is likely that dEAAT1 functions in glutamate uptake and as part of a transmitter recycling mechanism involving conversion of glial glutamate to glutamine—via glutamine synthetase (GS) —and transport of glutamine to presynaptic nerve endings for synthesis of glutamate transmitter. Indeed, it is known that GS and dEAAT1 are expressed in astrocytes of the fly central nervous system (Doherty et al., 2009; Freeman et al., 2003; Sinakevitch et al., 2010) although, to my knowledge, the transporters for glutamine retrieval by neurons have not been identified in Drosophila. Nonetheless, it is likely that the basic mechanism of glutamate retrieval is conserved between flies and mammals. Similar to the Ebony story and amine neurotransmitter retrieval, it may be that glutamate retrieval by glia is important for circadian behavior. Evidence suggests, for example, that glutamatergic neurotransmission from dorsal clock neurons (DN) to small ventral lateral clock neurons (s-LNv) is important for the function of the fly pacemaker circuitry (Hamasaka et al., 2007).
VIP regulates astrocyte molecular clocks
Although studies of glia-to-neuron communication have not been reported for the mammalian SCN, there is evidence from an analysis of cultured astrocytes that VIP—known to be present in the SCN and essential for neuronal synchrony (Aton et al., 2005)—might be important for the opposite: neuron-to-glia communication. Those studies utilized cortical astrocytes obtained from a mouse per2-luc strain to show that VIP can shift the phase of the glial PER-based oscillator in a dose- and time-of-day-dependent manner (Marpegan et al., 2009). The same studies indicated that daily administration of VIP can entrain the astrogial clock such that peak clock gene (Per) expression occurs at a time of day that is predicted by a phase-response curve representing the response of the clock to the peptide. As VIP is localized and released from neurons within the core of the SCN (the mammalian clock), the work suggests that neuronal release of VIP, in vivo, may function to synchronize oscillators present in adjacent astrocytes. Of note, it has also been shown that cultured chick astrocytes possess high-affinity melatonin receptors and that rhythmic melatonin administration, at physiological levels, leads to diurnal glucose uptake rhythms in such cultures (Adachi et al., 2002). Thus, multiple factors may synchronize astrocyte rhythms in vivo.
Regarding neuropeptide control of glial rhythmicity, it is of interest that certain Ebony-containing glia are close to PDF neuronal projections within the CNS, suggesting regulation by the neuropeptide (Suh and Jackson, 2007). However, Suh and Jackson showed that Ebony transcriptional cycling was normal in a fly pdf null mutant lacking the neuropeptide. Interestingly, the receptor for PDF is broadly expressed in the clock circuitry and may be present in certain glial cells (Im and Taghert, 2010), but these glia do not correspond to the Ebony cells; rather, they appear to be the fenestrated glia of the optic lamina which express the aforementioned VMAT glial variant. Thus, it will be of interest to determine whether VMAT within those laminar glial cells is regulated in a PDF-dependent manner.
CONCLUSION AND FUTURE DIRECTIONS
Studies have demonstrated that Drosophila glial cells and mammalian astrocytes contain PER-based circadian oscillators. These findings and additional observations summarized in this review suggest that glial cells may have similar functions in the circadian systems of Drosophila and mammals. I suggest that answering the several questions posed below may provide additional advances in our understanding of the glial regulation of behavior.
Are glial cell circadian oscillators required for normal behavioral rhythmicity, or do they regulate other physiological events?
Furthermore, are reciprocal interactions between glial and neuronal cells important for circadian behavior in diverse species including mammals and insects?
And if neuron-glia interactions are important, what are the neuronal cell types modulated by glia: pacemaker neurons or other neurons of the circadian circuitry?
Finally, does the glial release of transmitters or other signaling molecules contribute to dynamic glia-to-neuron interactions that are necessary for normal behavioral rhythmicity?
Much is known about how mammalian astrocytes of the adult brain support and regulate neurons, and this foundation of knowledge has facilitated studies of neuron-glia communication in the mammalian clock system. In contrast, there is more limited information about neuron-glia communication (gliotransmission) in the adult Drosophila brain. However, the recent identification of astrocyte glia and other glial cell classes in the adult fly brain (Awasaki et al., 2008; Doherty et al., 2009) will facilitate genetic approaches in this model to flesh out the picture of insect gliotransmission and determine its relevance for circadian behavior. The major strength of the Drosophila model is the ease with which conditional genetic manipulations can be performed in live, behaving animals, and this advantage will surely lead to the identification of molecules and pathways relevant for the glial regulation of behavior. Such findings will likely translate into novel insights about astrocyte-neuron interactions in mammals.
Whereas knowledge about gliotransmission is currently limited in Drosophila, genetic approaches in this model have begun to identify glial-specific factors such as Ebony which participate, in vivo, in the regulation of circadian behavior. Similar findings have not yet been reported for the mammalian circadian system. In this regard, one wonders whether cycling Ebony-like factors and glial-based monoamine neurotransmitter retrieval mechanisms might be important for mammalian pacemaker function. Although there is not an obvious ebony ortholog in the vertebrate genome, there is a mouse non-ribosomal peptide synthetase with similarity to Ebony, as note previously (Jackson and Haydon, 2008). Furthermore, there is a vertebrate carnosine synthetase (synthetic enzyme for β-alanyl-L-histidine or β-alanyl-N-methyl-L-histidine dipeptides) that may be expressed in glia (Jackson and Haydon, 2008). Thus, it is possible that such enzymes participate in amine recycling or produce β-alanyl-containing gliotransmitters that are relevant for circadian behavior.
In ending, I wish to remind the reader of the seminal neurogenetic approaches of Seymour Benzer, Ron Konopka (Konopka and Benzer, 1971) and other investigators following their lead that resulted in the description of the first circadian mutants in any organism (Drosophila melanogaster). That work opened up the field of circadian molecular biology and led to explicit biochemical models for animal clocks in species ranging from flies to humans. A similar approach will undoubtedly have significant impact for an understanding of glia-neuron interactions in the circadian systems of insects and mammals.
Acknowledgments
Grant sponsor: NIH; Grant number: R01 HL59873; Grant sponsor: Tufts Center for Neuroscience Research (NIH); Grant number: P30 NS047243.
The authors thanks Dr. Fanny Ng (Tufts University School of Medicine) for her help with preparation of this review and Drs. Ng and Laurie Newby for reading the manuscript.
References
- Adachi A, Natesan AK, Whitfield-Rucker MG, Weigum SE, Cassone VM. Functional melatonin receptors and metabolic coupling in cultured chick astrocytes. Glia. 2002;39:268–278. doi: 10.1002/glia.10109. [DOI] [PubMed] [Google Scholar]
- Andretic R, Chaney S, Hirsh J. Requirement of circadian genes for cocaine sensitization in Drosophila. Science. 1999;285:1066–1068. doi: 10.1126/science.285.5430.1066. [DOI] [PubMed] [Google Scholar]
- Andretic R, van SB, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15:1165–1175. doi: 10.1016/j.cub.2005.05.025. [DOI] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasaki T, Lai SL, Ito K, Lee T. Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J Neurosci. 2008;28:13742–13753. doi: 10.1523/JNEUROSCI.4844-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainton RJ, Tsai LT, Schwabe T, DeSalvo M, Gaul U, Heberlein U. moody encodes two GPCRs that regulate cocaine behaviors and blood-brain barrier permeability in Drosophila. Cell. 2005;123:145–156. doi: 10.1016/j.cell.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Beaule C, Swanstrom A, Leone MJ, Herzog ED. Circadian modulation of gene expression, but not glutamate uptake, in mouse and rat cortical astrocytes. PLoS One. 2009;4:e7476. doi: 10.1371/journal.pone.0007476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becquet D, Girardet C, Guillaumond F, Francois-Bellan AM, Bosler O. Ultrastructural plasticity in the rat suprachiasmatic nucleus. Possible involvement in clock entrainment. Glia. 2008;56:294–305. doi: 10.1002/glia.20613. [DOI] [PubMed] [Google Scholar]
- Bridges CB, Morgan TH. The third-chromosome group of mutant characters of Drosophila melanogaster. Publs Carnegie Instn. 1923;327:1–251. [Google Scholar]
- Bringmann A, Pannicke T, Grosche J, Francke M, Wiedemann P, Skatchkov SN, Osborne NN, Reichenbach A. Muller cells in the healthy and diseased retina. Prog Retin Eye Res. 2006;25:397–424. doi: 10.1016/j.preteyeres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Brown AM, Ransom BR. Astrocyte glycogen and brain energy metabolism. Glia. 2007;55:1263–1271. doi: 10.1002/glia.20557. [DOI] [PubMed] [Google Scholar]
- Burg MG, Sarthy PV, Koliantz G, Pak WL. Genetic and molecular identification of a Drosophila histidine decarboxylase gene required in photoreceptor transmitter synthesis. EMBO J. 1993;12:911–919. doi: 10.1002/j.1460-2075.1993.tb05732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafferty P, Auld VJ. No pun intended: Future directions in invertebrate glial cell migration studies. Neuron Glia Biol. 2007;3:45–54. doi: 10.1017/S1740925X07000634. [DOI] [PubMed] [Google Scholar]
- Castel M, Belenky M, Cohen S, Wagner S, Schwartz WJ. Light-induced c-Fos expression in the mouse suprachiasmatic nucleus: Immunoelectron microscopy reveals co-localization in multiple cell types. Eur J Neurosci. 1997;9:1950–1960. doi: 10.1111/j.1460-9568.1997.tb00762.x. [DOI] [PubMed] [Google Scholar]
- Crews ST. Axon-glial interactions at the Drosophila CNS midline. Cell Adh Migr. 2010;4:67–71. doi: 10.4161/cam.4.1.10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker A, Shahidullah M, Levitan IB, Sehgal A. Identification of a neural circuit that underlies the effects of octopamine on sleep: wake behavior. Neuron. 2010;65:670–681. doi: 10.1016/j.neuron.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty J, Logan MA, Tasdemir OE, Freeman MR. Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci. 2009;29:4768–4781. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworak M, McCarley RW, Kim T, Kalinchuk AV, Basheer R. Sleep and brain energy levels: ATP changes during sleep. J Neurosci. 2010;30:9007–9016. doi: 10.1523/JNEUROSCI.1423-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TN, Meinertzhagen IA. The functional organisation of glia in the adult brain of Drosophila and other insects. Prog Neurobiol. 2010;90:471–497. doi: 10.1016/j.pneurobio.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells influence on circadian behavioral rhythms. J Neurosci. 1992;12:3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields RD, Burnstock G. Purinergic signalling in neuron-glia interactions. Nat Rev Neurosci. 2006;7:423–436. doi: 10.1038/nrn1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38:567–580. doi: 10.1016/s0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- Freeman MR, Doherty J. Glial cell biology in Drosophila and vertebrates. TINS. 2006;29:82–90. doi: 10.1016/j.tins.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Gerhold LM, Wise PM. Vasoactive intestinal polypeptide regulates dynamic changes in astrocyte morphometry: Impact on gonadotropin-releasing hormone neurons. Endocrinology. 2006;147:2197–2202. doi: 10.1210/en.2005-1262. [DOI] [PubMed] [Google Scholar]
- Girardet C, Blanchard MP, Ferracci G, Leveque C, Moreno M, Francois-Bellan AM, Becquet D, Bosler O. Daily changes in synaptic innervation of VIP neurons in the rat suprachiasmatic nucleus: Contribution of glutamatergic afferents. Eur J Neurosci. 2010;31:359–370. doi: 10.1111/j.1460-9568.2009.07071.x. [DOI] [PubMed] [Google Scholar]
- Gordon GR, Mulligan SJ, MacVicar BA. Astrocyte control of the cerebrovasculature. Glia. 2007;55:1214–1221. doi: 10.1002/glia.20543. [DOI] [PubMed] [Google Scholar]
- Gorska-Andrzejak J, Keller A, Raabe T, Kilianek L, Pyza E. Structural daily rhythms in GFP-labelled neurons in the visual system of Drosophila melanogaster. Photochem Photobiol Sci. 2005;4:721–726. doi: 10.1039/b417023g. [DOI] [PubMed] [Google Scholar]
- Gorska-Andrzejak J, Salvaterra PM, Meinertzhagen IA, Krzeptowski W, Gorlich A, Pyza E. Cyclical expression of Na+/K+-ATPase in the visual system of Drosophila melanogaster. J Insect Physiol. 2009;55:459–468. doi: 10.1016/j.jinsphys.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa MM, Florian C, Fellin T, Munoz JR, Lee SY, Abel T, Haydon PG, Frank MG. Astrocytic modulation of sleep homeostasis and cognitive consequences of sleep loss. Neuron. 2009;61:213–219. doi: 10.1016/j.neuron.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa MM, Haydon PG. Integrated brain circuits: Astrocytic networks modulate neuronal activity and behavior. Annu Rev Physiol. 2010;72:335–355. doi: 10.1146/annurev-physiol-021909-135843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaka Y, Nassel DR. Mapping of serotonin, dopamine, and histamine in relation to different clock neurons in the brain of Drosophila. J Comp Neurol. 2006;494:314–330. doi: 10.1002/cne.20807. [DOI] [PubMed] [Google Scholar]
- Hamasaka Y, Rieger D, Parmentier ML, Grau Y, Helfrich-Forster C, Nassel DR. Glutamate and its metabotropic receptor in Drosophila clock neuron circuits. J Comp Neurol. 2007;505:32–45. doi: 10.1002/cne.21471. [DOI] [PubMed] [Google Scholar]
- Hardie SL, Zhang JX, Hirsh J. Trace amines differentially regulate adult locomotor activity, cocaine sensitivity, and female fertility in Drosophila melanogaster. Dev Neurobiol. 2007;67:1396–1405. doi: 10.1002/dneu.20459. [DOI] [PubMed] [Google Scholar]
- Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Haydon PG, Blendy J, Moss SJ, Jackson FR. Astrocytic control of synaptic transmission and plasticity: A target for drugs of abuse? Neuropharmacology. 2009;56:83–90. doi: 10.1016/j.neuropharm.2008.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C. The circadian clock in the brain: A structural and functional comparison between mammals and insects. J Compar Physiol A-Neuroethol Sensory Neural Behav Physiol. 2004;190:601–613. doi: 10.1007/s00359-004-0527-2. [DOI] [PubMed] [Google Scholar]
- Hirsh J, Riemensperger T, Coulom H, Iche M, Coupar J, Birman S. Roles of dopamine in circadian rhythmicity and extreme light sensitivity of circadian entrainment. Curr Biol. 2010;20:209–214. doi: 10.1016/j.cub.2009.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta Y, Benzer S. Abnormal electroretinograms in visual mutants of Drosophila. Nature. 1969;222:354–356. doi: 10.1038/222354a0. [DOI] [PubMed] [Google Scholar]
- Hovemann BT, Ryseck RP, Walldorf U, Stortkuhl KF, Dietzel ID, Dessen E. The Drosophila ebony gene is closely related to microbial peptide synthetases and shows specific cuticle and nervous system expression. Gene. 1998;221:1–9. doi: 10.1016/s0378-1119(98)00440-5. [DOI] [PubMed] [Google Scholar]
- Im SH, Taghert PH. PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J Comp Neurol. 2010;518:1925–1945. doi: 10.1002/cne.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson FR, Haydon PG. Glial cell regulation of neurotransmission and behavior in drosophila. Neuron Glia Biol. 2008;4:11–17. doi: 10.1017/S1740925X09000027. [DOI] [PubMed] [Google Scholar]
- Karatsoreos IN, Silver R. Minireview: The neuroendocrinology of the suprachiasmatic nucleus as a conductor of body time in mammals. Endocrinology. 2007;148:5640–5647. doi: 10.1210/en.2007-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavialle M, Servière J. Circadian fluctuations in GFAP distribution in the Syrian hamster suprachiasmatic nucleus. Neuroreport. 1993;4:1243–1246. doi: 10.1097/00001756-199309000-00008. [DOI] [PubMed] [Google Scholar]
- Lebestky T, Chang JS, Dankert H, Zelnik L, Kim YC, Han KA, Wolf FW, Perona P, Anderson DJ. Two different forms of arousal in Drosophila are oppositely regulated by the dopamine D1 receptor ortholog DopR via distinct neural circuits. Neuron. 2009;64:522–536. doi: 10.1016/j.neuron.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebovitz RM, Takeyasu K, Fambrough DM. Molecular characterization and expression of the (Na+ + K+)-ATPase alpha-subunit in Drosophila melanogaster. EMBO J. 1989;8:193–202. doi: 10.1002/j.1460-2075.1989.tb03364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marpegan L, Krall TJ, Herzog ED. Vasoactive intestinal polypeptide entrains circadian rhythms in astrocytes. J Biol Rhythms. 2009;24:135–143. doi: 10.1177/0748730409332042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA, Sidiropoulou K, Vitaterna M, Takahashi JS, White FJ, Cooper DC, Nestler EJ. Regulation of dopaminergic transmission and cocaine reward by the Clock gene. Proc Natl Acad Sci USA. 2005;102:9377–9381. doi: 10.1073/pnas.0503584102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RY, Eichler VB. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972;42:201–206. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- Newby LM, Jackson FR. Drosophila ebony mutants have altered circadian activity rhythms but normal eclosion rhythms. J Neurogenet. 1991;7:85–101. doi: 10.3109/01677069109066213. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–R93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- Pak WL, Grossfield J, White NV. Nonphototactic mutants in a study of vision of Drosophila. Nature. 1969;222:351–354. doi: 10.1038/222351a0. [DOI] [PubMed] [Google Scholar]
- Parker RJ, Auld VJ. Roles of glia in the Drosophila nervous system. Semin Cell Dev Biol. 2006;17:66–77. doi: 10.1016/j.semcdb.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Shy D, Hartenstein V. Morphogenesis and proliferation of the larval brain glia in Drosophila. Dev Biol. 2005;283:191–203. doi: 10.1016/j.ydbio.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Prolo LM, Takahashi JS, Herzog ED. Circadian rhythm generation and entrainment in astrocytes. J Neurosci. 2005;25:404–408. doi: 10.1523/JNEUROSCI.4133-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser RA, Edgar DM, Heller HC, Miller JD. A possible glial role in the mammalian circadian clock. Brain Res. 1994;643:296–301. doi: 10.1016/0006-8993(94)90036-1. [DOI] [PubMed] [Google Scholar]
- Pyza E, Cymborowski B. Circadian rhythms in behaviour and in the visual system of the blow fly, Calliphora vicina. J Insect Physiol. 2001;47:897–904. [Google Scholar]
- Pyza E, Gorska-Andrzejak J. Involvement of glial cells in rhythmic size changes in neurons of the housefly’s visual system. J Neurobiol. 2004;59:205–215. doi: 10.1002/neu.10307. [DOI] [PubMed] [Google Scholar]
- Pyza E, Meinertzhagen IA. Monopolar cell axons in the first optic neuropil of the housefly, Musca domestica L, undergo daily fluctuations in diameter that have a circadian basis. J Neurosci. 1995;15:407–418. doi: 10.1523/JNEUROSCI.15-01-00407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Comparing clockworks: Mouse versus fly. J Biol Rhythms. 2000;15:357–364. doi: 10.1177/074873000129001459. [DOI] [PubMed] [Google Scholar]
- Richardt A, Kemme T, Wagner S, Schwarzer D, Marahiel MA, Hovemann BT. Ebony, a novel nonribosomal peptide synthetase for beta-alanine conjugation with biogenic amines in Drosophila. J Biol Chem. 2003;278:41160–41166. doi: 10.1074/jbc.M304303200. [DOI] [PubMed] [Google Scholar]
- Richardt A, Rybak A, Strortkuhl KF, Meinertzhagen LA, Hovemann B. Ebony protein in the Drosophila nervous system: Optic neuropile expression in glial cells. J Comp Neurol. 2002;452:93–102. doi: 10.1002/cne.10360. [DOI] [PubMed] [Google Scholar]
- Rival T, Soustelle L, Cattaert D, Strambi C, Iche M, Birman S. Physiological requirement for the glutamate transporter dEAAT1 at the adult Drosophila neuromuscular junction. J Neurobiol. 2006;66:1061–1074. doi: 10.1002/neu.20270. [DOI] [PubMed] [Google Scholar]
- Rival T, Soustelle L, Strambi C, Besson MT, Iche M, Birman S. Decreasing glutamate buffering capacity triggers oxidative stress and neuropil degeneration in the Drosophila brain. Curr Biol. 2004;14:599–605. doi: 10.1016/j.cub.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Romero-Calderon R, Uhlenbrock G, Borycz J, Simon AF, Grygoruk A, Yee SK, Shyer A, Ackerson LC, Maidment NT, Meinertzhagen IA, Hovemann BT, Krantz DE. A glial variant of the vesicular monoamine transporter is required to store histamine in the Drosophila visual system. PLoS Genet. 2008;4:e1000245. doi: 10.1371/journal.pgen.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage D, Ganem J, Guillaumond F, Laforge-Anglade G, Francois-Bellan AM, Bosler O, Becquet D. Influence of the corticosterone rhythm on photic entrainment of locomotor activity in rats. J Biol Rhythms. 2004;19:144–156. doi: 10.1177/0748730403261894. [DOI] [PubMed] [Google Scholar]
- Schubiger M, Feng Y, Fambrough DM, Palka J. A mutation of the Drosophila sodium pump α subunit gene results in bang-sensitive paralysis. Neuron. 1994;12:373–381. doi: 10.1016/0896-6273(94)90278-x. [DOI] [PubMed] [Google Scholar]
- Schwartz WJ, Gainer H. Suprachiasmatic nucleus: Use of 14C-labeled deoxyglucose uptake as a functional marker. Science. 1977;197:1089–1091. doi: 10.1126/science.887940. [DOI] [PubMed] [Google Scholar]
- Shinohara K, Honma S, Katsuno Y, Honma K. Circadian release of excitatory amino acids in the suprachiasmatic nucleus culture is Ca2+-independent. Neurosci Res. 2000;36:245–250. doi: 10.1016/s0168-0102(99)00131-5. [DOI] [PubMed] [Google Scholar]
- Silies M, Edenfeld G, Engelen D, Stork T, Klambt C. Development of the peripheral glial cells in Drosophila. Neuron Glia Biol. 2007;3:35–43. doi: 10.1017/S1740925X07000622. [DOI] [PubMed] [Google Scholar]
- Silver R, Schwartz WJ. The suprachiasmatic nucleus is a functionally heterogeneous timekeeping organ. Methods Enzymol. 2005;393:451–465. doi: 10.1016/S0076-6879(05)93022-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard M, Nedergaard M. The neurobiology of glia in the context of water and ion homeostasis. Neuroscience. 2004;129:877–896. doi: 10.1016/j.neuroscience.2004.09.053. [DOI] [PubMed] [Google Scholar]
- Sinakevitch I, Grau Y, Strausfeld NJ, Birman S. Dynamics of glutamatergic signaling in the mushroom body of young adult Drosophila. Neural Dev. 2010;5:10. doi: 10.1186/1749-8104-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanagel R, Pendyala G, Abarca C, Zghoul T, Sanchis-Segura C, Magnone MC, Lascorz J, Depner M, Holzberg D, Soyka M, Schreiber S, Matsuda F, Lathrop M, Schumann G, Albrecht U. The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nature Med. 2005;11:35–42. doi: 10.1038/nm1163. [DOI] [PubMed] [Google Scholar]
- Spindler SR, Ortiz I, Fung S, Takashima S, Hartenstein V. Drosophila cortex and neuropile glia influence secondary axon tract growth, pathfinding, and fasciculation in the developing larval brain. Dev Biol. 2009;334:355–368. doi: 10.1016/j.ydbio.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci USA. 1972;69:1583–1586. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart AE, Borycz J, Meinertzhagen IA. The dynamics of signaling at the histaminergic photoreceptor synapse of arthropods. Prog Neurobiol. 2007;82:202–227. doi: 10.1016/j.pneurobio.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Suh J, Jackson FR. Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron. 2007;55:435–447. doi: 10.1016/j.neuron.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Salvaterra PM. Two Drosophila nervous system antigens, Nervana 1 and 2, are homologous to the β subunit of Na+,K+-ATPase. Proc Natl Acad Sci USA. 1995;92:5396–5400. doi: 10.1073/pnas.92.12.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Xu P, Wang W, Salvaterra PM. In vivo modification of Na(+),K(+)-ATPase activity in Drosophila. Comp Biochem Physiol B Biochem Mol Biol. 2001;130:521–536. doi: 10.1016/s1096-4959(01)00470-5. [DOI] [PubMed] [Google Scholar]
- Swanson RA, Liu J, Miller JW, Rothstein JD, Farrell K, Stein BA, Longuemare MC. Neuronal regulation of glutamate transporter subtype expression in astrocytes. J Neurosci. 1997;17:932–940. doi: 10.1523/JNEUROSCI.17-03-00932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodosis DT, Poulain DA, Oliet SH. Activity-dependent structural and functional plasticity of astrocyte-neuron interactions. Physiol Rev. 2008;88:983–1008. doi: 10.1152/physrev.00036.2007. [DOI] [PubMed] [Google Scholar]
- Van den Pol AN, Finkbeiner SM, Cornell-Bell AH. Calcium excitability and oscillations in suprachiasmatic nucleus neurons and glia in vitro. J Neurosci. 1992;12:2648–2664. doi: 10.1523/JNEUROSCI.12-07-02648.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P, Kula-Eversole E, Pyza E. Circadian control of dendrite morphology in the visual system of Drosophila melanogaster. PLoS One. 2009;4:e4290. doi: 10.1371/journal.pone.0004290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womac AD, Burkeen JF, Neuendorff N, Earnest DJ, Zoran MJ. Circadian rhythms of extracellular ATP accumulation in suprachiasmatic nucleus cells and cultured astrocytes. Eur J Neurosci. 2009;30:869–876. doi: 10.1111/j.1460-9568.2009.06874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright TRF. The genetics of biogenic amine metabolism, sclerotization, and melanization in Drosophila melanogaster. Adv Gene. 1987;24:127–223. [PubMed] [Google Scholar]
- Wulff K, Gatti S, Wettstein JG, Foster RG. Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat Rev Neurosci. 2010;11:589–599. doi: 10.1038/nrn2868. [DOI] [PubMed] [Google Scholar]
- Yagita K, Yamanaka I, Emoto N, Kawakami K, Shimada S. Real-time monitoring of circadian clock oscillations in primary cultures of mammalian cells using Tol2 transposon-mediated gene transfer strategy. BMC Biotechnol. 2010;10:3. doi: 10.1186/1472-6750-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gozen O, Watkins A, Lorenzini I, Lepore A, Gao Y, Vidensky S, Brennan J, Poulsen D, Won PJ, Li JN, Robinson MB, Rothstein JD. Presynaptic regulation of astroglial excitatory neurotransmitter transporter GLT1. Neuron. 2009;61:880–894. doi: 10.1016/j.neuron.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MW, Kay SA. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Lin F, Zheng X, Sehgal A. Serotonin modulates circadian entrainment in Drosophila. Neuron. 2005;47:115–127. doi: 10.1016/j.neuron.2005.05.027. [DOI] [PubMed] [Google Scholar]
- Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS, the visual system of Drosophila. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178:1147–1155. doi: 10.1534/genetics.107.088658. [DOI] [PMC free article] [PubMed] [Google Scholar]