Abstract
This perspective gives an overview of recent developments in surface-enhanced Raman scattering (SERS) for biosensing. We focus this review on SERS papers published in the last 10 years and to specific applications of detecting biological analytes. Both intrinsic and extrinsic SERS biosensing schemes have been employed to detect and identify small molecules, nucleic acids, lipids, peptides, and proteins, as well as for in vivo and cellular sensing. Current SERS substrate technologies along with a series of advancements in surface chemistry, sample preparation, intrinsic/extrinsic signal transduction schemes, and tip-enhanced Raman spectroscopy are discussed. The progress covered herein shows great promise for widespread adoption of SERS biosensing.
I. Introduction
Rapid and reliable detection of a diverse set of biomolecules, such as metabolites, pharmaceuticals, nucleic acids, amino acids, proteins and peptides require analytical techniques capable of label-free chemical identification. Though Raman scattering seems an unlikely signal transduction mechanism for this task based on its inherently small scattering cross-section, a series of fundamental and technological advancements in the past three decades have made Raman a viable option for biosensing.1–6 Specifically, the advent of surface-enhanced Raman scattering (SERS) has facilitated Raman spectroscopic detection of numerous biomolecules using relatively simple laboratory equipment and even field-portable devices.7–11
What is SERS and how is it used?
Raman scattering is an inelastic process wherein incident photons either gain energy from or lose energy to the vibrational and rotational motion of the analyte molecule. The resulting Raman spectra consist of bands corresponding to vibrational or rotational transitions specific to the molecular structure, and therefore provide chemical “fingerprints” to identify the analyte. However, this is a feeble phenomenon, as only approximately 1 in 106–1010 photons are scattered inelastically.12–14 Typical Raman scattering cross-sections are between 10−31 and 10−29 cm2/molecule. In contrast, typical fluorescence dyes have cross-sections of ~10−15 cm2/molecule. It should be noted that resonant Raman scattering can dramatically increase the cross-section. For example, the resonant Raman cross-section of rhodamine 6G (R6G) at λ=532 nm can be as high as 10−23 cm2/molecule.15
Between the time of discovery (1928) and the 1960s, Raman measurements were largely limited to neat solvents. The range of accessible analytes and analyte concentrations was improved upon invention of the laser in the 1960s, but weak signals still limited the utility of this phenomenon for chemical analysis. This changed in the 1970s when Jeanmaire and Van Duyne reported, following Fleischmann's initial observation16, that molecular adsorption onto or near a roughened noble metal surface led to drastically increased Raman signal intensity due to electromagnetic and chemical enhancement mechanisms.17, 18
Fleischmann and coworkers originally reported intense vibrational spectra of pyridine, sodium carbonate, formic acid, and potassium formate adsorbed to redox-cycled silver electrodes as well as pyridine adsorbed to copper electrodes.16, 19, 20 Jeanmaire and Van Duyne further examined factors such as surface features and potential of the electrode, solution analyte concentration, and electrolyte composition of the solution, that affect the intensity of the Raman bands of adsorbed molecules.17 A series of subsequent experiments confirmed that noble metal films with roughened surfaces or nanoscale patterns can dramatically increase Raman scattering signals of analytes and produce enhancement factors (EFs) of 104–108 over normal Raman scattering.21, 22
The enhancement factors for SERS, as compared to normal Raman scattering, are attributed to two mechanisms: an electromagnetic mechanism and a chemical mechanism.23, 24 The chemical mechanism contributes to enhancement through chemisorption of the molecule to the noble metal surface, allowing the electrons from the molecule to interact with the electrons from the metal surface. These interactions lead to an enhancement of signal up to 102, but the chemical mechanism can vary between substrates, substrate adsorption sites, and adsorbed molecules.13, 25 The electromagnetic enhancement is a wavelength-dependent effect arising from the excitation of the localized surface plasmon resonance (LSPR). This collective oscillation of conduction electrons can occur in noble metal nanoparticles (NPs), sharp metal tips, or roughened metal surfaces, and enhances the incident electric field intensity |E|2 by 102–104 times in the vicinity of the metal surface (viz. 0–50 nm of the surface).13, 26 SERS enhancement factors ranging from 106 to 108 have been observed from a variety of substrates.13, 17, 27 Unfortunately, EF calculations are not always consistent from research group to research group; accordingly, care must be taken when comparing EF values from different sources. For single molecule SERS of R6G, enhancement factors of up to 1014 have been reported, but it was recently determined that these enhancements are partially attributable to the unusually large Raman cross-section of R6G. In this case, the surface or electromagnetic enhancement is actually ~108, with the remainder of the enhancement due to the Raman scattering cross-section of R6G and its resonance Raman contribution of ~106. 5
Although SERS detection does not require the adsorbate to be in direct contact with the metal surface, the EM enhancement sharply decreases as the distance between the adsorbate and the surface increases. For example, Van Duyne and coworkers observed from their silver film over nanospheres (AgFON) substrates that a 2.8 nm separation between the adsorbate and the Ag surface decreased the SERS intensity tenfold.28 Clearly, to measure intense SERS signals, analytes must dwell within a few nanometers of the substrate surface. For this reason, many SERS studies have been performed on molecules containing a thiol or amine group which can chemically adsorb to the metal surface.13 To take advantage of the sensitivity and selectivity of SERS for the detection of molecules unlikely to dwell within 2–4 nm of the nanoscale roughness features, an alkanethiol self-assembled monolayer (SAM) has been employed on the surface of the substrate to facilitate the approach and concentration of the analyte within the zone of enhancement.29, 30 Though this approach has not been broadly applied to biomolecule analytes, there are a few examples where a partition layer has been employed to model lipid bilayers and as a tether in aptamer-based sensors.31, 32
SERS detection of biomolecules has been accomplished in both intrinsic and extrinsic formats. In intrinsic SERS biosensing, the molecular signature for the analyte of interest, such as small molecule, DNA strand, or protein, is acquired directly. In extrinsic SERS, the analyte or interaction of interest is associated with a molecule with an intense and distinguishable Raman signature, traditionally a commercially available fluorescent dye, and it is the SERS spectrum of the tag that is used for sensing or quantification. In either format, SERS has unique advantages for biosensing.
Why is SERS a good candidate for biosensing?
Rapid label-free identification of small target analytes is of importance for broad applications ranging from biomarker detection to homeland security. SERS is particularly well-suited to these tasks because of the high sensitivity, the “fingerprinting” ability to produce distinct spectra from molecules similar in structure and function, and the elimination of expensive reagents or time-consuming sample preparation steps associated with other techniques such as polymerase chain reaction (PCR) or immunoassays. In addition, water has a very small Raman scattering cross-section, which leads to minimal background signal from aqueous samples.
In addition, extrinsic SERS detection can provide further advantages over conventional fluorescence-based assays: (1) Raman peaks typically have 10–100 times narrower spectral widths than fluorescence labels, minimizing the overlap between different labels and increasing multiplex capability; (2) when the laser excitation wavelength is matched with the substrate LSPR wavelength, strong SERS signal is achieved from any SERS-active molecule within the zone of electromagnetic enhancement, thus a single source can be used for multiple labels; and (3) SERS labels are not susceptible to photobleaching.33, 34
What are the potential limitations to using SERS for biosensing?
While SERS has the capacity for biosensor signal transduction, there have been several real or perceived limitations to widespread use. Though the SERS community has largely come to agreement about the mechanisms responsible for enhanced inelastic scattering, as detailed above, the impression remains that fundamental information is missing. This supposed knowledge gap may have delayed large-scale investment in SERS biosensing platforms by major instrument manufacturers and funding agencies. In the instance of single molecule SERS, it is true that there is a lack of fundamental mechanistic understanding; while the SERS community is exploring this phenomenon, only limited application of this very high EF detection scheme is possible. Another perception perpetuated in the literature is that SERS substrates do not have reproducible enhancement factors. In fact, there are many examples where a given substrate has been fabricated and used in multiple laboratories with little discrepancy.35–39 It is true that very slight changes to a nanostructured noble metal surface can lead to significant LSPR shifts and that the performance of substrates with narrow plasmons will be influenced by this shift. These shifts, and the resulting changes in enhancement factors, are well-understood based on electrodynamic theory. There are also many SERS substrates with broad localized surface plasmon resonances that are not nearly as sensitive to substrate changes.40–42 Finally, there is a perceived (but incorrect) limitation of SERS that Ag substrates, which generally have larger enhancement factors than substrates made from any other element, are handicapped due to the formation of an oxide layer. While it is true that an Ag2O layer forms on Ag, this layer is thin (viz. 2 nm) and known to be self-limiting. In addition, this oxide layer is displaced upon covalent attachment of molecules to the surface.43
There are some real limitations to the widespread adoption of SERS biosensors. First, based on the fundamental cross-sections of fluorescence and Raman scattering, even a small amount of fluorescence has the potential to mask SERS signals. Clearly, this problem can be ameliorated by using infrared excitation or metallic NPs/surfaces to quench fluorophores or by performing confocal SERS, where a smaller sample thickness is probed. However, if the fluorophore is not in direct contact with the SERS substrate, background fluroescence will result. A similar issue exists with elastic scatter, a process that is also much more efficient than Raman scattering. Elastic scatter becomes an issue when capturing very small wavenumber shift Raman bands or when trying to capture inelastically scattered photons through a complex sample (e.g. a biological cell). The ability to distinguish low wavenumber shift Raman bands is completely dependent on the filtering technology used to eliminate transmitted excitation light. When expanding SERS to detection in complex biological fluids, elastic scatter may occur in all directions from the various intracellular features (i.e organelles, cytoskeleton, etc), creating significant background signal. In addition, interfering species and non-specific binding can mask SERS signal from the analyte of interest. This can be addressed by using antibody-based detection and/or partition layers, as pioneered by Van Duyne.29, 30, 44 Finally, the practical issue of instrumentation cost limits use of SERS; while there is precedent for inexpensive Raman spectrometers,9 for the most part, the cost of a laser, optics, spectrograph, and detector is high and thus, prohibitive to many researchers.
What substrates have been used for SERS?
Inexpensive high-throughput fabrication of SERS substrates with reproducible and large Raman enhancement is a prerequisite for biosensor applications. To date, a large portion of SERS research has utilized colloidal Au/Ag NPs or roughened metal surfaces. Although high EFs have been observed from these substrates, practical applications require engineered SERS substrates that provide tunability as well as reproducibility because maximum SERS intensity is observed when the laser excitation wavelength is tuned near the plasmon resonance maximum of the substrates.45 Therefore, many groups have been developing new techniques for making nanospheres, nanoshells, nanogaps, nanoholes, and sharp tips with tailored optical properties. Table 1 summarizes a variety of engineered metallic structures that have been used for SERS detection. Of particular importance for the future of substrate engineering is the development of novel nanofabrication methods for precise, repeatable, and high-throughput reproduction of various metallic nanostructures.
Table 1.
Metallic Nanostructures for SERS
| Category | Fabrication methods | Features |
|---|---|---|
| Nanosphere | Nanosphere Lithography (NSL)7, 30, 35, 36, 67, 149, 150 Spin Coating38, 151, Nanocrescent152 | - Low-cost, solution-based batch processing - Easily combined with LSPR biosensing - Ultrastable AgFON substrates with high EFs |
| Nanoparticle & Nanoshell | -Nanoparticle90, 93, 95, 111, 115, 116, 118, 119, 153, 154 Nanoshell2, 62–64 | - Geometrically tunable plasmon resonance -Solution phase SERS measurements - Near-infrared SERS probes for intracellular spectroscopy |
| Nanogap | E-beam lithography155, ALD156, Electromigration, On-wire Lithography157, 158, Electrochemical Deposition159 | - Metal-insulator-metal geometry for extreme sub-wavelength energy confinement - Single molecule SERS using ultrasmall nanogaps - Key challenge is reproducible fabrication |
| Nanotip | Electrochemical Etching54, 160, 161, Metal Deposition on a Pulled Fiber52, 162, 163, Template Stripping145, 164, NSL Triangle165, 166 |
- Single molecule TERS demonstrated. - Combine SERS with nano-resolution of SPM |
| Nanowire | Langmuir-Blodgett167, Glancing Angle Deposition (GLAD)168, Anodic Aluminum Oxidation (AAO)169 | - Well-defined surfaces compared to colloidal NPs - Tunability of surface plasmon resonance -Broad plasmon for excitation wavelength flexibility |
| Nanohole | E-beam Lithography170, 171, NSL39, 172 FIB173, 174 | - Easily combined with SPR sensors - Geometrically tunable plasmon resonanc (hole shape, size and periodicity) -Potential for high-throughput array use |
For the most part, Au or Ag have been used for making SERS substrates, although other metals such as Al, In, Cu, and Ga can also support plasmon resonances in the UV-vis-NIR range. Among these, Al and Cu are of potential interest for SERS and plasmonic biosensing because of their abundance and low cost. Furthermore, aluminum has plasmon resonances in the UV regime and thus can extend the spectral range of Au- or Ag-based devices. Rapid formation of aluminum oxide or copper oxide, however, presents significant challenges to using Al or Cu for practical applications, since the unwanted oxide layers sharply degrade the detection sensitivity or dampen the LSPR.46, 47 Although the poor chemical stability is often cited as a major concern for Ag-based substrates, Van Duyne and coworkers demonstrated AgFON substrates (Fig. 1) with temporal stability exceeding 9 months by coating the Ag surfaces with a sub-1-nm alumina overlayer.48
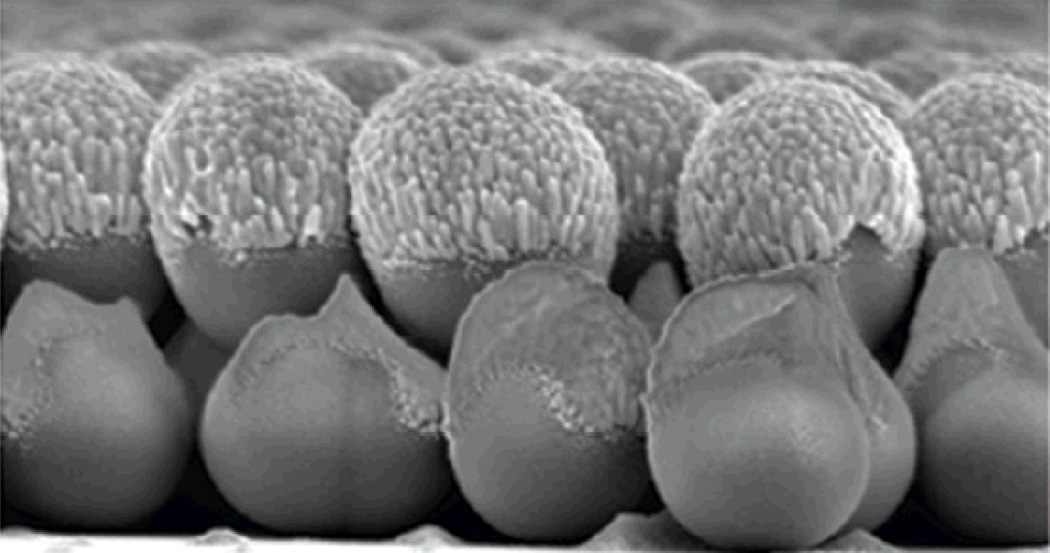
Fig. 1.
Scanning electron micrograph of a AgFON, a widely used substrate for SERS. Figure adapted from Ref. 175, reproduced with permission from the American Chemical Society.
Tip-enhanced Raman spectroscopy (TERS) is an emerging new branch of SERS, wherein the substrate is fabricated on or mounted to the probe of a scanning probe microscope (SPM).49 In this configuration, the apex of a laser-irradiated metal tip captures incident light and generates a strong LSPR for SERS.50 Typically, the tip is a metal-coated SPM tip or a thin silver or gold wire. Single-crystalline silver wires have also been used.51 The radius of the tip is much smaller than the diffraction-limited spot size, allowing ultra-high-resolution SERS imaging. This combination of SERS chemical fingerprinting and SPM imaging is advantageous because it can provide high sensitivity as well as contrast results with a lateral resolution down to a few tens of nanometers with a few seconds of acquisition time. Furthermore, TERS is a versatile technique that can extend the utility of SERS because the analyte does not need to be in direct contact with the SERS-active substrate. The presence of the tip is directly measurable, since when inserted, Bulgarevich and Futamata showed that the Raman signal from an isolated diamond particle can be enhaced 1000-fold.52 Theoretical modeling predicts that bringing a tip close to a metallic substrate can increase the local electric field intensity 5000-fold.53 When a gold tip is brought to within one nanometer of a gold surface coated with non-resonant benzenethiol molecules, EFs of 106 to 108 have been reported.54 In addition, a series of recent studies have established TERS as a promising detection technique with single molecule sensitivity.53–55
In the past decade, a variety of SERS and TERS substrates have been employed for detection of small molecules, DNA/aptamers, and proteins/peptides.56–64 While small molecule detection with SERS has been predominantly accomplished with intrinsic SERS, DNA/aptamers and proteins/peptides have both been detected directly with intrinsic and extrinsic formats. This review covers the exciting developments of the previous decade in SERS biosensing.
II. Small Molecule SERS Biosensing
Intrinsic
There are a number of examples in the recent literature where SERS has been applied for detection of biologically relevant small molecules. These small molecules range from antioxidants, like glutathione, to glucose, to small molecule markers for biowarfare agents such as anthrax. Recent work from Van Duyne’s group has demonstrated the use of SERS for anthrax biomarker detection.7 Silver film over nanosphere (AgFON) substrates were optimized for 750 nm Ti:Saph laser excitation and combined with a battery-powered portable Raman spectrometer. Calcium dipicolinate (CaDPA) - a biomarker for bacillus spores - was detected by SERS over the spore concentration range of 10−14 to 10−12 M by monitoring a peak at 1020 cm−1. Overall, using an 11 minute procedure with a 1 minute data acquisition time, their platform was capable of detecting ~2,600 anthrax spores, well below the anthrax infectious dose of ~10,000 spores. Importantly, the shelf life of prefabricated AgFON substrates in air exceeded 40 days. Their next step was to chemically functionalize the silver surface to enhance the analyte binding affinity as well as the stability of substrates.48 The AgFON substrates were coated with a sub-1-nm alumina layer deposited by atomic layer deposition (ALD), which simultaneously serves to stabilize the Ag surfaces of AgFON substrates and present the surface chemistry of alumina. Dipicolinic acid displays strong binding to the ALD alumina-modified AgFON, and this strong affinity for carboxylate groups makes the alumina-coated AgFON substrate an ideal candidate for bacillus spores detection. The limit of detection (LOD) was further improved to ~1,400 spores with a 10 second data collection time. More importantly, the alumina overlayer dramatically increased the shelf life of prefabricated substrates to at least 9 months. Although the controlled deposition of sub-1-nm alumina films is not a trivial task, ALD facilitates fabrication of functionalized SERS substrates while simultaneously protecting the metal surface against unwanted oxidation or environmental contamination.
In addition to detection of markers of exogenous species, like anthrax biomarkers, SERS can be employed to detect endogenous molecules like the antioxidant glutathone. Multiple schemes have been demonstrated for SERS detection of glutathione – a biologically important tripeptide that exists both in the reduced form (glutathione, GSH) and oxidized dimeric form (glutathione disulfide, GSSG) in tissues. Glutathione plays a role in the respiration of mammalian and plant tissues, protects cells against hydrogen peroxide, and serves as a cofactor for various enzymes. Glutathione is readily detectable with SERS by monitoing the C-S stretching band at 660 cm−1. Ozaki and coworkers mixed glutathione with Ag colloidal solution, heated (60–100°C) until dry, and acquired SERS spectra of dry films, which improved Raman signals from glutathione compared to samples that were not heat-treated.57 Increased aggregation of Ag NPs was observed with glutathione. The linear concentration range for glutathione detection was 100–800 nM, the LOD was 50 nM, and EF on these agglomerated Ag NPs was 7.5×106. In another study, Deckert and coworkers measured TERS spectra of oxidized glutathione (GSSG) immobilized on a thin gold nanoplate.58 The SERS spectra were mainly dominated by carboxyl bands at 1408 cm−1 and amide bands at 1627 cm−1. TERS measurements were performed in back reflection mode through a transparent 20 nm thick gold nanoplate, which was synthesized with a smooth top surface to facilitate homogeneous adsorption and orientation of GSSG molecules. The SERS spectra were recorded at numerous positions on the nanoplate and were consistent in all locations, leading to the conclusion that GSSG adsorbed to the gold in a consistent manner via the carboxyl terminus of the glutamyl, the sulfur of the cystinyl and the amide groups. Consistent immobilization of peptides in this manner will be important for future efforts in charaterizing and sequencing peptides with SERS.
In another example of endogenous small molecule detection, Gogotsi and coworkers used a glass substrate coated with gold NPs for SERS detection of nicotinic acid adenine dinucleotide phosphate (NAADP), a calcium secondary messenger that plays a crucial role for intracellular Ca2+ release.65 Analysis of standards was performed in a sample volume of 1 µL, and SERS detection of 100 µM NAADP was demonstrated. At these high concentrations, the adenine band at 733 cm−1 dominates the spectrum. Using principal component analysis, NAADP from cell extracts was detected in response to treatment with the agonists ATP, acetylcholine and histamine (acid extraction of NAADP from cultured breast cancer SkBr3 cells). This work suggests an interesting possibility of intracellular SERS detection of the calcium messengers, which could help elucidate the mechanisms of calcium signaling pathways in cells.
Among the many small molecule biological analytes intrinsically detectable with SERS, glucose stands out because of its intimate connection with diabetes, which according to the National Institutes of Health affects 10.7% of Americans over the age of 20 and 23.1% of those over the age of 60. Traditional electrochemical methods of glucose monitoring require blood samples to be taken. Though these samples have very small volumes, in some cases as low as 300 nL, they still require collection of blood from the patient.66 Many groups are working on minimally invasive optical techniques to detect and monitor glucose, and SERS is at the forefront of these efforts. In order to be a viable glucose monitoring method, SERS must prove to be accurate and reliable in the clinically relevant concentration range. According to the guidelines set by the National Institute of Diabetes and Digestive and Kidney Diseases (an institute of the NIH) fasting glucose levels below 50 mg/dL (2.7 mM) indicate severe hypoglycemia and potential brain function impairment, while glucose concentrations of 70 – 99 mg/dL are considered normal. Concentrations over 100 mg/dL are indicative of a pre-diabetic condition and a reading of greater than 126 mg/dL (7.0 mM) generally results in a diagnosis of diabetes. Because SERS is compatible with aqueous solutions and it can discriminate interferants by spectral characteristics, it is an attractive method for glucose monitoring in complex biological fluids.
A large challenge, however, is the minimal adsorption of glucose to bare SERS-active substrates such as roughened silver. To overcome this challenge, Van Duyne’s group implemented a SAM of decanethiol on a AgFON substrate.60 The SAM functions as a partition layer that concentrates glucose near the AgFON surface. Without the partition layer, glucose was undetectable, but with it was detectable at concentrations lower than 5 mM by monitoring the appearance of vibrational bands at 1123 and 1064 cm−1. By employing a partial least squares leave one out (PLS-LOO) method of analysis, they demonstrated quantitative detection of glucose over a large, clinically relevant concentration range.
Building on this initial work, the Van Duyne group switched to an ethylene glycol-terminated SAM as the partition layer.67 In this work, a SAM of (1-mercaptoundeca-11-yl)tri(ethylene glycol) (EG3) was formed on a AgFON and was exposed to glucose in aqueous humor containing bovine serum albumin (BSA), a model interferant. EG3 is known to resist protein adsorption and improve biocompatibility, and in this work was shown to form a stable monolayer on the AgFONs in saline for 3 days. They showed quantitative detection of glucose over large and physiologically relevant concentration ranges (0 – 4,500 mg/dL, 0 – 250 mM and 0 – 450 mg/dL, 0 – 25 mM, respectively), reversibility of the glucose sensor, and that exposure to BSA did not hamper glucose detection.
To further improve the performance of partition layer glucose sensors, a mixed monolayer of decanethiol and mercaptohexanol (DT/MH) was formed on an AgFON and employed for real-time sensing.68 The DT/MH monolayer was shown to be stable for 10 days, and was used as a partition layer for quantitative analysis of glucose with less calibration error than observed for glucose detection using the EG3 SAM. Moreover, this sensing strategy proved useful for real-time detection of physiological concentrations of glucose (0 – 450 mg/dL) in a complex biological milieu, bovine plasma. In a flow cell setup, they demonstrated glucose sensing and departitioning with time constants of 25 and 28 seconds, respectively.
After optimizing the partition layer the next steps were to provide a more stable surface for SAM formation, improve chemometric analysis, and shift SERS resonances to near-IR wavelengths to facilitate the use of lower cost lasers.69 This was accomplished by replacing the AgFON with a AuFON and using a shorter chain length version of EG3 as the partition layer. Using a AuFON not only resulted in a SAM layer that was stable for at least 11 days, but also red-shifted the SERS resonance, lowering the biological autofluorescence and allowing greater biological tissue depth penetration. Accurate glucose detection was possible over a larger concentration range (10 – 800 mg/dL, 0.5 – 44 mM), making this sensing strategy applicable to a more diverse set of diabetes patients.
To demonstrate multianalyte sensing capabilities with partition layer-modified substrates, Van Duyne and coworkers used AgFONs with DT/MH mixed SAMs to detect both glucose and lactate, which is an important indicator of potential mortality in intensive care patients.70 Like earlier studies on glucose alone, they showed both partitioning and departitioning of lactate from the SAM layer. Upon partitioning into the SAM, lactate bands at 1463, 1422, 1272, 1134, 1094, 1051, 936 and 868 cm−1 were readily apparent. Using PLS-LOO methods, they demonstrated quantitative analysis of lactate in the concentration range of 10 – 240 mg/dL. Sequential injection of lactate and glucose into a flow cell was used to demonstrate the capability of the sensor to discriminate between the two analytes.
Another category of biomolecules intrinsically detectable with SERS are lipids. In biology, lipids are crucial as structural elements of cell membranes, as a form of energy storage as fats in adipose tissue and as important signaling molecules. Lipids display a large structural diversity, from amphiphilic structures with glycerol backbones like phospholipids to multiple ring structures like steroids. Groups have employed Raman microscopy for investigations of lipid bilayers,61 but SERS for studying lipids is a newly emerging area. Most applications of SERS to lipid sensing have focused on investigations of phospholipid bilayers and their properties, as well as their interactions with various molecules of interest.
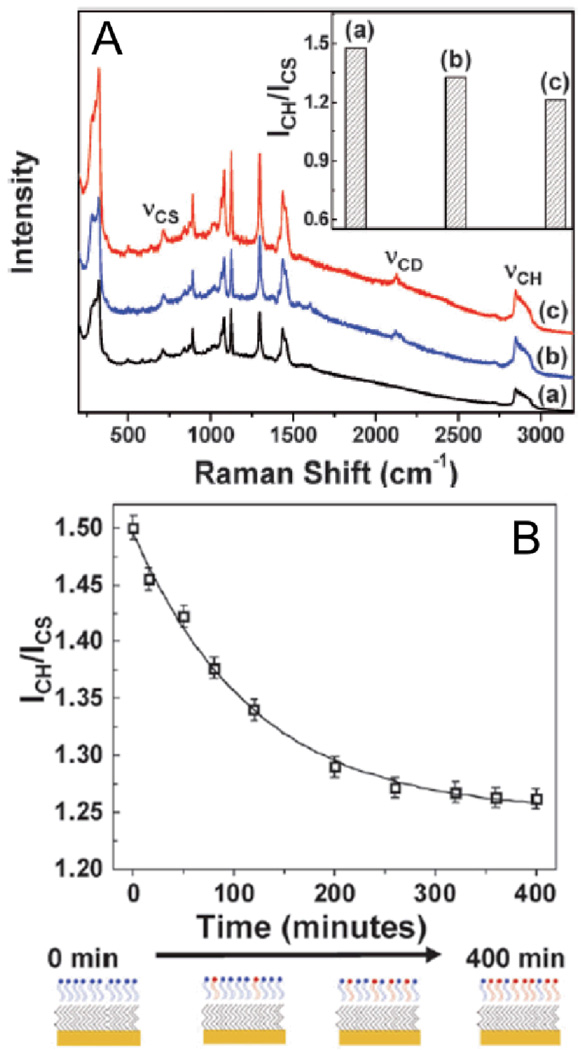
One study by Halas and coworkers investigated the transfer of phospholipids from vesicles (spherical phospholipid bilayers 85–100 nm in diameter) to hybrid lipid bilayers (HBLs) on gold nanoshell supports.71 The vesicles were composed of deuterated 1,2-dimyristoyl-sn-glycero-3-phosphocholine (D-DMPC) whereas the HBLs were a monolayer of DMPC spread over a SAM of dodecanethiol. To determine whether D-DMPC was transferred from the vesicles to the HBLs on the nanoshells, they monitored the intensity of C-H stretch at 2850 cm−1 normalized to the intensity of the C-S stretch at 710 cm−1 (ICH/ICS). When D-DMPC vesicles were mixed with HBL-coated nanoshells, a significant decrease in ICH/ICS was observed, indicating transfer of lipids from vesicles to HBLs. (Fig. 2A) By monitoring ICH/ICS as a function of time, the rate constant for lipid transfer was determined. A plot of ICH/ICS versus time was fit to a first order exponential curve to give a rate constant of K = 1.3 × 10−4 s−1. (Fig. 2B)
Fig. 2.
(A) SERS spectra of (a) a hybrid bilayer formed with DMPC, (b) a hybrid bilayer formed with DMPC incubated for 2 hours with deuterated-DMPC vesicles, and (c) a hybrid bilayer formed with deuterated-DMPC. The decrease in ICH/ ICS (2850 and 710 cm−1, respectively) for the three different systems, as shown in the inset, clearly demonstrates exchange/transfer of lipids. (B) Kinetics of the transfer of deuterated lipids from vesicles to hybrid bilayers as obtained by monitoring the change in ICH/ ICS. The line is a first order exponential fit to the data points. Accompanied is a schematic of the plausible changes in hybrid bilayer composition. Figure adapted from Ref. 71, reproduced by permission of The Royal Society of Chemistry.
In another study by Halas and coworkers, the interaction of a small molecule drug (ibuprofen) with HBL-coated nanoshells was investigated.72 The interaction of ibuprofen with lipid bilayers in the gastrointestinal tract has been suggested as one of the mechanisms of observed side effects, such as gastrointestinal bleeding. When HBL nanoshells were incubated with ibuprofen, ring modes at 803, 1185, 1205, and 1610 cm−1 were present due to ibuprofen partitioning into the HBL, and the intensity of the peaks increased with increased ibuprofen concentration. By monitoring the peak at 1610 cm−1, it was determined that ibuprofen partitioned into the HBL with isotherm-like behavior. Their results also indicate that ibuprofen partitioning into a deuterated-HBL disrupted the order of the HBL. This was determined by monitoring the carbon-deuterium stretch as a function of ibuprofen concentration. As the ibuprofen concentration was increased, the carbon-deuterium stretch intensity decreased.
In order to interface lipid bilayers with solid substrates, many groups employ tethered lipid bilayers. In these systems, a lipid molecule with a longer linking group is deposited on a surface as a SAM and then a second lipid monolayer is deposited to form a bilayer. In many cases, the ordering of the lipid SAM can be determined by SERS. In one study, a dipalmitoylphosphatidylethanolamine-mercaptopropionamine (DPPE-MPA) SAM was used as the lipid tether and its organization on a roughened polycrystalline gold substrate was assessed with SERS.73 In this work, they observed a significant difference in the conformation of the linking moiety of the DPPE-MPA monolayer in air compared to aqueous solution. In air the trans conformation was determined to be the dominant for the S-C-C tether due to a large peak at 720 cm−1. Conversely, in aqueous solution the gauche form of the tether predominated as judged by a large peak at 643.5 cm−1. These results were confirmed electrochemically and gave insight into the formation of tethered lipid bilayers for biosensing applications.
SERS for the study of lipids is a developing field and expanding the types of lipids studied could have impacts in membrane and lipid biology. For example, lipids such as sphingosine-1-phosphate and platelet activating factor are known to function as cellular signalling molecules, but can be challenging to detect. With partition layer schemes, lipid signalling molecules could be detected in complex mixtures based on their spectroscopic profiles.
Extrinsic
The limited use of extrinsic SERS for small molecules is due in large part to the ease of obtaining the structural vibrations and rotations directly from small molecules as opposed to more complex systems like DNA and proteins, which are discussed later in this paper. However, some extrinsic small molecule SERS detection schemes do exist. Also focused on anthrax biosensing, Chung and coworkers used Au NPs modified with a 16 amino acid peptide or antibody coupled to the Raman reporter 5, 5’-dithiobis(succinimidyl-2-nitrobenzoate) (DSMB) to detect a different anthrax biomarker, protective antigen (PA).74 In this example, either the peptide or antibody which binds to PA and DSMB is detected by its characteristic band at 1336 cm−1. The peptide binding partner was shown to be as efficient as an antibody binding partner, and LODs for their biomarker were in the low fM range. As discussed in the intrinsic SERS section, SERS detection can be employed to detect endogenous species like glutathione as well as previously mentioned examples of exogenous species. Ozaki and coworkers developed an extrinsic SERS method for detection of glutathione.75 In this ‘reversed reporting agent’ scheme, SERS signal of reporting agent-capped Ag colloids is reduced upon addition of glutathione, which induces aggregation of the Ag colloids. Various reporting agents were tested, but 5 µM R6G was the only viable candidate for glutathione sensing. The LOD using this scheme was 1 µM, which is much higher than for their heat-induced SERS sensing of glutathione.
SERS has been applied broadly to small molecule and lipid sensing due to the small number of vibrational bands associated with the analyte. This field of study would benefit from substrates with higher EFs for improved LODs. More tailoring of substrate surface chemistry to the desired analytes, making detection from complex mixtures easier, reducing biofouling of the substrate, and minimizing sample preparation time. While intrinsic and extrinsic small molecule sensing employ relatively easy detection schemes, more complex biological molecules like DNA and aptamers present new SERS sensing challenges.
III. DNA/Aptamer SERS Biosensing
Intrinsic
Intrinsic SERS and TERS measurements can also be employed beyond small molecule detection for DNA and aptamer biosensing. Recent advances in achieving low limits of detection and good reproducibility make SERS a useful tool for either single or double-stranded thiolated DNA oligomer detection. For example, Halas and coworkers used Au nanoshells bound to glass substrates to obtain SERS spectra of DNA, which were dominated by adenine vibrational bands at 729 cm−1.63 Moreover, they observed changes in the dsDNA spectrum upon interaction with cisplatin and transplatin, cis and trans forms of common chemotherapy agent, revealing an opportunity for SERS to contribute to pharmaceutical research. Critical to the achievement of highly reproducible DNA spectra in this work was a thermal pretreatment that promoted extended linear conformation of ssDNA and dsDNA on the Au nanoshell substrate. The report of adenine as the dominant base was not particularly surprising since this is the only DNA base showing single-molecule detection to date, suggesting a large Raman scattering cross-section.76–78 An alternate approach to thermal pretreatment for achieving label-free, target-specific and highly sensitive SERS of DNA employs an electrokinetic preconcentration method, electrophoresis. Lee and coworkers used negatively charged plate electrodes as SERS substrates for detection of positively charged adenine.79 They illustrated that application of a constant electric field, 0.6 V cm−1 for adenine measurement, resulted in a 51-fold amplification in signal over open circuit detection of adenine. Although, this measurement combination is limited owing to pH sensitivity and substrate contamination issues, it is a promising method for label-free analysis of charged biomolecules. Other intrinsic SERS studies of DNA have focused on detection of changes in conformation rather than individual base detection. Neumann and coworkers demonstrated high-specificity detection of DNA and lower-specificity detection of small molecules upon using SERS to examine conformational changes caused by interactions between an aptamer and analyte molecules. In this case, they used Au nanoshells immobilized on quartz substrates which display either an anti-platelet-derived growth factor (anti-PDGF) aptamer for PDGF detection or an anti-cocaine aptamer for cocaine detection. Comparison of the conformational changes indicated using SERS and the standard technique of circular dischroism (CD) showed high correlation in this work. In addition, comparison between substrate incubation with specific (PDGF) versus non-specific (lysozyme) target analyte confirmed the high specifity of this biosensing scheme. On the contrary, the anti-cocaine aptamer showed non-specific binding to both caffeine and benzocaine, suggesting that more work needs be done for aptamer based SERS sensing of specific small molecule targets. However, this limitation for aptamer-target specificity could be exploited for identification of multiple targets by a single aptamer.80
TERS has also been proposed for direct DNA and RNA sequencing, identification of biomacromolecules, and characterization of single viruses at the molecular level. Bailo and Deckert used TERS for direct, label-free detection of a single stranded RNA cytosine homopolymer.81 All spectra obtained along the length of a 20-nm-long single strand of RNA showed spectral features of cytosine with little variation in band intensities or positions. Moreover, as deduced from this experiment, it is possible to find the sequence of RNA with controlled movement of the TERS probe from base to base. Deckert and coworkers have also demonstrated that TERS is a powerful tool for direct detection of single virus particles belonging to different species.82 All TERS virus spectra measured in this work showed bands that were attributed to spectral features of single tobacco mosaic virus (TMV) due to interaction between the coat proteins and RNA with the Ag-coated AFM tip. They obtained EFs of 106, and they claimed that higher EFs are achievable if the excitation wavelength overlaps with plasmon absorption profile of the nanoroughened Ag on the AFM tip.
Even though there are certainly challenges related to fabrication of uniform, highly sensitive and reproducible SERS substrates for biosensing, there has been clear progress on the application of SERS and TERS to DNA, aptamer, and single biomolecule detection.
Extrinsic
While there are relatively few examples of intrinsic SERS for DNA and aptamer detection, DNA and aptamer monitoring via extrinsic SERS has been employed extensively. As shown in Scheme 1, a common detection scheme for DNA binding events is to functionalize a Au or Ag NP with a reporter molecule (usually a fluorescent dye) and a single stranded piece of DNA. Upon hybridization with a complementary strand of DNA, typically bound to another Au or Ag surface, the SERS or surface-enhanced resonant Raman scattering (SERRS) signal of the reporter molecule is observed. Many groups have employed this DNA detection scheme to achieve various goals: (1) Vo-Dinh and coworkers used an immobilized DNA capture strand on a Ag surface for the detection of the breast cancer gene (BRCA1);83 (2) Moskovits and coworkers used a sandwich assay between ssDNA attached to a flat Ag layer and a Ag NP labeled with the complementary DNA strand to detect hybridization;84 (3) Mirkin and coworkers used on-wire lithographically produced Ag nanorods with etched gaps for the detection of DNA binding;85 and (4) Moskovits and coworkers detected a protein after a Au NP functionalized with dsDNA bound and the SERRS signal was further enhanced by electroless Ag plating onto the complex.86
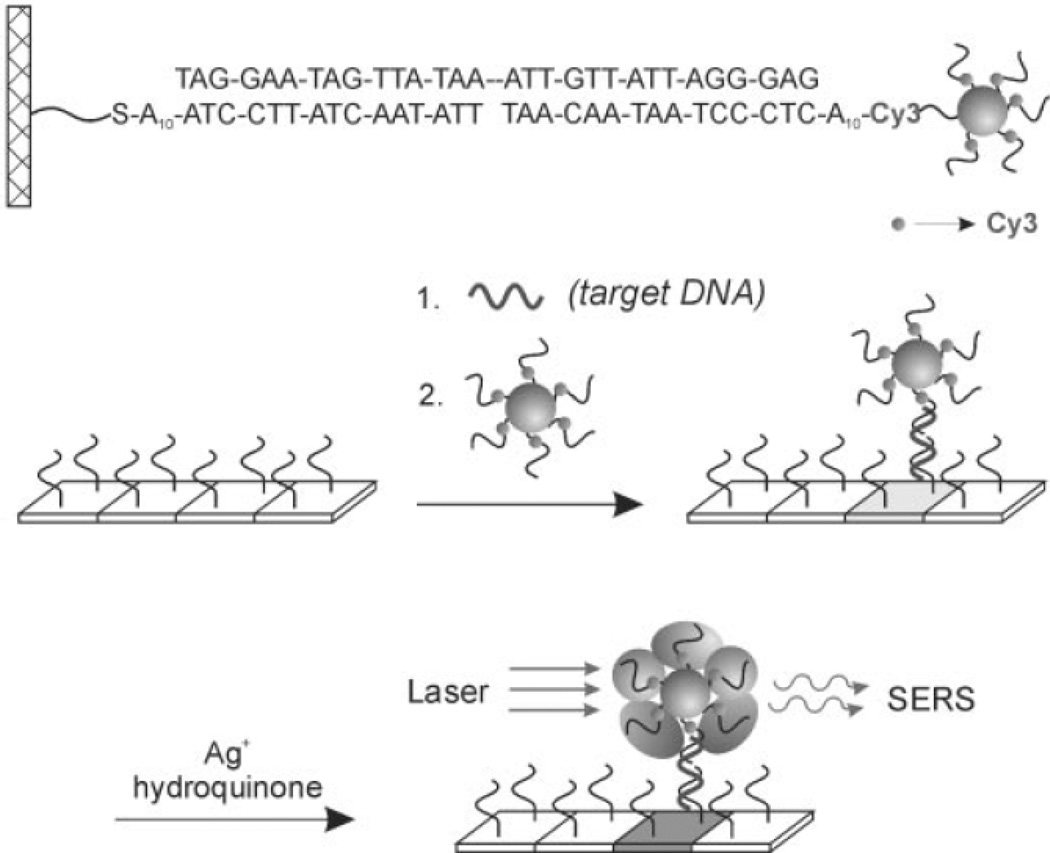
Scheme 1.
A SERS probe consisting of a 13-nm-diameter Au nanoparticle functionalized with a Raman dye-labelled oligonucleotide. A three-component sandwich assay is used in a microarray format and the Raman probe detected, after Ag enhancing, by SERS. Figure adapted from Ref. 96, reproduced with permission from the American Academy for the Advancement of Science.
The intentional creation of DNA strands with single nucleotide polymorphisms allows researchers to evaluate how specific their assays are for the complementary ssDNA capture strands. Graham and coworkers found that DNA hybridization was observed only when a fully complementary DNA sequence was added to their DNA-functionalized Ag NPs,87 proving that they could detect single nucleotide polymorphisms when present in the DNA mixtures. They were also able to modulate the SERRS signal by heating or cooling the analyte solutions to influence extent of DNA hybridization. The LOD of these hybridization assays can be further improved by functionalizing the Au or Ag surface. Graham and coworkers also employed commercially available Klarite gold substrates for the detection of dye-labeled oligonucleotides,88 and they had the most reproducible results when they functionalized the Au surface with a bidentate SAM of a ssDNA capture ligand, resulting in a LOD of 10−7 M. Along with tailoring the surface chemistry of the Klarite substrates for DNA hybridization and attachment, Graham and coworkers also investigated the surface chemistry that would best allow DNA to attach to Ag NPs.89 Through their investigations, they determined the number of amine modifications needed so that the overall strand has a positive charge and would attach most efficiently to a Ag NP; this strategy yielded a LOD of around 10−12 M. With qualitative detection of DNA hybridization clearly feasible, Graham and coworkers also began pushing for quantitative detection of DNA. The researchers optimized oligonucleotide detection conditions by employing 8 commercially available fluorescent dyes attached to Ag NPs.90 After optimization of Ag NP dilution and aggregating agent, calibration curves were created for each dye to facilitate quantitative SERRS detection. With this approach, they calculated LODs for optimized conditions with all 8 dyes; in the best case, a LOD of 0.5 fM was achieved.
Using multiple SERS tags for the labeling or detection of DNA associated with disease has been covered in a review by Natan and coworkers.34 This review identified the great potential of a SERS multiplex sensing platform, as was demonstrated in 2006 by Mirkin and coworkers.91 This work used various DNA sequences immobilized on glass beads, hybridized with a Au NP, and labeled with a complementary ssDNA and reporter molecule.91 The multiple Raman labels were created by using single and quantitative mixtures of fluorescent dyes attached to the Au NP. They labeled DNA sequences for Hepatitis A, Hepatitis B, HIV, Ebola virus, Variola virus (smallpox), Bacillus anthracis, Francisella tularensis, and hog cholera segment with the Raman labels. The bead-NP complexes were then further enhanced with Ag plating to increase the SERS signal. Similarly, Graham and coworkers used 5 different DNA sequences, each labeled with a different fluorescent dye and a Ag NP.92 They labeled a probe for human papillomavirus with R6G, the VT2 gene of E.coli 157 with ROX, and a universal primer with FAM, CY5.5, and BODIPY TR-X. They found LODs from 10−11 and 10−12, and since DNA sequence choice does not affect the SERRS signal, the detection scheme can be generalized to any target. The linear response of these probes at biologically relevant concentrations with all 5 probes simultaneously indicates a promising future for DNA detection with multiplex labels.
DNA-based SERS sensors are not limited to only DNA detection; by using aptamers generated to detect other biomarkers, these schemes can be broadly adapted. For example, Lee and coworkers fabricated an aptamer-based SERRS sensor using gold NPs bound to a glass slide coated with a thrombin-binding aptamer; a methlyene blue reporter molecule facilitated detection of thrombin binding events.93 This simple detection scheme had a LOD of 100 pM, which was lower than the disassociation constant of 25 nM for thrombin. A LOD lower than the disassociation constant indicates that thrombin has a higher affinity for the substrate bound thrombin-binding aptamer.
While the formation of DNA-NP complexes is generally used for the detection of specific binding events, the complex formed between DNA, or DNA-like molecules, and NPs can also be used to create SERS hotspots, or small volumes with extremely large electromagnetic fields. Graham and coworkers used fluorescent dye and locked nucleic acid-labeled Ag NPs for controlled aggregation of NPs to induce SERRS hotspots.94 When the probes were combined with appropriate target DNA sequences they took one of three different conformations, head to head, tail to tail, or tail to head, all of which caused enhancement of SERRS signal. DNA hybridization can also be achieved with peptide nucleic acids (PNA), as demonstrated by Moskovits and coworkers, who exploited the electrostatic interactions between surface bound PNA and target DNA to create SERS hotspots.95 The DNA-PNA hybrid in this work had a net negative charge, which then allowed a positively charged Ag NP to attach to the DNA-PNA hybrid. Upon exposure to R6G, Raman hot spots could be observed where the DNA-PNA complex had formed.
Detailed above are multiple ways DNA hybridization is detected or exploited; a similar scheme can also be employed for RNA detection. Mirkin and coworkers accomplished this by starting with a typical sandwich assay where DNA immobilized on a slide was hybridized with a complementary DNA strand with a dye-functionalized Au NP.96 (Fig. 3) As detailed in aforementioned examples, this complex was further enhanced with a Ag plating bath to increase the SERS signal. The group then went further and immobilized two different RNA strands that can bind to the same DNA capture strand but have single nucleotide polymorphisms. After stringent washing to denature and remove the imperfect DNA/RNA complexes, they were able to detect only perfectly matched RNA strands.
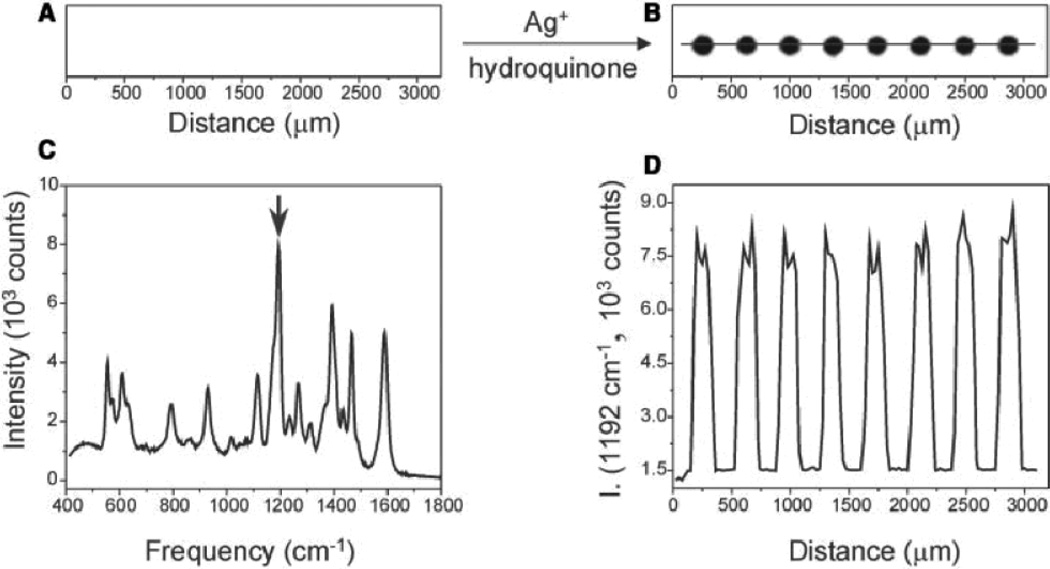
Fig. 3.
A flatbed scanner image of Raman probes (A) before and (B) after Ag enhancement. (C) A SERS spectrum of one of the Ag spots. (D) Raman intensity of the 1192 cm−1 as the laser is scanned across the chip from left to right. Figure adapted from Ref. 96, reproduced with permission from the American Academy for the Advancement of Science.
Gene detection using Au or Ag NPs functionalized with ssDNA and a SERS or SERRS reporter molecule has also been demonstrated. Vo-Dinh and coworkers used a scheme similar to that described above to detect a SERS signal from the HIV-I gene based on its hybridization to DNA labeled with a NP-reporter molecule complex.97 In another paper, Vo-Dinh and coworkers utilized a Au NP fuctionalized with a hairpin ssDNA sequence for the detection of HIV-I gene, which, upon the addition of the complementary DNA sequence caused SERS signal to decrease in the presence of the target, however they did see a 10% decrease in signal when non-complementary DNA was present.98 Most recently, this group was able to detect the presence of two target genes indicative of breast cancer with this hairpin detection scheme.99
While the intrinsic work on SERS for DNA has been limited, the future of this field and its applications to DNA sequencing look very promising due to the opportunity for label-free and target-specific detection. Recently, Halas and coworkers demonstrated label-free SERS detecton of DNA. Replacement of adenine with 2-aminopurine in the capture DNA strand allowed for the adenine band of the target DNA to be monitored and hybridization efficiency monitored.100 In most cases, it is the low EFs of the substrates and the inability to create zones of enhancement small enough to see SERS signal from one nucleotide base at a time that have been the limiting factors. While multiplexing SERS tags and quantitative extrinsic SERS is interesting, we will eventually reach a limit as to how many tags we can discriminate within a mixture. The area where extrinsic SERS is the most promising is in the gene detection schemes proposed by Vo-Dinh, Graham and Mirkin. The recent emergence of some hypenated extrinsic SERS techniques has opened doors into more forensic science and genotyping areas of interest. The SERS-melting (either electrochemical or thermal) combination demonstrated by Bartlett and coworkers allowed them to effectively detect short tandem repeats and single nucleotide polymorphisms of DNA bound to an Au sphere segment void substrate.101, 102 Batt and coworkers utilized the combination of ligase detection reaction with SERS to quantitatively detect single nucleotide polymorphisms in oncogneic K-Ras with a 10 pM detection limit.103 The ability to detect the presence of diseases quickly, with little sample preparation and without the need to amplify the amount of DNA present, is one of the most exciting directions for further work using extrinsic SERS.
IV. Protein/Enzyme/Peptide/Antibody SERS Biosensing
Intrinsic
While intrinsic SERS measurements are relatively straightforward for small molecules, lipids, and DNA where each target has a small number of vibrational modes, it is also feasible to perform SERS biosensing on more complicated, larger molecules such as peptides, proteins, enzymes, and antibodies.104 Unlike the more tractable small molecules, band assignments for spectra from these larger species are often based on general motifs seen through the molecules rather than localized vibrational modes. One commonly used example is the characteristic aromatic amino acid bands present at ~950 cm−1, for the C-COO− stretch, and ~1400 cm−1, for the COO− symmetric stretch. Another commonly used spectral feature is the broad amide (CO-NH) I and III bands, at 1600–1700 cm−1 and 1200–1350 cm−1, respectively, used in intrinsic peptide and protein analysis to describe both primary and secondary structure characteristics.105
SERS sensing of the simplest amino acid structures, peptides, has been pursued both from a fundamental perspective, with the goal of assigning Raman bands to particular amino acids, and from an applied perspective, to sense particular biomarkers. Proniewicz and coworkers used a series of homodipeptides on Ag colloids to assign Raman bands for Cys, Gly, Leu, Met, Phe, and Pro.106 Their results also yield insight about adsorption orientation of the amino acids, specifically resolving a previous controversy about Gly-Gly adsorption to show that it initially adsorbs through its C-terminus but rearranges to adsorb through its N-terminus with time. Hartgerink and coworkers also used dipeptides, where different aromatic amino acids were linked to cysteine, to demonstrate that one amino acid can be distinguished from another.64 In this case, Au nanoshells were the SERS substrate and the cysteine was employed to promote covalent attachment of the peptide to the nanoshell. Comparison of the measured SERS spectra directly to normal Raman spectra showed little shift in the Stokes Raman bands but significant broadening of the SERS peaks. As a proof-of-concept for measurement of more complicated peptides, this paper also includes the prediction and measured SERS spectrum of a 19 amino acid cell-penetrating peptide known as penetratin, and this spectrum is dominated by the aromatic features from 3 amino acids within the molecule. With a more applied goal, Ozaki and coworkers have employed SERS to sense bombesin, a 14 amino acid neurotransmitter that is a tumor marker.107 This group's work characterizes the Ag colloid adsorption behavior of bombesin and bombesin fragments and compares the measured spectra with density functional theory predictions. While this is a significant first step toward making SERS a viable biosensor for bombesin, much work must still be done to determine sensor limits of detection as well as sensing in the presence of interfering species. In parallel with the peptide work detailed above, many groups are also pursuing SERS detection of more complicated amino acid structures such as proteins.
Moskovits and coworkers recently demonstrated protein SERS spectra after sandwiching double cysteine mutants of the small protein FynSH3, a common model protein for kinetic and thermodynamic studies, between two Ag NPs.108 If this protein remains in its native state between two NPs, the distance between the linked NPs would be 2.3 nm, creating a small gap and large electromagnetic enhancement. The captured SERS spectra clearly show evidence of the cysteine linkage to the Ag NP as well as bands for the aromatic amino acids histidine, tryptophan, and phenylalanine. The band intensities varied from second to second during collection, suggesting that the spectrum was sensitive to protein conformation/orientation. In some cases, they saw momentary spectra indicating graphitic carbon (protein degradation), quickly replaced by a new amino acid spectrum, suggesting that they were measuring spectra from single or small numbers of protein molecules within an electromagnetic hot spot. In another recent example, Ozaki and coworkers adapted traditional Western Blot analysis of proteins to include SERS and SERRS detection.109 First, they separated a protein mixture using gel electrophoresis followed by electroblotting onto nitrocellulose to immobilize proteins. After applying Ag NP stain, they recorded confocal SERRS or SERS of 2 model proteins, myoglobin and bovine serum albumin, respectively. While they could distinguish these two proteins using amide and aromatic amino acid bands, there was, unfortunately, no clear linear relationship between the amount of protein and SERS band intensities. Another concern was that the SERS spectra after Ag NP staining did not look like the bulk Raman spectra, perhaps because only a portion of the protein is within the large electromagnetic fields responsible for SERS. Deckert and coworkers also used myoglobin as a model protein to demonstrate SERS detection following free flow electrophoresis separation with isotachophoretic focusing.110 Using a microfluidic platform, they were able to focus a Ag NP/myoglobin mixture to elute from only 2 channels. Though the myoglobin concentration was quite high (410 µM) and there were some issues with Ag NPs clogging the microfluidic channels, this work shows the promise of SERS detection within "hyphenated techniques" and on microfluidic platforms.
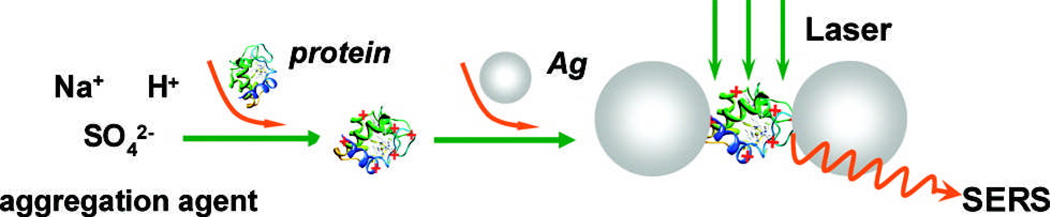
While previous examples focus mainly on identifying proteins or protein components, SERS has also been used to look at an important sub-class of proteins, enzymes. Ozaki and coworkers measured the intrinsic SERS spectra of the enzymes lysozyme, ribonuclease B, avidin, catalase, hemoglobin, and cytochrome c with low µg/mL - ng/mL limits of detection.111 They accomplished this by mixing the enzymes first with acidified sulfate and then colloidal silver before capturing NIR SERS spectra, as shown in Fig. 4. The acidified solutions and sulfate greatly enhanced the detection limits for proteins examined, and again, the spectra were mostly characterized based on amide and aromatic amino acid band locations and intensity. In a distinct effort to perform single enzyme analysis, Käll and coworkers used immobilized protein/NP aggregates (with 90-nm-diameter Ag NPs) to measure single molecule SERRS spectra of horseradish peroxidase (HRP) at various points in its enzymatic cycle.112 They found that Ag NPs alone denatured the protein but that the NPs could be made biocompatible simply by adding H2O2 before adding enzyme. In 1 second SERS collections with low concentration HRP, the SERRS spectra fluctuate, and the authors suggest that these fluctuations correspond to the various forms of HRP found within the catalytic cycle.
Fig. 4.
Detection scheme for label-free protein sensing with SERS. The presence of protein induces the aggregation of Ag nanoparticles, which is then used to produce a strong SERS signal. Figure adapted from Ref. 111, reproduced by permission of the American Chemical Society.
While the protein spectra measured here are promising and there are clear spectral differences between different mutants and different proteins, ab initio identification of proteins from measured SERS spectra is still not possible. This may be feasible with advanced chemometric or peak deconvolution techniques but recent focus in this area has been mainly on using extrinsic SERS for protein, and even antibody, biosensing.
Extrinsic
To address the challenges of intrinsic SERS detection of proteins, Raman reporter molecules have been used when specific protein spectra are not necessary, and the goal is to quantitatively detect either individual proteins or total protein content. For extrinsic SERS detection of proteins, reporter molecules must (1) maintain stability during tagging and measurement and (2) generate a robust and consistent Raman spectrum under the conditions of the SERS measurement.34 For protein detection, commonly used SERS reporter molecules include 5,5'-dithiobis(succinimidyl-2-nitrobenzoate) (DSNB) with intensity monitored at 1336 cm−1, 113–116 4-mercaptobenzoic acid (MBA) with intensity monitored at 1585 cm−1,117 4-nitrobenzenethiol (4-NBT) with intensity monitored at 1336 cm−1, 2-methoxybenzenethiol (2-MeOBT) with intensity monitored at 1037 cm−1, 3-methoxybenzenethiol (3-MeOBT) with intensity monitored at 992 cm−1, and 2-napthalenethiol (NT) with intensity monitored at 1384 cm−1.118 Also used as extrinsic Raman labels for SERS are 4,4'-bipyridine (BiPy), with strong bands present at 1609, 1227, 1291 cm−1, thiophenol (TP), with strong bands at 994 and 1570 cm−1, and p-aminothiophenol (PATP), with strong bands at 390, 1077, and 1578 cm−1.119 Fluorescein isothiocyanate (FITC) and malachite green isothiocyanate (MGITC), which are widely used fluorescence tags, have also been used as common Raman reporter molecules for SERRS and SERS detection of proteins. FITC intensity is monitored at 1630 cm−1 following excitation at 514.5 nm, and MGITC intensity is monitored at 1615 cm−1 when excitation at 647.4 nm is used.120, 121
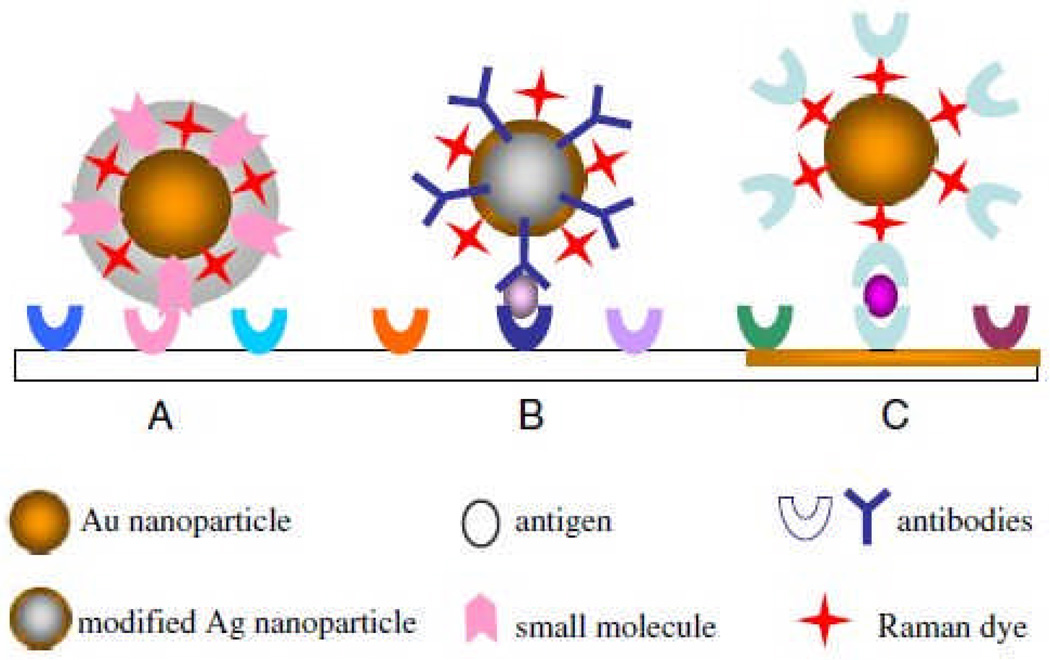
Many of the aforementioned extrinsic Raman labels have been used for immunoasssays, which rely on the specificity of the interaction between antibodies and their corresponding antigens.44 Multiple formats have been employed for SERS-based immunoassays, including the use of silver island films, colloids in solution, immobilized colloids, and modified colloids as probe molecules.44
An immunoassay using SERS was first reported in 1989 by Tarcha and colleagues.122 In this early SERS-based immunoassay, SERS was employed for the detection of thyroid stimulating hormone. The sandwich format used a silver island film as the substrate to which a capture antibody was attached, followed by incubation with antigen, then addition of another antibody conjugated to p-dimethylaminoazobenzene. Since then, SERS-based immunoassays have largely utilized colloids as SERS substrates to address the challenge of reproducibly manufacturing island films with nanoscale roughness.123 The use of colloids as SERS substrates also offers versatility in immunoassays, including the possibility for multiplexing and extension into in vitro and in vivo SERS.44 A common format involves a sandwich assay similar to those used in aforementioned SERS DNA detection schemes: antibodies are immobilized, then exposed to antigen, followed by exposure to gold NPs conjugated to antibodies as well as probe molecules. Variations of this format for SERS-based immunoassays are shown in Fig. 5. Using the sandwich structure, Hepatitis B virus surface antigen has been detected using murine monoclonal and polyclonal antibodies.117 To improve the LOD of the virus surface antigen to 0.5 µg/mL, a silver staining step was incorporated into the immunoassay as well.117 Feline calcivirus and Mycobacterium avium subsp. paratuberculosis have been detected using a sandwich format as well.115, 116 Porter and coworkers optimized the conditions for feline calcivirus detection and detected virus concentrations as low as 1×106 viruses/mL, or 70 captured viruses.115 Porter and coworkers were also able to detect 200 ng/mL (1000 bacteria/mL) of Mycobacterium avium subsp. paratuberculosis in a milk matrix.116
Fig. 5.
Schematic of SERS immunoassay. Multiple formats are available for detection: (A) shows a Au NP core coated with a shell into which extrinsic Raman labels and antibodies are immobilized, (B) shows a Au-coated Ag NP probe conjugated to both extrinsic Raman labels and capture antibodies, and (C) shows antibodies conjugated to a Au NP using extrinsic Raman labels. Figure adapted from Ref. 104, reproduced by permission of The Royal Society of Chemistry.
To evaluate the potential of SERS-based immunoassays to replace or augment current immunoassay technology, it is necessary to compare the LODs of the aforementioned SERS immunoassays to the LODs of widespread immunoassays. Fluorescence and microcantilever detection of viruses have been achieved at concentrations on the order of 105 viruses/mL while microbial detection with ELISA can be achieved with 104 cells/mL.115, 124, 125,126 Electrochemical methods were employed for detecting E. Coli O517:H7 or Salmonella at concentrations as low as 6×102 or 5×103 colony forming units (living bacterial cells/mL), respectively.127, 128
Multiplexing is also possible with SERS immunoassays. Tian and colleagues used two approaches to multiplex a sandwich-format SERS immunoassay: immobilized antibodies were exposed to the target analytes (mouse or human IgG), then to (1) gold NPs with antibodies and 2 reporter molecules attached or (2) gold and gold/silver bimetallic NPs with antibodies and 1 reporter molecule attached.119 With the first detection scheme, 50 µg/mL mouse and human IgG were detected simultaneously. With their second detection scheme, the multianalyte assay was not successful due to overlapping peaks of the Raman reporter label on gold and gold/silver bimetallic NPs, but mouse and human IgG were detected individually at 100 µg/mL.119 Porter and colleagues developed a tetraplexed assay to detect IgG from four species using four Raman reporters by employing mixed monolayers of extrinsic Raman labels on gold NPs.118
Direct detection of enzymes using SERS has been mentioned previously, but direct detection is difficult due to enzyme denaturation and loss of activity.129 Indirect detection of enzyme activity, in which an enzyme produces a SERRS-active dye as a product, have been used for enzyme-based immunoassays. Extrinsic SERS detection of proteins has also been accomplished by modifying colloids to produce NP probes.130, 131 Using gold NPs and electroless silver plating, Mirkin and colleagues coated NPs with a hydrophilic oligonucleotide, Raman dye label and a small molecule to detect protein-small molecule interactions.130 In another probe design to detect protein-protein interactions, they coated NPs with antibodies and labeled them with oligonucleotide and a Raman dye.130 A similar probe design using Coomassie Blue was developed by Ozaki and coworkers.131
Though many SERS-based immunoassays have used colloids as substrates, single-walled carbon nanotubes (SWNT) have been used by Dai and coworkers to achieve lower LODs. They used functionalized SWNTs as multicolor Raman tags for highly sensitive detection of protein interactions on microarrays.132 They used a sandwich-assay scheme wherein an analyte (antibody) from a serum sample was captured by immobilized proteins in a microarray, followed by incubation of SWNTs conjugated to goat anti-mouse antibody that specifically bind to the capture analyte. The strong SERS signal produced by the SWNT tag enabled protein detection sensitivity down to 1 fM, demonstrating great potential for extrinsic SERS detection toward applications in proteomics and autoimmune disease research.
The use of Raman labels for protein detection is not limited to immunoassays. A protein concentration assay developed by Han et al., uses the SERS signal of Coomassie Brilliant Blue dye adsorbed non-specifically to silver colloids to monitor the total protein concentration.131 The SERS signal of Coomassie Brilliant Blue displayed a linear and inverse relationship to protein concentration over a bovine serum albumin concentration range of 10−5 to 10−9 g/mL.131 Clearly, extrinsic SERS has enabled protein sensing ex vitro; in fact, a similar approach can be adopted to perform SERS detection inside cells as well.
Direct SERS detection of proteins is limited by the spectral similarities of many proteins. Indirect SERS detection of protein concentration through immunoassays is promising, but the use of antibodies and detection limits on the order of ELISA assays with fluorescence detection prevent SERS-based immunoassays from replacing the more widespread enzyme-based immunoassays for clinical and diagnostic use.
V. Cellular and in vivo Sensing
Intrinsic
In a relatively small number of cases, SERS biosensing of proteins, DNA, and small molecules has been extended from the aforementioned examples toward cellular and in vivo systems. However, there are only a few where direct, intrinsic, SERS detection is performed in cells: this is due to the complex biological environment which can mask signals from analytes of interest or cause fluoresence. The Van Duyne group first demonstrated in vivo application of intrinsic SERS by measuring the glucose concentration from the interstitial fluid of a rat.133 A SAM-functionalized AgFON substrate was surgically implanted under the skin of a rat such that it was in contact with the interstitial fluid and optically addressable through a glass window placed along the midline of the rat's back. The SERS spectra from glucose were acquired through the window using a 785 nm Ti:sapphire laser with a power of 50 mW for 2 minutes. The glucose concentrations obtained from the implanted AgFON sensor matched the data from a commercial glucometer. With further refinements and miniaturization in the system, SERS biosensors could help the treatment and care of diabetics or other conditions that would benefit from time lapse monitoring. SERS spectra have also been successfully measured from endosomes in live cells (rat renal proximal tubule cells and mouse macrophages) using gold NPs.134 Both cell lines were exposed for 30 minutes to gold NPs, which were then localized in the endosomes of the cells. In this case, cells were raster-scanned with a low-power (2 mW) NIR excitation laser for 1 sec to prevent interference from normal Raman scattering and possible change in the live cells due to laser illumination. The strongest SERS signal was acquired 120 minutes after exposure to NPs, when gold aggregates formed in the lysosomes. Gold aggregates in both cell lines produced tentatively assigned bands in the spectra that indicate the presence of proteins, lipids, carbohydrates, and nucleotides chemical nano-environments of lysosomes. In addition, the spectral signature of adenosine phosphate was detected in macrophage endosomes, which indicates differences in the endosomes of the two cell lines. To date, intrinsic SERS of the cellular millieu provides general information about the types of biomolecules present, but provides limited information about specific biomarkers of interest. The use of extrinsic Raman labels in the examples that follow are one approach to overcome this weakness, showing increased specificity for biomolecules of interest.
Extrinsic
The use of extrinisic SERS in cellular and in vivo sensing is extensive due to the ability of extrinsic Raman labels to overcome background signals from a complex biological matrix and associate with specific biomolecules. Many groups have used extrinsic SERS for mapping the local pH in cells based on the important role pH plays in regulating cellular function.135–137 Kneipp and coworkers measured pH-dependent SERS spectra of p-mercaptobenzoic acid (pMBA), a pH-sensitive Raman tag, on aggregated gold NPs.135 The accessible pH range of SERS and surface-enhanced hyper-Raman scattering (SEHRS) were 5.5–8 and 2–8, respectively, but the high laser power (~ 10 mW) and long collection time (10 sec) required for SERS were not suitable for scanning live cells. Moskovits and coworkers also showed pH dependence of SERS spectra using pMBA-functionalized NP clusters and mapped local pH in live HeLa cells.137 In this work, Ag clusters were linked using bifunctional hexamethylenediamine (HMD) molecules and encapsulated by polyvinylpyrrolidone (PVP) to prevent aggregation during MBA tag infusion. The SERS-active clusters were also coated by dye-labeled streptavidin with BSA to track the distribution of Ag NP clusters in cells and correlate the fluorescence and SERS pH maps. The MBA molecules infused through the polymer coat into junctions between NPs and facilitated measurement of pH values inside live HeLa cells using a relatively low laser power (1.1 mW) and short integration time (250 ms). These pH mapping probes could be used to understand intracellular interactions, including endocytotic pathways. Vo-Dinh and co-workers used a Ag-coated sub-micron sized fiber-optic probe functionalized by pMBA for similar purposes.138 The probe was physically inserted into a live cell using a micromanipulator. The intracellular pH value was determined by comparing SERS intensity of pMBA bands to a calibration curve obtained from standard pH solutions ranging from pH 6.0 to 7.5.
Clearly, it is feasible to perform extrinsic SERS detection in cells using a single extrinsic Raman labels but there is also significant interest in multiplex biomarker detection. In fact, Gambhir and coworkers have demonstrated multiplexed extrinisic SERS.139 They used 10 SERS-NP complexes for multiplex imaging, and each particle consisted of a unique Raman reporter molecule layer adsorbed onto a 60-nm-diameter Au core coated with silica, making the total diameter about 120 nm. Each molecular layer shows distinguishable SERS spectra, and five of them were used for in vivo SERS imaging in a nude mouse. A mixture of four kinds of unique NPs of varying concentrations was injected either intravenously or subcutaneously into the mouse where a linear correlation of SERS signal with the concentrations was measured.
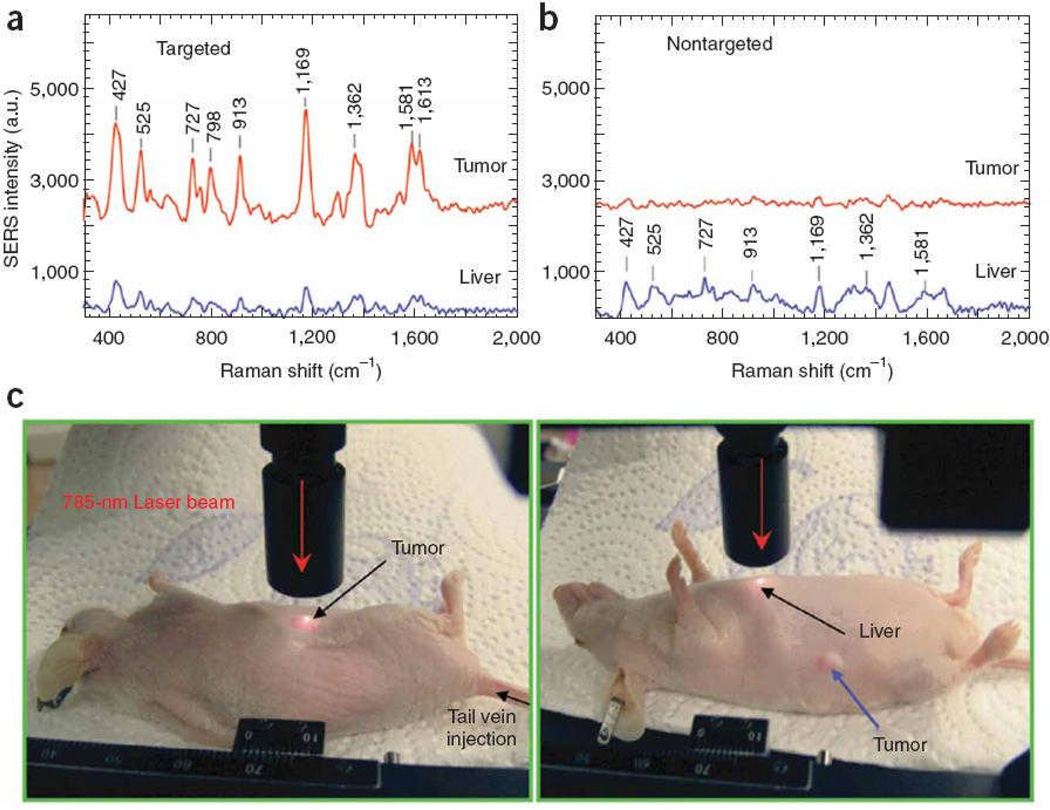
SERS has also been utilized for extrinsic detection of cancer markers in a live cell. Oh and coworkers used Au/Ag core-shell NPs where R6G Raman tags were adsorbed on the gold surface with a BSA layer.140 Subsequently, the NPs were conjugated with IgG antibodies that selectively bind to phopholipase Cγ1 biomarker proteins (PCγ1) on HEK293 (human embryonic kidney) cells. While no normal Raman signal from R6G was measured from a control cell, the cancer cell showed significant Raman signal, based on the 1650 cm−1, R6G peak, that correlated well with quantum dot-labeled fluorescence images. Moving from imaging of cancer markers in live cells to tumor targeting, Nie and coworkers used PEGylated gold NPs as extrinsic SERS labels with tumor-targeting ligand to identify tumors both in vitro and in vivo.141 A core size of 60–80 nm diameter was chosen to position the LSPR peaks within the ‘water window’ (630–785 nm) where the optical absorption of water is minimal. SERS tags were over 200 times brighter than NIR-emitting quantum dots. They could measure SERS spectra at targeted tumor sites up to 2 cm below the skin. After injecting NPs into the tail vein, SERS spectra could be obtained from a targeted tumor site with some non-specific biodistribution into liver and spleen, but not into brain, muscle or other major organs. (Fig. 6) Moreover, Stone and coworkers proposed deep Raman spectroscopy with citrate-reduced Ag conjugated nanoparticles for detecting low concentration of molecules through tissues of up to 25 mm thick. It demonstrated a potential use of SERS nanoparticles for detecting target molecules deeply buried within tissues.142
Fig. 6.
In vivo cancer marker detection using surface enhanced Raman with scFv-antibody conjugated gold nanoparticles that recognize the tumor marker. (a) SERS spectra obtained from the tumor (red) and liver (blue) by using targeted nanoparticles and (b) non-targeted nanoparticles. (c) Photographs showing a laser beam focusing on tumor or liver sites. In vivo SERS spectra were obtained with a 785 nm laser at 20 mW and 2 sec integration. Figure adapted from Ref. 141, reproduced by permission of The Nature Publishing Group.
Because of the high sensitivity, label-free detection and multiplex capability, applications of SERS targeting in live cells, tissues and in vivo detection will continue to expand. At the same time, many hurdles need to be overcome, which include the stability as well as toxicity of SERS tags, and the background noise resulting from the structural similarity of many proteins and molecules.
VI. Perspective
This review article summarized recent advances in harnessing SERS toward rapid in vitro and in vivo detection of a wide range of biological analytes. Theoretical and experimental work in the last decade have vastly expanded our understanding of SERS mechanisms and the ability to control plasmon resonances in nanostructured metals. Researchers have employed noble metal particles, shells, tips, gaps, wires and holes to demonstrate SERS substrates with tuned plasmon resonances and high EFs. Going forward, widespread applications of SERS biosensing will hinge on the ability to mass-produce reliable substrates with well-defined nanoscale patterns, precisely tailored LSPR bands, large and reproducible EFs, and highly specific surface chemistry. One likely scheme for practical use of these substrates will begin with SPR/LSPR based screening, where a plasmon shift indicates a “hit”, followed by SERS interrogation to characterize the chemical nature of the newly associated species. Toward this aim, SERS substrates fabricated with the NSL method have already demonstrated inexpensive wafer-scale processing, EFs of ~108, and an excellent temporal stability.48 Various techniques to fabricate ultrathin metallic nanogaps will further enhance our understanding of deep subwavelength optical confinement toward single-molecule SERS, nonlinear spectroscopy and plasmonics.143 In addition, TERS will open up an exciting new avenue, since it empowers SERS to probe a small number of molecules with a nanoscale imaging resolution. Furthermore, TERS will be particularly useful in the cases where target analytes cannot be brought into contact with the substrates, e.g. probing of cell membrane molecules. Several fabrication challenges should be addressed before TERS can be used as a reliable and widely available technique. Nanoscale roughness in the substrate supporting the sample can also significantly affect the TERS signals, leading to large imaging artifacts.54 In this regard, making ultrasmooth substrates with roughness less than 1 nm may be required.144 Furthermore, recently developed methods of template stripping patterned metals can create ultrasmooth patterned surfaces with tunable optical properties if synergistic interaction of the substrate and TERS probe is required.145 The fabrication of smooth features as well as ultrasharp (<10 nm radius) pyramidal tips will likely advance both SPM and TERS. 146
While substrates like the ones described above will be of great use to intrinsic and extrinsic small molecule detection, immunoassays, and development of DNA arrays for clinical settings, many cellular and in vivo applications of SERS require further development of nanoparticle substrates in suspension. Recently, Kotov and coworkers developed gold lace nanoshells with SERS hotspots on the nanoparticle surface producing EFs of 102 larger than traditional Au spheres of the same diameter.147 Such improvements over widely used Au nanoparticles will allow for more sensitive SERS detection within cells and in vivo. Au nanoparticles are an especially important substrate for intracellular or in vivo SERS based on significantly better biocompatability than their Ag counterparts.
Another technique that allows SERS to probe for molecules without being in contact with the substrate, like TERS, is the shell-isolated nanoparticle-enhanced Raman spectroscopy (SHINERS) pioneered by Tian and coworkers.148 In this method, a spherical gold core is coated with a thin oxide layer (~2 nm of SiO2 or Al2O3) to yield a particle that can be dusted onto an area of interest for SERS detection. The group demonstrated SHINERS use in the monitoring of biological targets and food safety, and their results show promise for surface-based and portable applications of SERS biosensing. Rapid progress in nanofabrication on both wafer and nanoparticle scale, in conjunction with advances in surface chemistry, microfluidics, modeling as well as fundamental understanding of SERS mechanisms, show great promise for the future of SERS biosensing.
VII. Conclusions
SERS research is a truly multi-disciplinary field that has benefited from dynamic interactions among chemists, physicists and engineers. New and exciting developments presented herein show a bright future for the widespread adoption of SERS and will facilitate the transition of SERS from research laboratories to real-world diagnostics, biomedical sensing, and field applications.
Contributor Information
Sang-Hyun Oh, Email: sang@umn.edu.
Christy L. Haynes, Email: chaynes@umn.edu.
Notes and references
- 1.Kneipp K, Kneipp H, Itzkan I, Dasari RR, Feld MS. Journal of Physics-Condensed Matter. 2002;14:R597–R624. [Google Scholar]
- 2.Jackson JB, Halas NJ. Proc. of the Natl. Acad. of Sci. of the U.S.A. 2004;101:17930–17935. doi: 10.1073/pnas.0408319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes CL, McFarland AD, Van Duyne RP. Anal. Chem. 2005;77:338A–346A. [Google Scholar]
- 4.Qian XM, Peng XH, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, Yang L, Young AN, Wang MD, Nie SM. Nat. Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 5.Wustholz KL, Brosseau CL, Casadio F, Van Duyne RP. Phys. Chem. Chem. Phys. 2009;11:7350–7359. doi: 10.1039/b904733f. [DOI] [PubMed] [Google Scholar]
- 6.Larmour IA, Faulds K, Graham D. Chem. Sci. 2010;1:151–160. [Google Scholar]
- 7.Zhang XY, Young MA, Lyandres O, Van Duyne RP. J. Am. Chem. Soc. 2005;127:4484–4489. doi: 10.1021/ja043623b. [DOI] [PubMed] [Google Scholar]
- 8.Mohs AM, Mancini MC, Singhal S, Provenzale JM, Leyland-Jones B, Wang MD, Nie SM. Anal. Chem. 2010;82:9058–9065. doi: 10.1021/ac102058k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young MA, Stuart DA, Lyandres O, Glucksberg MR, Van Duyne RP. Can. J. Chem. 2004;82:1435–1441. [Google Scholar]
- 10.Quang LX, Lim C, Seong GH, Choo J, Do KJ, Yoo SK. Lab Chip. 2008;8:2214–2219. doi: 10.1039/b808835g. [DOI] [PubMed] [Google Scholar]
- 11.Yan F, Vo-Dinh T. Sens. Actuators, B. 2007;121:61–66. [Google Scholar]
- 12.Raman CV, Krishnan KS. Nature. 1928;121:501–504. [Google Scholar]
- 13.McCreery, RL . Raman Spectroscopy for Chemical Analysis. Wiley-Interscience; 2000. [Google Scholar]
- 14.Singh R. Physics in Perspective. 2002;4:399–420. [Google Scholar]
- 15.Shim S, Stuart CM, Mathies RA. ChemPhysChem. 2008;9:697–699. doi: 10.1002/cphc.200700856. [DOI] [PubMed] [Google Scholar]
- 16.Fleischmann M, Hendra PJ, McQuillan AJ. Chem. Phys. Lett. 1974;26:163–166. [Google Scholar]
- 17.Jeanmaire DL, Van Duyne RP. J. Electroanal.Chem. 1977;84:1. [Google Scholar]
- 18.Albrecht MG, Creighton JA. J. Am. Chem. Soc. 1977;99:5215–5217. [Google Scholar]
- 19.Paul R, McQuillan A, Hendra P, Fleischmann M. Journal of Electroanal. Chem. 1975:248–249. [Google Scholar]
- 20.McQuillan A, Hendra P, Fleischmann M. J. Electroanal‥ Chem. 1975:933–944. [Google Scholar]
- 21.Liao PF, Bergman JG, Chemla DS, Wokaun A, Melngailis J, Hawryluk AM, Economou NP. Chem. Phys. Lett. 1981;82:355–359. [Google Scholar]
- 22.Howard RE, Liao PF, Skocpol WJ, Jackel LD, Craighead HG. Science. 1983;221:117–121. doi: 10.1126/science.221.4606.117. [DOI] [PubMed] [Google Scholar]
- 23.Kambhampati P, Foster M, Campion A. J. of Chem. Phys. 1999:551–558. [Google Scholar]
- 24.Schatz G, Young M, Van Duyne R. Surface-Enhanced Raman Scattering: Physics and Applications. 2006:19–45. [Google Scholar]
- 25.Zhao LL, Jensen L, Schatz GC. Nano Lett. 2006;6:1229–1234. doi: 10.1021/nl0607378. [DOI] [PubMed] [Google Scholar]
- 26.Haes AJ, Haynes CL, McFarland AD, Schatz GC, Van Duyne RR, Zou SL. MRS Bulletin. 2005;30:368–375. [Google Scholar]
- 27.Aroca R. Surface-Enhanced Vib. Spectrosc. West Sussex: John Wiley & Sons Ltd; 2006. [Google Scholar]
- 28.Camden JP, Dieringer JA, Zhao J, Van Duyne RP. Acc. of Chem. Res. 2008;41:1653–1661. doi: 10.1021/ar800041s. [DOI] [PubMed] [Google Scholar]
- 29.Kennedy BJ, Spaeth S, Dickey M, Carron KT. J. Phys. Chem. B. 1999;103:3640–3646. [Google Scholar]
- 30.Bantz KC, Haynes CL. Vib. Spectrosc. 2009;50:29–35. [Google Scholar]
- 31.Chen LX, Choo JB. Electrophoresis. 2008;29:1815–1828. doi: 10.1002/elps.200700554. [DOI] [PubMed] [Google Scholar]
- 32.Levin CS, Kundu J, Barhoumi A, Halas NJ. Analyst. 2009;134:1745–1750. doi: 10.1039/b909080k. [DOI] [PubMed] [Google Scholar]
- 33.Grubisha DS, Lipert RJ, Park HY, Driskell J, Porter MD. Anal. Chem. 2003;75:5936–5943. doi: 10.1021/ac034356f. [DOI] [PubMed] [Google Scholar]
- 34.Doering W, Piotti M, Natan M, Freeman R. Adv. Mater. 2007:3100–3108. [Google Scholar]
- 35.Dick LA, McFarland AD, Haynes CL, Van Duyne RP. J. Phys. Chem. B. 2002;106:853–860. [Google Scholar]
- 36.Stropp J, Trachta G, Brehm G, Schneider S. J. Raman Spectrosc. 2003;34:26–32. [Google Scholar]
- 37.Fang Y, Seong NH, Dlott DD. Science. 2008;321:388–392. doi: 10.1126/science.1159499. [DOI] [PubMed] [Google Scholar]
- 38.Liu XF, Sun CH, Linn NC, Jiang B, Jiang P. J. Phys. Chem. C. 2009;113:14804–14811. [Google Scholar]
- 39.Lee SH, Bantz KC, Lindquist NC, Oh S-H, Haynes CL. Langmuir. 2009;25:13685–13693. doi: 10.1021/la9020614. [DOI] [PubMed] [Google Scholar]
- 40.Leverette CL, Jacobs SA, Shanmukh S, Chaney SB, Dluhy RA, Zhao YP. Appl. Spectrosc. 2006;60:906–913. doi: 10.1366/000370206778062084. [DOI] [PubMed] [Google Scholar]
- 41.Sockalingum GD, Beljebbar A, Morjani H, Angiboust JF, Manfait M. Biospectroscopy. 1998;4:S71–S78. doi: 10.1002/(SICI)1520-6343(1998)4:5+<S71::AID-BSPY8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 42.Bantz KC, Haynes CL. Langmuir. 2008;24:5862–5867. doi: 10.1021/la800103b. [DOI] [PubMed] [Google Scholar]
- 43.Erol M, Han Y, Stanley S, Stafford C, Du H, Sukhishvili S. J. Am. Chem. Soc. 2009:7480–7481. doi: 10.1021/ja807458x. [DOI] [PubMed] [Google Scholar]
- 44.Xu SP, Ji XH, Xu WQ, Zhao B, Dou XM, Bai YB, Ozaki Y. Journal of Biomedical Optics. 2005;10:03112. doi: 10.1117/1.1915487. [DOI] [PubMed] [Google Scholar]
- 45.Tripp RA, Dluhy RA, Zhao YP. Nano Today. 2008;3:31–37. [Google Scholar]
- 46.Chan GH, Zhao J, Hicks EM, Schatz GC, Van Duyne RP. Nano Lett. 2007;7:1947–1952. [Google Scholar]
- 47.Chan GH, Zhao J, Schatz GC, Van Duyne RP. J. Phys. Chem. C. 2008;112:13958–13963. [Google Scholar]
- 48.Zhang XY, Zhao J, Whitney AV, Elam JW, Van Duyne RP. J. Am. Chem. Soc. 2006;128:10304–10309. doi: 10.1021/ja0638760. [DOI] [PubMed] [Google Scholar]
- 49.Pettinger B. Surface-Enhanced Raman Scattering: Physics and Applications. 2006;103:217–240. [Google Scholar]
- 50.Lal S, Link S, Halas NJ. Nat. Photonics. 2007;1:641–648. [Google Scholar]
- 51.You Y, Purnawirman NA, Hu H, Kasim J, Yang H, Du C, Yu T, Shen Z. J. Raman Spectrosc. 2010;41:1156–1162. [Google Scholar]
- 52.Bulgarevich DS, Futamata M. Applied Spectroscopy. 2004;58:757–761. doi: 10.1366/0003702041389292. [DOI] [PubMed] [Google Scholar]
- 53.Roth RM, Panoiu NC, Adams MM, Osgood RM, Neacsu CC, Raschke MB. Optics Express. 2006;14:2921–2931. doi: 10.1364/oe.14.002921. [DOI] [PubMed] [Google Scholar]
- 54.Zhang WH, Cui XD, Yeo BS, Schmid T, Hafner C, Zenobi R. Nano Lett. 2007;7:1401–1405. doi: 10.1021/nl070616n. [DOI] [PubMed] [Google Scholar]
- 55.Steidtner J, Pettinger B. Phys. Rev. Lett. 2008;100:236101. doi: 10.1103/PhysRevLett.100.236101. [DOI] [PubMed] [Google Scholar]
- 56.Bishnoi SW, Rozell CJ, Levin CS, Gheith MK, Johnson BR, Johnson DH, Halas NJ. Nano Lett. 2006;6:1687–1692. doi: 10.1021/nl060865w. [DOI] [PubMed] [Google Scholar]
- 57.Huang GG, Han XX, Hossain MK, Ozaki Y. Anal. Chem. 2009;81:5881–5888. doi: 10.1021/ac900392s. [DOI] [PubMed] [Google Scholar]
- 58.Deckert-Gaudig T, Bailo E, Deckert V. Phys. Chem. Chem. Phys. 2009;11:7360–7362. doi: 10.1039/b904735b. [DOI] [PubMed] [Google Scholar]
- 59.Singhal R, Bhattacharyya S, Orynbayeva Z, Vitol E, Friedman G, Gogotsi Y. Nanotechnology. 2010;21:015304. doi: 10.1088/0957-4484/21/1/015304. [DOI] [PubMed] [Google Scholar]
- 60.Shafer-Peltier K, Haynes C, Glucksberg M, Van Duyne R. J. Am. Chem. Soc. 2003;125:588–593. doi: 10.1021/ja028255v. [DOI] [PubMed] [Google Scholar]
- 61.Fox C, Uibel R, Harris J. J. Phys. Chem. B. 2007:11428–11436. doi: 10.1021/jp0735886. [DOI] [PubMed] [Google Scholar]
- 62.Levin C, Kundu J, Janesko B, Scuseria G, Raphael R, Halas N. J. Phys. Chem. B. 2008;112:14168–14175. doi: 10.1021/jp804374e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barhoumi A, Zhang D, Tam F, Halas NJ. J. Am. Chem. Soc. 2008;130:5523–5529. doi: 10.1021/ja800023j. [DOI] [PubMed] [Google Scholar]
- 64.Wei F, Zhang DM, Halas NJ, Hartgerink JD. J. Phys. Chem. B. 2008;112:9158–9164. doi: 10.1021/jp8025732. [DOI] [PubMed] [Google Scholar]
- 65.Vitol EA, Brailoiu E, Orynbayeva Z, Dun NJ, Friedman G, Gogotsi Y. Anal. Chem. 2010;82:6770–6774. doi: 10.1021/ac100563t. [DOI] [PubMed] [Google Scholar]
- 66.Heller A, Feldman B. Chemical Reviews. 2008;108:2482–2505. doi: 10.1021/cr068069y. [DOI] [PubMed] [Google Scholar]
- 67.Yonzon CR, Haynes CL, Zhang XY, Walsh JT, Van Duyne RP. Anal. Chem. 2004;76:78–85. doi: 10.1021/ac035134k. [DOI] [PubMed] [Google Scholar]
- 68.Lyandres O, Shah NC, Yonzon CR, Walsh JT, Glucksberg MR, Van Duyne RP. Anal. Chem. 2005;77:6134–6139. doi: 10.1021/ac051357u. [DOI] [PubMed] [Google Scholar]
- 69.Stuart DA, Yonzon CR, Zhang XY, Lyandres O, Shah NC, Glucksberg MR, Walsh JT, Van Duyne RP. Anal. Chem. 2005;77:4013–4019. doi: 10.1021/ac0501238. [DOI] [PubMed] [Google Scholar]
- 70.Shah NC, Lyandres O, Walsh JT, Glucksberg MR, Van Duyne RP. Anal. Chem. 2007;79:6927–6932. doi: 10.1021/ac0704107. [DOI] [PubMed] [Google Scholar]
- 71.Kundu J, Levin CS, Halas NJ. Nanoscale. 2009;1:114–117. doi: 10.1039/b9nr00063a. [DOI] [PubMed] [Google Scholar]
- 72.Levin CS, Kundu J, Janesko BG, Scuseria GE, Raphael RM, Halas NJ. J. Phys. Chem. B. 2008;112:14168–14175. doi: 10.1021/jp804374e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krysinski P, Zebrowska A, Michota A, Bukowska J, Becucci L, Moncelli M. Langmuir. 2001;17:3852–3857. [Google Scholar]
- 74.Ryu K, Haes AJ, Park HY, Nah S, Kim J, Chung H, Yoon MY, Han SH. J. Raman Spectrosc. 2010;41:121–124. [Google Scholar]
- 75.Huang GG, Hossain MK, Han XX, Ozaki Y. Analyst. 2009;134:2468–2474. doi: 10.1039/b914976g. [DOI] [PubMed] [Google Scholar]
- 76.Domke K, Zhang D, Pettinger B. J. Am. Chem. Soc. 2007;129:6708–6709. doi: 10.1021/ja071107q. [DOI] [PubMed] [Google Scholar]
- 77.Green M, Liu FM, Cohen L, Kollensperger P, Cass T. Faraday Discuss. 2006;132:269–280. doi: 10.1039/b506636k. [DOI] [PubMed] [Google Scholar]
- 78.Bell SEJ, Sirimuthu NMS. J. Am. Chem. Soc. 2006;128:15580–15581. doi: 10.1021/ja066263w. [DOI] [PubMed] [Google Scholar]
- 79.Cho HS, Lee B, Liu GL, Agarwal A, Lee LP. Lab Chip. 2009;9:3360–3363. doi: 10.1039/b912076a. [DOI] [PubMed] [Google Scholar]
- 80.Neumann O, Zhang DM, Tam F, Lal S, Wittung-Stafshede P, Halas NJ. Anal. Chem. 2009;81:10002–10006. doi: 10.1021/ac901849k. [DOI] [PubMed] [Google Scholar]
- 81.Bailo E, Deckert V. Angew. Chem. Int. Ed. 2008;47:1658–1661. doi: 10.1002/anie.200704054. [DOI] [PubMed] [Google Scholar]
- 82.Cialla D, Deckert-Gaudig T, Budich C, Laue M, Moller R, Naumann D, Deckert V, Popp J. J. Raman Spectrosc. 2009;40:240–243. [Google Scholar]
- 83.Pal A, Isola N, Alarie J, Stokes D, Vo-Dinh T. Faraday Discuss. 2006;132:293–301. doi: 10.1039/b506341h. [DOI] [PubMed] [Google Scholar]
- 84.Braun G, Lee SJ, Dante M, Nguyen T-Q, Moskovits M, Reich N. J. Am. Chem. Soc. 2007;129:6378–6379. doi: 10.1021/ja070514z. [DOI] [PubMed] [Google Scholar]
- 85.Banholzer M, Qin L, Millstone J, Osberg K, Mirkin C. Nat. Protoc. 2009;4:838–848. doi: 10.1038/nprot.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bonham A, Braun G, Pavel I, Moskovits M, Reich N. J. Am. Chem. Soc. 2007;129:14572–14573. doi: 10.1021/ja0767837. [DOI] [PubMed] [Google Scholar]
- 87.Graham D, Thompson DG, Smith WE, Faulds K. Nat. Nanotechnol. 2008;3:548–551. doi: 10.1038/nnano.2008.189. [DOI] [PubMed] [Google Scholar]
- 88.Stokes R, Macaskill A, Dougan J, Hargreaves P, Stanford H, Smith W, Faulds K, Graham D. Chem. Comm. 2007;27:2811–2813. doi: 10.1039/b705873j. [DOI] [PubMed] [Google Scholar]
- 89.Faulds K, McKenzie F, Graham D. Analyst. 2007;132:1100–1102. doi: 10.1039/b711744b. [DOI] [PubMed] [Google Scholar]
- 90.Faulds K, Smith WE, Graham D. Anal. Chem. 2004;76:412–417. doi: 10.1021/ac035060c. [DOI] [PubMed] [Google Scholar]
- 91.Jin R, Cao Y, Thaxton C, Mirkin C. Small. 2006;2:375–380. doi: 10.1002/smll.200500322. [DOI] [PubMed] [Google Scholar]
- 92.Faulds K, McKenzie F, Smith WE, Graham D. Angew. Chem. Int. Ed. 2007;46:1829–1831. doi: 10.1002/anie.200604265. [DOI] [PubMed] [Google Scholar]
- 93.Cho H, Baker BR, Wachsmann-Hogiu S, Pagba CV, Laurence TA, Lane SM, Lee LP, Tok JBH. Nano Lett. 2008;8:4386–4390. doi: 10.1021/nl802245w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McKenzie F, Graham D. Chem. Comm. 2009;38:5757–5759. doi: 10.1039/b913140j. [DOI] [PubMed] [Google Scholar]
- 95.Fabris L, Dante M, Braun G, Lee S, Reich N, Moskovits M, Nguyen T, Bazan G. J. Am. Chem. Soc. 2007;129:6086–6087. doi: 10.1021/ja0705184. [DOI] [PubMed] [Google Scholar]
- 96.Cao Y, Jin R, Mirkin C. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 97.Vo-Dinh T, Yan F, Wabuyele M. J. Raman Spectrosc. 2005;36:640–647. [Google Scholar]
- 98.Wabuyele M, Vo-Dinh T. Anal. Chem. 2005;77:7810–7815. doi: 10.1021/ac0514671. [DOI] [PubMed] [Google Scholar]
- 99.Wang H, Vo-Dinh T. Nanotechnology. 2009;20:065101. doi: 10.1088/0957-4484/20/6/065101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barhoumi A, Halas NJ. J. Am. Chem. Soc. 2010;132:12792–12793. doi: 10.1021/ja105678z. [DOI] [PubMed] [Google Scholar]
- 101.Corrigan DK, Gale N, Brown T, Bartlett PN. Angew. Chem. Int. Ed. 2010;49:5917–5920. doi: 10.1002/anie.201001389. [DOI] [PubMed] [Google Scholar]
- 102.Mahajan S, Richardson J, Brown T, Bartlett PN. J. Am. Chem. Soc. 2008;130:15589–15601. doi: 10.1021/ja805517q. [DOI] [PubMed] [Google Scholar]
- 103.Lowe AJ, Huh YS, Strickland AD, Erickson D, Batt CA. Anal. Chem. 2010;82:5810–5814. doi: 10.1021/ac100921b. [DOI] [PubMed] [Google Scholar]
- 104.Han X, Zhao B, Ozaki Y. Anal. Bioanal. Chem. 2009:1719–1727. doi: 10.1007/s00216-009-2702-3. [DOI] [PubMed] [Google Scholar]
- 105.Socrates G. Infrared and Raman Characteristic Group Frequencies: Tables and Charts. New York: Wiley; 2001. [Google Scholar]
- 106.Podstawka E, Ozaki Y, Proniewicz LM. Appl. Spectrosc. 2004;58:570–580. doi: 10.1366/000370204774103408. [DOI] [PubMed] [Google Scholar]
- 107.Podstawka E, Ozaki Y. Biopolymers. 2008;89:807–819. doi: 10.1002/bip.21017. [DOI] [PubMed] [Google Scholar]
- 108.Pavel I, McCarney E, Elkhaled A, Morrill A, Plaxco K, Moskovits M. J. Phys. Chem. C. 2008;112:4880–4883. doi: 10.1021/jp710261y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Han X, Jia H, Wang Y, Lu Z, Wang C, Xu W, Zhao B, Ozaki Y. Anal. Chem. 2008;80:2799–2804. doi: 10.1021/ac702390u. [DOI] [PubMed] [Google Scholar]
- 110.Becker M, Budich C, Deckert V, Janasek D. Analyst. 2009;134:38–40. doi: 10.1039/b816717f. [DOI] [PubMed] [Google Scholar]
- 111.Han X, Huang G, Zhao B, Ozaki Y. Anal. Chem. 2009;81:3329–3333. doi: 10.1021/ac900395x. [DOI] [PubMed] [Google Scholar]
- 112.Bjerneld E, Foldes-Papp Z, Kall M, Rigler R. J. Phys. Chem. B. 2002;106:1213–1218. [Google Scholar]
- 113.Driskell J, Uhlenkamp J, Lipert R, Porter M. Anal. Chem. 2007;79:4141–4148. doi: 10.1021/ac0701031. [DOI] [PubMed] [Google Scholar]
- 114.Narayanan R, Lipert RJ, Porter MD. Anal. Chem. 2008;80:2265–2271. doi: 10.1021/ac7026436. [DOI] [PubMed] [Google Scholar]
- 115.Driskell J, Kwarta K, Lipert R, Porter M, Neill J, Ridpath J. Anal. Chem. 2005;77:6147–6154. doi: 10.1021/ac0504159. [DOI] [PubMed] [Google Scholar]
- 116.Yakes B, Lipert R, Bannantine J, Porter M. Clinical and Vaccine Immunology. 2008;15:227–234. doi: 10.1128/CVI.00334-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu S, Ji X, Xu W, Li X, Wang L, Bai Y, Zhao B, Ozaki Y. Analyst. 2004;129:63–68. doi: 10.1039/b313094k. [DOI] [PubMed] [Google Scholar]
- 118.Wang G, Park H, Lipert R. Anal. Chem. 2009;81:9643–9650. doi: 10.1021/ac901711f. [DOI] [PubMed] [Google Scholar]
- 119.Cui Y, Ren B, Yao J, Gu R, Tian Z. J. Raman Spectrosc. 2007;38:896–902. [Google Scholar]
- 120.Han XX, Cai LJ, Guo J, Wang CX, Ruan WD, Han WY, Xu WQ, Zhao B, Ozaki Y. Anal. Chem. 2008;80:3020–3024. doi: 10.1021/ac702497t. [DOI] [PubMed] [Google Scholar]
- 121.Hwang H, Chon H, Choo J, Park J. Anal. Chem. 2010;82:7603–7610. doi: 10.1021/ac101325t. [DOI] [PubMed] [Google Scholar]
- 122.Rohr TE, Cotton T, Fan N, Tarcha PJ. Anal. Biochem. 1989;182:388–398. doi: 10.1016/0003-2697(89)90613-1. [DOI] [PubMed] [Google Scholar]
- 123.Porter M, Lipert R, Siperko L, Wang G, Narayanana R. Chem. Soc. Rev. 2008;37:1001–1011. doi: 10.1039/b708461g. [DOI] [PubMed] [Google Scholar]
- 124.Donaldson K, Kramer M, Lim D. Biosens. & Bioelectron. 2004;20:322–327. doi: 10.1016/j.bios.2004.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ilic B, Yang Y, Craighead H. Appl. Phys. Lett. 2004;85:2604–2606. [Google Scholar]
- 126.Principles of Bacterial Detection: Biosensors, Recognition Receptors, and Microsystems. Springer Science; 2008. [Google Scholar]
- 127.Ruan C, Wang H, Li Y. Transactions of the ASAE. 2002;45:249–255. [Google Scholar]
- 128.Che Y, Li Y, Slavik M, Paul D. J. of Food Prot. 2000;63:1043–1048. doi: 10.4315/0362-028x-63.8.1043. [DOI] [PubMed] [Google Scholar]
- 129.Larmour IA, Faulds K, Graham D. Chem. Sci. 1:151–160. [Google Scholar]
- 130.Cao YC, Jin RC, Nam JM, Thaxton CS, Mirkin CA. J. Am. Chem. Soc. 2003;125:14676–14677. doi: 10.1021/ja0366235. [DOI] [PubMed] [Google Scholar]
- 131.Han X, Xie Y, Zhao B, Ozaki Y. Anal. Chem. 2010;82:4325–4328. doi: 10.1021/ac100596u. [DOI] [PubMed] [Google Scholar]
- 132.Chen Z, Tabakman SM, Goodwin AP, Kattah MG, Daranciang D, Wang XR, Zhang GY, Li XL, Liu Z, Utz PJ, Jiang KL, Fan SS, Dai HJ. Nat. Biotechnol. 2008;26:1285–1292. doi: 10.1038/nbt.1501. [DOI] [PubMed] [Google Scholar]
- 133.Stuart DA, Yuen JM, Lyandres NSO, Yonzon CR, Glucksberg MR, Walsh JT, Van Duyne RP. Anal. Chem. 2006;78:7211–7215. doi: 10.1021/ac061238u. [DOI] [PubMed] [Google Scholar]
- 134.Kneipp J, Kneipp H, McLaughlin M, Brown D, Kneipp K. Nano Lett. 2006;6:2225–2231. doi: 10.1021/nl061517x. [DOI] [PubMed] [Google Scholar]
- 135.Kneipp J, Kneipp H, Wittig B, Kneipp K. Nano Lett. 2007;7:2819–2823. doi: 10.1021/nl071418z. [DOI] [PubMed] [Google Scholar]
- 136.Kneipp J, Kneipp H, Wittig B, Kneipp K. J. Phys. Chem. C. 2010;114:7421–7426. [Google Scholar]
- 137.Pallaoro A, Braun GB, Reich NO, Moskovits M. Small. 2010;6:618–622. doi: 10.1002/smll.200901893. [DOI] [PubMed] [Google Scholar]
- 138.Scaffidi JP, Gregas MK, Seewaldt V, Vo-Dinh T. Anal. Bioanal. Chem. 2009;393:1135–1141. doi: 10.1007/s00216-008-2521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zavaleta CL, Smith BR, Walton I, Doering W, Davis G, Shojaei B, Natan MJ, Gambhir SS. Proc. Natl Acad. Sci. U. S. A. 2009;106:13511–13516. doi: 10.1073/pnas.0813327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee S, Kim S, Choo J, Shin SY, Lee YH, Choi HY, Ha SH, Kang KH, Oh CH. Anal. Chem. 2007;79:916–922. doi: 10.1021/ac061246a. [DOI] [PubMed] [Google Scholar]
- 141.Qian XM, Peng XH, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, Yang L, Young AN, Wang MD, Nie SM. Nat. Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 142.Stone N, Faulds K, Graham D, Matousek P. Anal. Chem. 2010;82:3969–3973. doi: 10.1021/ac100039c. [DOI] [PubMed] [Google Scholar]
- 143.Im H, Bantz K, Lindquist N, Haynes C, Oh S. Nano Lett. 2010;10:2231–2236. doi: 10.1021/nl1012085. [DOI] [PubMed] [Google Scholar]
- 144.Hegner M, Wagner P, Semenza G. Surf. Sci. 1993;291:39–46. [Google Scholar]
- 145.Nagpal P, Lindquist NC, Oh SH, Norris DJ. Science. 2009;325:594–597. doi: 10.1126/science.1174655. [DOI] [PubMed] [Google Scholar]
- 146.Lindquist N, Nagpal P, Lesuffleur A, Norris D, Oh S. Nano Lett. 2010;10:1369–1373. doi: 10.1021/nl904294u. [DOI] [PubMed] [Google Scholar]
- 147.Yang M, Alvarez-Puebla R, Kim H-S, Aldeanueva-Potel P, Liz-Marzan LM, Kotov NA. Nano Lett. 2010;10:4013–4019. doi: 10.1021/nl101946c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li JF, Huang YF, Ding Y, Yang ZL, Li SB, Zhou XS, Fan FR, Zhang W, Zhou ZY, Wu DY, Ren B, Wang ZL, Tian ZQ. Nature. 2010;464:392–395. doi: 10.1038/nature08907. [DOI] [PubMed] [Google Scholar]
- 149.Haynes CL, Van Duyne RP. J. Phys. Chem. B. 2001;105:5599–5611. [Google Scholar]
- 150.Wang H, Levin CS, Halas NJ. J. Am. Chem. Soc. 2005;127:14992–14993. doi: 10.1021/ja055633y. [DOI] [PubMed] [Google Scholar]
- 151.Hulteen JC, Vanduyne RP. Journal of Vacuum Science & Technology a-Vacuum Surfaces and Films. 1995;13:1553–1558. [Google Scholar]
- 152.Lu Y, Liu GL, Kim J, Mejia YX, Lee LP. Nano Lett. 2005;5:119–124. doi: 10.1021/nl048232+. [DOI] [PubMed] [Google Scholar]
- 153.Cao YWC, Jin RC, Mirkin CA. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 154.Lim DK, Jeon KS, Kim HM, Nam JM, Suh YD. Nat. Mater. 2010;9:60–67. doi: 10.1038/nmat2596. [DOI] [PubMed] [Google Scholar]
- 155.Gunnarsson L, Bjerneld EJ, Xu H, Petronis S, Kasemo B, Kall M. Appl. Phys. Lett. 2001;78:802–804. [Google Scholar]
- 156.Im H, Bantz KC, Lindquist NC, Haynes CL, Oh SH. Nano Lett. 2010;10:2231–2236. doi: 10.1021/nl1012085. [DOI] [PubMed] [Google Scholar]
- 157.Banholzer MJ, Qin LD, Millstone JE, Osberg KD, Mirkin CA. Nat. Protoc. 2009;4:838–848. doi: 10.1038/nprot.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chen XD, Yeganeh S, Qin LD, Li SZ, Xue C, Braunschweig AB, Schatz GC, Ratner MA, Mirkin CA. Nano Lett. 2009;9:3974–3979. doi: 10.1021/nl9018726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gopinath A, Boriskina SV, Premasiri WR, Ziegler L, Reinhard BM, Dal Negro L. Nano Lett. 2009;9:3922–3929. doi: 10.1021/nl902134r. [DOI] [PubMed] [Google Scholar]
- 160.Pettinger B, Picardi G, Schuster R, Ertl G. Single Molecules. 2002;3:285–294. [Google Scholar]
- 161.Guieu V, Talaga D, Servant L, Sojic N, Lagugne-Labarthet F. J. Phys. Chem. C. 2009;113:874–881. [Google Scholar]
- 162.Stockle RM, Suh YD, Deckert V, Zenobi R. Chem. Phys. Lett. 2000;318:131–136. [Google Scholar]
- 163.Vo-Dinh T, Kasili P, Wabuyele M. Nanomed. Nanotechnol. Biol. Med. 2006;2:22–30. doi: 10.1016/j.nano.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 164.Lindquist NC, Nagpal P, Lesuffleur A, Norris DJ, Oh SH. Nano Lett. 2010;10:1369–1373. doi: 10.1021/nl904294u. [DOI] [PubMed] [Google Scholar]
- 165.Haynes CL, Van Duyne RP. J. Phys. Chem. B. 2003;107:7426–7433. [Google Scholar]
- 166.Chan GH, Zhao J, Hicks EM, Schatz GC, Van Duyne RP. Nano Lett. 2007;7:1947–1952. [Google Scholar]
- 167.Tao A, Kim F, Hess C, Goldberger J, He RR, Sun YG, Xia YN, Yang PD. Nano Lett. 2003;3:1229–1233. [Google Scholar]
- 168.Shanmukh S, Jones L, Driskell J, Zhao YP, Dluhy R, Tripp RA. Nano Lett. 2006;6:2630–2636. doi: 10.1021/nl061666f. [DOI] [PubMed] [Google Scholar]
- 169.Yao JL, Tang J, Wu DY, Sun DM, Xue KH, Ren B, Mao BW, Tian ZQ. Surface Science. 2002;514:108–116. [Google Scholar]
- 170.Yu QM, Guan P, Qin D, Golden G, Wallace PM. Nano Lett. 2008;8:1923–1928. doi: 10.1021/nl0806163. [DOI] [PubMed] [Google Scholar]
- 171.Reilly TH, Chang SH, Corbman JD, Schatz GC, Rowlen KL. J. Phys. Chem. C. 2007;111:1689–1694. [Google Scholar]
- 172.Masson JF, Murray-Methot MP, Live LS. Analyst. 2010;135:1483–1489. doi: 10.1039/c0an00053a. [DOI] [PubMed] [Google Scholar]
- 173.Brolo AG, Arctander E, Gordon R, Leathem B, Kavanagh KL. Nano Lett. 2004;4:2015–2018. [Google Scholar]
- 174.Anema JR, Brolo AG, Marthandam P, Gordon R. J. Phys. Chem. C. 2008;112:17051–17055. [Google Scholar]
- 175.Casadio M, Leona JR, Van Duyne RP. Acc. Chem. Res. 2010;43:782–791. doi: 10.1021/ar100019q. [DOI] [PubMed] [Google Scholar]