Abstract
Background. Using multinational collections of methicillin-susceptible Staphylococcus aureus (MSSA) isolates from infective endocarditis (IE) and soft tissue infections (STIs), we sought to (1) validate the finding that S. aureus in clonal complex (CC) 30 is associated with hematogenous complications and (2) test the hypothesis that specific genetic characteristics in S. aureus are associated with infection severity.
Methods. IE and STI isolates from 2 cohorts were frequency matched by geographic origin. Isolates underwent spa typing to infer CC and multiplex polymerase chain reaction for presence of virulence genes.
Results. 114 isolate pairs were genotyped. IE isolates were more likely to be CC30 (19.5% vs 6.2%; P = .005) and to contain 3 adhesins (clfB, cna, map/eap; P < .0001 for all) and 5 enterotoxins (tst, sea, sed, see, and sei; P ≤ .005 for all). CC30 isolates were more likely to contain cna, tst, sea, see, seg, and chp (P < .05 for all).
Conclusions. MSSA IE isolates were significantly more likely to be CC30 and to possess a distinct repertoire of virulence genes than MSSA STI isolates from the same region. The genetic basis of this association requires further study.
Staphylococcus aureus is the most common cause of both infective endocarditis (IE) [1] and soft tissue infection (STI) [2] in the industrialized world. The frequency of S. aureus as a human pathogen is thought to be due in part to its diverse armamentarium of virulence-associated genes. Although substantial evidence suggests that clinical manifestations of S. aureus are influenced by the genetic characteristics of the infecting strain [3–7], the association between S. aureus genes and severity of illness is incompletely understood.
Previously, we demonstrated a significant association between specific S. aureus isolates genotypes and infection severity [4]. We used multilocus sequence typing (MLST) to show that clonal complex (CC) 5 and CC30 were significantly associated with the presence of IE and bone and joint infection among 371 clinically well-characterized S. aureus isolates from a single geographical region. However, these findings must be confirmed prior to being considered broadly generalizable.
The current investigation seeks to externally validate these previously observed associations between bacterial genotype and infection severity in S. aureus. To do this, we used bacterial isolates from 2 large multinational cohorts of patients with distinct forms of staphylococcal disease: IE and STI.
PATIENTS AND METHODS
Patients and Settings
IE Isolates.
IE isolates were obtained from the Microbiological Repository of the International Collaboration on Endocarditis–Prospective Cohort Study (ICE-PCS) [8]. The Microbiological Repository [1, 9, 10] contains >1300 bloodstream isolates from prospectively identified patients with definite IE from 16 countries, obtained between June 2000 and September 2006. Bloodstream isolates from all patients with definite methicillin-susceptible S. aureus (MSSA) IE were eligible for inclusion in this study.
STI Isolates.
STI isolates were obtained from the ATLAS (Assessment of TeLAvancin in complicated Skin and skin structure infections) clinical trial, which included 2 methodologically identical, double-blind, randomized, active-controlled, parallel-group, multinational, phase 3 studies investigating the efficacy and safety of telavancin versus vancomycin for the treatment of gram-positive STI. The study designs of the ATLAS trials have been published in detail elsewhere [11, 12]. In brief, nonpregnant adult (≥18 years) patients were enrolled from 129 participating centers in 21 countries from September 2004 through June 2006. Included patients were diagnosed with complicated skin and skin structure infections (caused by a suspected or confirmed gram-positive organism) that warranted ≥7 days of parenteral antibiotic therapy. Bacterial isolates were obtained from all patients at baseline by needle aspiration or surgical procedure. In this study, the analysis population was defined as microbiologically evaluable patients with complicated skin and skin structure infections due to monomicrobial MSSA.
The study was approved by the Duke University Medical Center institutional review board.
Definitions
IE Isolates.
Definite IE was defined according to modified Duke criteria [13]. Healthcare-associated IE was defined as development of first signs/symptoms consistent with IE >48 hours after hospitalization. Also included are patients with IE diagnosed within 48 hours of admission with extensive healthcare contact as reflected by any of the following criteria: (1) received intravenous therapy, wound care, or specialized nursing care at home within the previous 30 days; (2) attended a hospital or hemodialysis clinic or received intravenous chemotherapy within the previous 30 days; (3) was hospitalized in an acute care hospital for 2 or more days within the previous 90 days; or (4) resided in a nursing home or long-term care facility [14]. Community-acquired IE was defined as IE diagnosed within 48 hours of admission in a patient not fulfilling the criteria for healthcare-associated infection. Infections were considered to be injection drug use associated if the patient actively used these substances at the time of IE diagnosis and was admitted from the community without an alternate presumed source. Vancomycin therapy was defined as being present if it was identified by the investigator as the predominant antibiotic used in the treatment of the infection.Persistent bacteremia was defined as >3 days of bacteremia despite receipt of an antibiotic to which the isolate was susceptible in vitro [9]. An intracardiac abscess was defined as a thickened area or mass with a heterogeneous echogenic or echolucent appearance by echocardiography, or the presence of pus by direct visualization at the time of surgery [15]. The remaining clinical, echocardiographic, and outcome variables were defined as reported elsewhere [1].
STI Isolates.
STI was defined, as previously described [11], by the presence of 1 of the following conditions: cellulitis, a major abscess requiring surgical drainage, an infected wound or ulcer, or an infected burn. Purulent drainage and/or collection or ≥3 of the following signs or symptoms also were required for inclusion: erythema, heat and/or localized warmth, fluctuance, swelling and/or induration, pain and/or tenderness to palpation, fever (temperature >38°C), white blood cell count >10000 cells/mm3, or >15% bands. Renal impairment was defined as estimated baseline creatinine clearance of ≤50 mL/min. For this study, healthcare association for the STI cohort was defined as a patient with any of the following criteria: (1) hospitalization within the previous 6 months; (2) chronic illness; (3) nursing home residence; and (4) recent surgical procedure.
Laboratory Methods and Susceptibility Definition
Multiplex Polymerase Chain Reaction.
Genomic DNA was prepared as previously described [3]. Bacterial determinants including adhesins, toxins, agr group I–IV, and other genes were screened by multiplex polymerase chain reaction (PCR) as described before [3, 7]. All negative calls on the multiplex PCR were confirmed by uniplex PCR.
Spa Typing.
Spa typing and MLST were performed as previously described [4, 16]. PCR oligonucleotide primers for the 7 MLST targets and spa were described previously [4]. Samples were sequenced at the Duke University sequencing laboratory. For spa typing, eGenomics software (http://tools.egenomics.com/) was used to scan the primary sequence to help identify the orders and names of each repeat. The spa type number is representative of the repeat organization. Clonal complexes for the isolates were identified via repeats pattern recognition from existing spa type and CC database provided by Drs Barry Kreiswirth and José Mediavilla that were previously confirmed via MLST [16]. Isolates whose spa type did not map to a known CC underwent MLST typing. For MLST, the sequence chromatograms for unique alleles were deposited in the MLST database (http://www.mlst.net). Alleles at the 7 loci (arcC, aroE, glpF, gmk, pta, tpi, and yqiL) were used to identify a unique sequence type (ST). MLST allele names and STs were derived from http://www.mlst.net. CCs were assigned to groups of isolates sharing 6 of 7 alleles by using the eBURST algorithm (http://eburst.mlst.net) [17].
Sample Selection
In order to minimize confounding introduced by the highly clonal nature of methicillin-resistant S. aureus causing STI (eg, USA300 clone) [18], we limited our comparison to MSSA isolates. To address the confounding effect introduced by the intrinsic genetic differences of geographically distinct bacteria, we frequency matched IE and STI MSSA isolates within strata of their geographic origin (North America, South America, Europe/Middle East/Africa, or Australia/New Zealand). Equal numbers of IE and STI isolates were randomly selected within each stratum and constitute the final study population for the investigation.
Statistics
Simple descriptive statistics were used to describe patient characteristics and the genetic profile of the bacterial isolates. Statistics were presented as frequency counts and percentages for categorical factors and as medians (interquartile range) for continuous variables. The statistical significance of association between variables was calculated using the Kruskal-Wallis test for continuous measures and with Fisher exact test for cross-classifications of categorical data. Significance levels of multiplex PCR results were corrected for multiple tests using the false discovery rate (FDR) procedure [19]. FDR thresholds of 10% were reported to balance the type I and type II error probabilities. Genetic diversity was estimated using Simpson’s index of diversity [20]. Since the sample counts within matching strata were substantial, unconditional logistic regression models were used to assess the association between infection type (IE vs STI) and S. aureus characteristics [21]. An α = .05 significance level was required for predictors to remain in the model. For all tests, a P value <.05 was considered statistically significant. All statistical analyses were performed using SAS version 9.2 (SAS Institute).
RESULTS
Clinical Characteristics of IE and STI Patients
Isolates from prospectively enrolled patients from 7 countries with definite MSSA IE (n = 114) and STI (n = 114) were included in this study. Baseline characteristics and outcomes in IE patients were similar across geographic regions (Table 1). As expected, STI patients differed from IE patients in several demographic characteristics, including gender, presence of fever, renal impairment, and healthcare association (Table 2).
Table 1.
Clinical Characteristics and Outcomes of Patients With Definite Methicillin-Susceptible Staphylococcus aureus Endocarditis by Geographic Region
| Characteristics, no. (%) | North America n = 26 (23) | Europe & Middle East n = 76 (68) | Australia & New Zealand n = 10 (9) | P value |
| Male sex | 21 (80.8) | 62 (81.6) | 5 (50) | .091 |
| Age, y, median (interquartile range) | 57 (49–67) | 61 (40–71) | 55 (28–77) | .975 |
| Type of IE | .153 | |||
| Native | 15 (57.7) | 52 (75.4) | 10 (100) | … |
| Prosthetic | 7 (26.9) | 10 (14.5) | 0 (0) | … |
| Other | 4 (15.4) | 7 (10.1) | 0 (0) | … |
| Diabetes mellitus | 8 (30.8) | 8 (10.5) | 0 (0) | .022 |
| Renal impairment | 4 (15.4) | 4 (5.3) | 0 (0) | .223 |
| Implantable cardioverter defibrillator present | 2 (7.7) | 1 (1.3) | 0 (0) | .241 |
| Congenital heart disease | 3 (11.5) | 2 (2.7) | 1 (10) | .119 |
| Place of acquisition | .028 | |||
| Community | 15 (62.5) | 52 (68.4) | 9 (90) | … |
| Injection drug use associated | 3 (11.5) | 21 (27.6) | 3 (30) | … |
| Healthcare associateda | 9 (37.5) | 24 (31.6) | 1 (10) | … |
| Unknown | 2 (7.7) | 0 (0.0) | 0 (0.0) | … |
| New/worsening murmur | 7 (26.9) | 33 (44) | 3 (30) | .570 |
| Echocardiographic findings | ||||
| Aortic or mitral | 16 (61.5) | 46 (61.3) | 6 (60) | 1.000 |
| Tricuspid or pulmonic | 3 (11.5) | 21 (28) | 4 (40) | .108 |
| New regurgitation | 13 (50) | 41 (53.9) | 7 (70) | .573 |
| Intracardiac vegetations | 22 (84.6) | 72 (94.7) | 9 (90) | .212 |
| Vancomycin therapy | 6 (24) | 8 (10.5) | 1 (10) | .183 |
| Complications | ||||
| Stroke | 4 (15.4) | 13 (17.1) | 2 (20) | 1.000 |
| Congestive heart failure | 11 (42.3) | 21 (27.6) | 3 (30) | .398 |
| Persistent bacteremia | 6 (23.1) | 5 (6.6) | 1 (10) | .051 |
| Intracardiac abscess | 4 (15.4) | 8 (10.8) | 1 (10) | .804 |
| Surgery | 9 (34.6) | 29 (38.2) | 4 (40) | .953 |
| Embolization | 5 (19.2) | 25 (32.9) | 4 (40) | .335 |
| In-hospital death | 4 (15.4) | 22 (28.9) | 3 (30) | .376 |
NOTE. Two of 114 patients were excluded from geographical analysis because they originated from South America. None of the P values were statistically significant after multiple comparisons adjustment with a false discovery rate of 10%. IE, infective endocarditis.
Includes patients with both nosocomial infection and nonnosocomial healthcare-associated infections.
Table 2.
Comparison of Baseline Clinical Characteristics of Patients With Endocarditis or Soft Tissue Infections Due to Methicillin-Susceptible Staphylococcus aureus
| Characteristics, no. (%) | Infective endocarditis (n = 114) | Soft tissue infection (n = 114) | P value |
| Age, y, median (interquartile range) | 59 (42–71) | 56 (41–69) | .58 |
| Male sex | 90 (78.9) | 63 (55.3) | <.001 |
| Presence of fever | 102 (96.2) | 17 (15.0) | <.001 |
| Predisposing conditions | |||
| Diabetes | 17 (14.9) | 29 (25.4) | .069 |
| Renal impairment | 8 (7) | 21 (18.4) | .016 |
| Healthcare associationa | 34 (30.4) | 95 (83.3) | <.001 |
NOTE. a Healthcare contact for infective endocarditis isolates included patients with both nosocomial infection and nonnosocomial healthcare-associated infections. For soft tissue infection isolates, healthcare association was defined as a patient with any of the following criteria: (1) hospitalization within previous 6 months; (2) chronic illness; (3) nursing home residence; and (4) recent surgical procedure.
Geographical Variation of IE and STI Isolates
The distribution of virulence genes in both IE and STI isolates differed significantly by geographic region (Table 3 and 4). Results were unchanged when isolates from Australia/New Zealand were omitted (results not shown).
Table 3.
Genotypic Characteristics of Methicillin-Susceptible Staphylococcus aureus Bloodstream Isolates From Patients With Endocarditis According to Geographic Region
| Genes, no. (%) | North America n = 26 (23) | Europe & Middle East n = 76 (68) | Australia & New Zealand n = 10 (9) | P value |
| Adhesins | ||||
| fnbA | 26 (100) | 76 (100.0) | 8 (80) | .007a |
| fnbB | 8 (30.8) | 5 (6.8) | 2 (20) | .006a |
| clfA | 26 (100) | 75 (98.7) | 10 (100) | 1.000 |
| clfB | 26 (100) | 76 (100.0) | 10 (100) | … |
| cna | 18 (69.2) | 67 (88.2) | 7 (70) | .041 |
| spa | 24 (92.3) | 75 (98.7) | 10 (100) | .241 |
| sdrC | 14 (53.8) | 33 (43.4) | 3 (30) | .385 |
| sdrD | 17 (65.4) | 45 (59.2) | 8 (80) | .450 |
| sdrE | 15 (57.7) | 37 (48.7) | 7 (70) | .389 |
| bbp | 24 (92.3) | 67 (88.2) | 8 (80) | .516 |
| ebpS | 26 (100.0) | 76 (100.0) | 10 (100.0) | … |
| map/eap | 15 (57.7) | 40 (52.6) | 8 (80) | .290 |
| Toxins | ||||
| eta | 8 (30.8) | 17 (22.4) | 2 (20) | .683 |
| etb | 0 (0.0) | 5 (6.6) | 0 (0) | .581 |
| tst | 23 (88.5) | 73 (96.1) | 9 (90) | .217 |
| sea | 16 (61.5) | 50 (65.8) | 6 (60) | .860 |
| seb | 2 (7.7) | 0 (0.0) | 2 (20) | .004a |
| sec | 9 (34.6) | 16 (21.1) | 4 (40) | .224 |
| sed | 9 (34.6) | 12 (15.8) | 3 (30) | .098 |
| see | 0 (0.0) | 28 (36.8) | 3 (30) | <.001a |
| seg | 16 (61.5) | 57 (75) | 5 (50) | .163 |
| seh | 4 (15.4) | 8 (10.5) | 1 (10) | .800 |
| sei | 24 (92.3) | 66 (86.8) | 10 (100) | .626 |
| sej | 2 (7.7) | 1 (1.3) | 1 (10) | .118 |
| pvl | 0 (0.0) | 20 (26.3) | 1 (10) | .003a |
| hlg | 26 (100) | 76 (100) | 10 (100) | … |
| Other putative virulence genes | ||||
| efb | 26 (100) | 75 (98.7) | 10 (100) | 1.000 |
| icaA | 26 (100) | 68 (89.5) | 10 (100) | .182 |
| chp | 24 (92.3) | 61 (80.3) | 7 (70) | .213 |
| v8 | 17 (65.4) | 40 (52.6) | 8 (80) | .211 |
| AGR group | ||||
| agr I | 8 (30.8) | 24 (31.6) | 4 (40) | .894 |
| agr II | 9 (34.6) | 26 (34.2) | 4 (40) | … |
| Agr III | 9 (34.6) | 21 (27.6) | 2 (20) | … |
| Agr IV | 0 (0.0) | 5 (6.6) | 0 (0) | … |
NOTE. Two of 114 patients were excluded from geographical analysis because they originated from South America.
Statistically significant after adjustment for multiple comparisons using a false discovery rate of 10%.
Table 4.
Genotypic Characteristics of Methicillin-Susceptible Staphylococcus aureus Isolates From Patients With Soft Tissue Infection Isolates by Geographic Region
| Genes, no. (%) | North America n = 26 (23) | Europe & Middle East n = 76 (68) | Australia & New Zealand n = 10 (9) | P value |
| Adhesins | ||||
| fnbA | 26 (100) | 76 (100) | 10 (100) | … |
| fnbB | 10 (38.5) | 5 (6.6) | 1 (10) | <.001a |
| clfA | 26 (100) | 76 (100) | 10 (100) | … |
| clfB | 18 (69.2) | 14 (18.4) | 0 (0) | <.001a |
| cna | 7 (26.9) | 46 (60.5) | 7 (70) | .007a |
| spa | 26 (100) | 76 (100) | 10 (100) | … |
| sdrC | 13 (50) | 19 (25) | 3 (30) | .060 |
| sdrD | 18 (69.2) | 44 (57.9) | 8 (80) | .317 |
| sdrE | 23 (88.5) | 34 (44.7) | 6 (60) | <.001a |
| bbp | 25 (96.2) | 70 (92.1) | 7 (70) | .045 |
| ebpS | 26 (100) | 76 (100) | 10 (100) | … |
| map/eap | 9 (34.6) | 11 (14.5) | 0 (0) | .024 |
| Toxins | ||||
| eta | 5 (19.2) | 12 (15.8) | 2 (20) | .782 |
| etb | 0 (0) | 0 (0) | 0 (0) | … |
| tst | 8 (30.8) | 25 (32.9) | 4 (40) | .899 |
| sea | 8 (30.8) | 21 (27.6) | 7 (70) | .035 |
| seb | 2 (7.7) | 4 (5.3) | 3 (30) | .046 |
| sec | 3 (11.5) | 20 (26.3) | 5 (50) | .056 |
| sed | 1 (3.8) | 5 (6.6) | 0 (0) | 1.000 |
| see | 2 (7.7) | 2 (2.6) | 0 (0) | .499 |
| seg | 9 (34.6) | 52 (68.4) | 4 (40) | .005a |
| seh | 0 (0) | 5 (6.6) | 1 (10) | .299 |
| sei | 8 (30.8) | 48 (63.2) | 4 (40) | .01a |
| sej | 2 (7.7) | 4 (5.3) | 0 (0) | .799 |
| pvl | 14 (53.8) | 17 (22.4) | 0 (0) | .001a |
| hlg | 26 (100) | 76 (100) | 10 (100) | … |
| Other putative virulence genes | ||||
| efb | 25 (96.2) | 74 (97.4) | 10 (100) | 1.000 |
| icaA | 26 (100) | 73 (96.1) | 10 (100) | .675 |
| chp | 19 (73.1) | 53 (69.7) | 5 (50) | .420 |
| v8 | 23 (88.5) | 42 (55.3) | 6 (60) | .005a |
| AGR group | .067 | |||
| agr I | 19 (73) | 30 (39.5) | 7 (70) | … |
| agr II | 4 (15.4) | 15 (19.7) | 1 (10) | … |
| agr III | 2 (7.7) | 22 (28.9) | 2 (20) | … |
| agr IV | 1 (3.8) | 9 (11.8) | 0 (0.0) | … |
NOTE. Two of 114 patients from each group were excluded from geographical analysis because they originated from South America.
Statistically significant after adjustment for multiple comparisons using a false discovery rate of 10%.
Comparison of IE and STI Isolates
A total of 52 known and 24 new spa types were identified in the 114 IE isolates. Among the 114 STI isolates, 47 known and 21 new spa types were identified. The genetic diversity in the IE and STI groups was similar (Simpson’s index of diversity: 0.899 for IE vs 0.900 for STI; P = .985). Although the spa types represented in this study mapped to a total of 19 CCs, most isolates (157 of 226, 69%) were contained in 6 CCs (CC1, CC5, CC8, CC15, CC30, and CC45). IE isolates were more likely than geographically matched STI isolates to be CC30 (19.5% vs 6.2%; P = .005) (Table 5).
Table 5.
Distribution of Clonal Complexes Among Geographically Matched Methicillin-Susceptible Staphylococcus aureus Isolates From Patients With Endocarditis or Soft Tissue Infection
| Clonal complex, no. (%) | Infective endocarditis (n = 113) | Soft tissue infection (n = 113) | P value |
| CC1 | 7 (6.2) | 10 (8.8) | .615 |
| CC5 | 14 (12.4) | 6 (5.3) | .099 |
| CC8 | 9 (8) | 16 (14.2) | .203 |
| CC15 | 15 (13.3) | 10 (8.8) | .397 |
| CC30 | 22 (19.5) | 7 (6.2) | .005 |
| CC45 | 16 (14.2) | 25 (22.1) | .167 |
| Other | 30 (26.5) | 39 (34.5) | .248 |
NOTE. We were unable to obtain spa type sequence or MLST for 1 isolate from each group due to technical difficulties. CC, clonal complex.
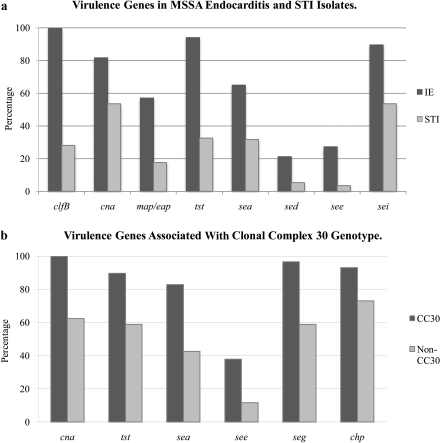
Next, we compared the prevalence of individual genes in the geographically matched IE and STI groups. Both sets of isolates contained almost 100% of certain genes (fnbA, clfA, spa, ebpS, hlg, efb) and low prevalence of others (etb, seb, sej). IE isolates were significantly more likely than STI isolates to contain 3 adhesins (clfB, cna, map/eap; P < .0001 for all after multiple comparisons adjustment) and 5 enterotoxins (tst, sea, sed, see, and sei; P ≤ .005 for all after multiple comparisons adjustment) (Table 6, Figure 1). Interestingly, the frequency of pvl did not differ significantly between IE and STI isolates (19.3% vs 27.2%, respectively; P = .209).
Table 6.
Genotypic Characteristics of Geographically Matched Methicillin-Susceptible Staphylococcus aureus Isolates From Patients With Endocarditis or Soft Tissue Infection
| Genes, no. (%) | Infective endocarditis (n = 114) | Soft tissue infection (n = 114) | P value |
| Adhesins | |||
| fnbA | 112 (98.2) | 110 (100) | .498 |
| fnbB | 17 (15.2) | 16 (14) | 1.000 |
| clfA | 113 (99.1) | 114 (100) | 1.000 |
| clfB | 114 (100) | 32 (28.1) | <.0001a |
| cna | 93 (81.6) | 61 (53.5) | <.0001a |
| spa | 111 (97.4) | 114 (100.0) | .247 |
| sdrC | 51 (44.7) | 35 (30.7) | .040 |
| sdrD | 71 (62.3) | 72 (63.2) | 1.000 |
| sdrE | 60 (52.6) | 65 (57) | .595 |
| bbp | 101 (88.6) | 104 (91.2) | .661 |
| ebpS | 114 (100) | 114 (100) | … |
| map/eap | 65 (57) | 20 (17.5) | <.0001a |
| Toxins | |||
| eta | 27 (23.7) | 19 (16.7) | .248 |
| etb | 5 (4.4) | 0 (0.0) | .060 |
| tst | 107 (93.9) | 37 (32.5) | <.0001a |
| sea | 74 (64.9) | 36 (31.6) | <.0001a |
| seb | 4 (3.5) | 9 (7.9) | .253 |
| sec | 29 (25.4) | 28 (24.6) | 1.000 |
| sed | 24 (21.1) | 6 (5.3) | <.001a |
| see | 31 (27.2) | 4 (3.5) | <.0001a |
| seg | 80 (70.2) | 66 (57.9) | .073 |
| seh | 13 (11.4) | 7 (6.1) | .241 |
| sei | 102 (89.5) | 61 (53.5) | <.0001a |
| sej | 4 (3.5) | 6 (5.3) | .748 |
| pvl | 22 (19.3) | 31 (27.2) | .209 |
| hlg | 114 (100) | 114 (100) | … |
| Other putative virulence genes | |||
| efb | 113 (99.1) | 111 (97.4) | .622 |
| icaA | 106 (93) | 111 (97.4) | .215 |
| chp | 94 (82.5) | 78 (68.4) | .021 |
| V8 | 66 (57.9) | 73 (64) | .415 |
| AGR group | .003a | ||
| agr I | 36 (31.6) | 57 (50) | … |
| agr II | 40 (35.1) | 20 (17.5) | … |
| agr III | 33 (28.9) | 27 (23.7) | … |
| agr IV | 5 (4.4) | 10 (8.8) | … |
NOTE. a Statistically significant after adjustment for multiple comparisons using a false discovery rate of 10%.
Next, we considered the possibility that patient-specific characteristics could influence apparent associations between bacterial genotype and infection severity. Because IE patients were significantly more likely to be male and less likely to have healthcare contact than STI patients (Table 2), we adjusted our analyses for these characteristics. In logistic regression models that included each of the genes individually and also adjusted for geographic region, sex, and healthcare association, cna (odds ratio [OR], 4.5; 95% confidence interval [CI], 2–9.6), map/eap (OR, 5.7; 95% CI, 2.7–11.8), tst (OR, 37; 95% CI, 13–104), sea (OR, 4.7; 95% CI, 2–9), sed (OR, 6.4; 95% CI, 2–19), see (OR, 13.4; 95% CI, 4–45), and sei (OR, 7.2; 95% CI, 3–16) were significantly more common among IE isolates, even after adjusting for multiple comparisons. Similarly, CC30 remained significantly more frequently associated with IE isolates than STI isolates (OR, 6.7; 95% CI, 2–19; adjusted P = .002). Because no IE isolates lacked clfB, no adjustment could be reported for that gene.
To consider the possibility that individual genes might be associated with IE due to the fact that they were more commonly present in the CC30 genotype, we compared the frequency of individual genes in CC30 and non-CC30 genetic backgrounds. After multiple comparisons adjustment, the adhesin cna (P ≤ .0001), enterotoxins tst, sea, see, seg (P < .01 for all), and chp (P = .043) were more frequently found in CC30 than in non-CC30 isolates (Figure 1; Supplementary Table 1).
Figure 1.
A, Specific virulence genes are significantly more common in methicillin-susceptible Staphylococcus aureus isolates from infective endocarditis (IE) compared with geographically matched methicillin-susceptible S. aureus isolates from soft tissue infection (STI). B, Specific virulence genes are significantly more common in clonal complex 30 (CC30) compared with non-CC30 methicillin-susceptible S. aureus. Genes listed are statistically significant after adjustment for multiple comparisons using a false discovery rate of 10%.
Based on this finding, we performed a logistic regression model to evaluate the independent association of the genes with IE phenotype. This model initially included all of the genes that were individually associated with IE. Genes were removed with backward elimination and only those significant at P < .05 were retained. After adjusting for geographic region, sex, healthcare association, and CC30 genotype, the adhesins sdrC (OR, 12.4; 95% CI, 2.4–64), cna (OR, 10.7; 95% CI, 2–56), and map/eap (OR, 5.9; 95% CI, 1.8–18) and the toxins tst (OR, 31.8; 95% CI, 8.7–116) and sei (OR, 10.9; 95% CI, 2.9–40) were independently associated with IE. When we confirmed these results using conditional regression, we obtained nearly identical parameter estimates and standard errors (results not shown).
DISCUSSION
The contribution of a particular strain of S. aureus to the severity of infection it causes is poorly understood. In the current study, we used isolates from 2 large international cohorts of patients with MSSA IE and STI. We validated our previous finding that the CC30 genotype is associated with an increased risk for IE [4]. We also demonstrated that MSSA isolates causing IE have a distinct genetic repertoire and that some of these genes are independently associated with IE even after adjusting for CC30 genotype.
The findings of this report provide strong evidence that bacterial genotype is associated with clinical phenotype. Several previous observations have noted a difference in the genetic contents of isolates from different S. aureus clinical presentations, including resolving versus persistent bacteremia [22, 23], bacteremia with and without hematogenous complications (such as IE) [4, 24], and invasive disease and carrier states [25]. More specifically, CC30 has been associated with increased hematogenous complications [4], persistent S. aureus bacteremia [23], and invasive disease [26]. Interestingly, CC30 (previously known as phage type 80/81) has been historically recognized as a virulent strain [27], and accounts for several of the predominant nosocomial methicillin-resistant S. aureus (MRSA) strains today [28].
The current study suggests that S. aureus isolates in the CC30 genotype may be particularly effective in establishing hematogenous complications. Although the specific genetic determinants of this association are unknown, it seems logical that bacterial adhesins could facilitate hematogenous dissemination by binding to specific host proteins. Thus, it is interesting that the adhesin cna was independently associated with IE and was significantly more common in the CC30 genotype. It is known that cna codes for collagen binding protein, which mediates S. aureus binding to human collagen [23]. Although its role in IE is incompletely understood [29], cna has been associated with persistent MRSA bacteremia [23] and osteomyelitis [30]. The fact that cna was associated with both IE and the CC30 genotype in our study suggests that it may have an important role in the pathogenesis of staphylococcal IE.
Two other adhesins, clfB and map/eap, were also associated with IE. ClfB encodes fibrinogen-binding protein and mediates fibrinogen-dependent adhesion and clumping of S. aureus cells [31]. In contrast, map/eap encodes an extracellular fibrinogen binding protein secreted by S. aureus. It promotes host cell internalization and blocks neutrophil recruitment and resultant inflammatory response [32]. Although clfB has been linked to IE previously [29, 33], our study is the first to show an association between the presence of map/eap and IE.
We also found that certain enterotoxin genes were more likely to be present in IE isolates. Encoded in pathogenicity islands and prophages, enterotoxins and tst are known for their potent superantigenic properties [34]. Previous studies have found increased prevalence of enterotoxin genes in bacteremic S. aureus isolates [35] and increased concentration of serum antibodies against enterotoxins in patients with S. aureus infection [36], including IE [37]. The role of enterotoxins in the pathogenesis of IE is not intuitive. Thus, the significance of their increased prevalence among IE isolates is unknown. These findings could therefore reflect linkage disequilibrium with unidentified virulence genes and represent a “biomarker” of S. aureus isolates with an increased risk for IE rather than a causal association.
This study focused on 2 specific types of S. aureus infections. By contrast, previous studies compared “invasive” isolates from multiple types of infections with nasal carriage isolates [6, 38–40]. These studies found either no difference in genotype between nasal colonizing and invasive strains [6, 38, 40] or found that CC30 was actually more common in nasal carriage versus bacteremic isolates [39]. The heterogeneous infection types included within the “invasive” category in these prior analyses and the use of nasal carriage isolates as controls may in part explain the discrepancies with our results. Although our study demonstrates a significant association between genotype and IE, it is important to point out that almost all of the common CCs were found in both IE and STI. Hence, most S. aureus strains have the capacity to cause most types of infections [4, 25]. It is likely that other factors, such as host genetic characteristics, also influence the severity of S. aureus infections.
Our geographic comparison revealed several interesting patterns. A number of genes (fnbA, clfA, spa, ebps, hlg, and efb) were ubiquitous, similar to previous findings [7]. These genes are primarily contained within the core genome and encode essential functions of S. aureus. In contrast, several genes (clfB, cna, sdrE, seb, see, seg, sei, pvl, and v8) showed significant differences in frequency according to geographical variations. Many of these genes are located on mobile genetic elements, such as pathogenicity islands and bacteriophages, which propagate via horizontal transfer between strains of S. aureus [41]. The regional dissemination of mobile genetic elements among S. aureus strains likely contributes to the geographical variation in the genomes of S. aureus. These genetic changes can also occur in the core genome. Harris et al demonstrated such geographic clustering and intercontinental dissemination of the core genome of S. aureus clone ST239 via a novel sequencing technology employing single-nucleotide polymorphisms [42]. This regional variation of S. aureus clinical isolates necessitated our geographically matched analysis strategy.
Although pvl is associated with MRSA primary skin infections and necrotizing pneumonia [5, 43], few studies have looked at pvl in IE [44, 45]. Our study is one of the first to look at the prevalence of pvl in MSSA IE and to find a low prevalence. Bae et al reported similarly low prevalence in MRSA IE [9]. Prior studies also noted low prevalence of pvl in deep-seated infections, such as bacteremia and osteomyelitis [44, 45], and a lack of association with increased mortality [12, 24]. Overall, these findings suggest that pvl is not a primary virulence factor in the pathogenesis of IE.
Our study has several limitations. First, this is an observational study of patients with IE and STI, with the isolates originating from different studies and time periods. The ATLAS specimen repository consisted of isolates from patients enrolled in a registrational clinical trial of treatment for STI. Hence, our STI cohort may not be representative of the epidemiology of the regions included. As the ATLAS study was set up to optimize recruitment of MRSA STIs, the definition of healthcare association differed slightly between the STI and IE groups. Our sample sizes in certain subgroups limited our ability to reach statistically significant conclusions. In addition, our findings only suggested associations of specific genotype with clinical phenotype and cannot distinguish between causality and correlation. Last, our inferences were based on the presence of genetic elements in S. aureus. Thus, our findings may not reflect the actual expression of these genes in the form of proteins and toxins nor the allelic variations that are directly responsible for the virulence of specific S. aureus infections.
In conclusion, our comparisons revealed that the genetic repertoire of MSSA IE varies by geographic region and clinical infection type. Compared with MSSA isolates causing STI, IE isolates are more likely to belong to CC30 and to contain specific virulence genes. Future studies are required to better understand the association of IE with CC30 genotype, adhesins, and enterotoxins, and to see if this clonal association holds true for MRSA infections. Future genome comparisons, for example, via SNP hybridizations or whole genome sequencing, will also be needed to further elucidate core genomic differences among the clinical spectrum of S. aureus.
Supplementary Data
Supplementary data are available at The Journal of Infectious Diseases online.
Funding
This work was supported by grants from Cubist Pharmaceuticals (to C. W. W.), Theravance (to V. G. F.), the National Institutes of Health (K24-AI093969 and R01-AI068804 to V. G. F.), and the Barton F. Haynes Resident Research Award to J. N. from the Department of Medicine at Duke University Hospital.
Supplementary Material
Acknowledgments
We would like to thank Drs Barry Kreiswirth and José R. Mediavilla from the University of Medicine and Dentistry of New Jersey for providing us with valuable guidance on spa typing, and Dr Lauren M. McIntyre for her guidance on the use of the false discovery rate method for multiple comparisons adjustments.
ICE Micro Investigator Index 2010 (last updated 22 November 2010):
Australia: Eugene Athan, MD, Owen Harris, MBBS (Barwon Health); Tony M. Korman, MD (Monash Medical Centre); Despina Kotsanas, BS (Southern Health); Phillip Jones, MD, Porl Reinbott, Suzanne Ryan, MHS (University of New South Wales); Brazil: Claudio Querido Fortes, MD (Hospital Universitario Clementino Fraga Filho/UFRJ); Chile: Patricia Garcia, MD, Sandra Braun Jones, MD (Hospital Clinico-Escuela de Medicina, Universidad Catolica de Chile); Bruno Barsic, MD, PhD, Suzana Bukovski, MD (Univ. Hospital for Infectious Diseases); France: Christine Selton-Suty, MD, Neijla Aissa, MD, Thanh Doco-Lecompte, MD (CHU Nancy-Brabois); Francois Delahaye, MD, PhD, Francois Vandenesch, MD (Hopital Louis Pradel); Pierre Tattevin, MD, PhD (Pontchaillou University); Bruno Hoen, MD, PhD, Patrick Plesiat, MD (University Medical Center of Besançon); Greece: Helen Giamarellou, MD, PhD, Efthymia Giannitsioti, MD (Attikon University General Hospital); Italy: Emanuele Durante-Mangoni, MD, PhD, Marie Françoise Tripodi, MD, Riccardo Utili, MD, PhD, Roberta Casillo, MD, PhD, Susanna Cuccurullo, MSc (II Università di Napoli); Pierre Yves Donnio, PhD, Annibale Raglio, MD, Fredy Suter, MD (Ospedali Riuniti di Bergamo); Lebanon: Zeina Kanafani, MD, MS, Souha S.Kanj, MD (American University of Beirut Medical Center); New Zealand: Arthur Morris, MD (Diagnostic Medlab); David R. Murdoch, MD, MSc, DTM&H (University of Otago); Slovenia: Manica Mueller Premru, MD, PhD, Tatjana Lejko-Zupanc, MD, PhD (Medical Center Ljubljana); Spain: Manuel Almela, MD, Yolanda Armero, MD, Manuel Azqueta, MD, Ximena Castañeda, MD, Carlos Cervera, MD, Elisa De Lazzari, MS, Ana del Rio, MD, PhD, Carlos Falces, MD, Cristina Garcia-de-la-Maria, PhD, Jose M. Gatell, MD, PhD, Magda Heras, MD, PhD, Francesc Marco, MD, PhD, Carlos A. Mestres, MD, PhD, José M. Miró, MD, PhD, Asuncion Moreno, MD, PhD, Salvador Ninot, MD, Carlos Paré, MD, PhD, Joan Pericas, MD, Jose Ramirez, MD, PhD, Marta Sitges, MD (Hospital Clinic—IDIBAPS, University of Barcelona); Emilio Bouza, MD, PhD, Marta Rodríguez-Créixems, MD, PhD, Victor Ramallo, MD (Hospital General Universitario Gregorio Marañón); USA: Suzanne Bradley, MD (Ann Arbor VA Medical Center); Dannah Wray, MD, Lisa Steed, PhD, MHS, Robert Cantey, MD (Medical University of South Carolina); Gail Peterson, MD, Amy Stancoven, MD (UT-Southwestern Medical Center); Christopher Woods, MD, MPH, G. Ralph Corey, MD, L. Barth Reller, MD, Vance G. Fowler Jr, MD, MHS, Vivian H. Chu, MD, MHS (Duke University Medical Center).
ICE Coordinating Center: Khaula Baloch, MPH, Vivian H. Chu, MD, MHS, G. Ralph Corey, MD, Christy C. Dixon, Vance G. Fowler Jr, MD, MHS, Tina Harding, RN, BSN, Paul Pappas, MS, Lawrence P. Park, PhD, Thomas Redick, MPH, Marian Jones-Richmond, Judy Stafford, MS.
ICE Publications Committee: Kevin Anstrom, PhD, Eugene Athan, MD, Arnold S. Bayer, MD, Christopher H. Cabell, MD, MHS, Vivian H. Chu, MD, MHS, G. Ralph Corey, MD, Vance G. Fowler Jr, MD, MHS, Bruno Hoen, MD, PhD, A. W. Karchmer, MD, Josè M. Miró, MD, PhD, David R. Murdoch, MD, MSc, DTM&H, Daniel J. Sexton, MD, Andrew Wang, MD.
ICE Steering Committee: Arnold S. Bayer, MD, Christopher H. Cabell, MD, MHS, Vivian Chu, MD, MHS, G. Ralph Corey, MD, David T. Durack, MD, DPhil, Susannah Eykyn, MD, Vance G. Fowler Jr, MD, MHS, Bruno Hoen, MD, PhD, Josè M. Miró, MD, PhD, Phillipe Moreillon, MD, PhD, Lars Olaison, MD, PhD, Didier Raoult, MD, PhD, Ethan Rubinstein, MD, LLB, Daniel J. Sexton, MD.
Abstract presentation: This work was presented in part at the American Society for Microbiology General Meeting, Poster Session, San Diego, California, May 2010.
References
- 1.Fowler VG, Jr., Miro JM, Hoen B, et al. Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA. 2005;293:3012–21. doi: 10.1001/jama.293.24.3012. [DOI] [PubMed] [Google Scholar]
- 2.Daum RS. Clinical practice. Skin and soft-tissue infections caused by methicillin-resistant Staphylococcus aureus. N Engl J Med. 2007;357:380–90. doi: 10.1056/NEJMcp070747. [DOI] [PubMed] [Google Scholar]
- 3.Campbell SJ, Deshmukh HS, Nelson CL, et al. Genotypic characteristics of Staphylococcus aureus isolates from a multinational trial of complicated skin and skin structure infections. J Clin Microbiol. 2008;46:678–84. doi: 10.1128/JCM.01822-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fowler VG, Jr., Nelson CL, McIntyre LM, et al. Potential associations between hematogenous complications and bacterial genotype in Staphylococcus aureus infection. J Infect Dis. 2007;196:738–47. doi: 10.1086/520088. [DOI] [PubMed] [Google Scholar]
- 5.Gillet Y, Issartel B, Vanhems P, et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002;359:753–9. doi: 10.1016/S0140-6736(02)07877-7. [DOI] [PubMed] [Google Scholar]
- 6.Lindsay JA, Moore CE, Day NP, et al. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J Bacteriol. 2006;188:669–76. doi: 10.1128/JB.188.2.669-676.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peacock SJ, Moore CE, Justice A, et al. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect Immun. 2002;70:4987–96. doi: 10.1128/IAI.70.9.4987-4996.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabell CH, Abrutyn E. Progress toward a global understanding of infective endocarditis. Early lessons from the International Collaboration on Endocarditis investigation. Infect Dis Clin North Am. 2002;16:255–72. doi: 10.1016/s0891-5520(01)00007-1. [DOI] [PubMed] [Google Scholar]
- 9.Bae IG, Federspiel JJ, Miro JM, et al. Heterogeneous vancomycin-intermediate susceptibility phenotype in bloodstream methicillin-resistant Staphylococcus aureus isolates from an international cohort of patients with infective endocarditis: prevalence, genotype, and clinical significance. J Infect Dis. 2009;200:1355–66. doi: 10.1086/606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murdoch DR, Corey GR, Hoen B, et al. Clinical presentation, etiology, and outcome of infective endocarditis in the 21st century: the International Collaboration on Endocarditis-Prospective Cohort Study. Arch Intern Med. 2009;169:463–73. doi: 10.1001/archinternmed.2008.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stryjewski ME, Graham DR, Wilson SE, et al. Telavancin versus vancomycin for the treatment of complicated skin and skin-structure infections caused by gram-positive organisms. Clin Infect Dis. 2008;46:1683–93. doi: 10.1086/587896. [DOI] [PubMed] [Google Scholar]
- 12.Bae IG, Tonthat GT, Stryjewski ME, et al. Presence of genes encoding the panton-valentine leukocidin exotoxin is not the primary determinant of outcome in patients with complicated skin and skin structure infections due to methicillin-resistant Staphylococcus aureus: results of a multinational trial. J Clin Microbiol. 2009;47:3952–7. doi: 10.1128/JCM.01643-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durack DT, Lukes AS, Bright DK. New criteria for diagnosis of infective endocarditis: utilization of specific echocardiographic findings. Duke Endocarditis Service. Am J Med. 1994;96:200–9. doi: 10.1016/0002-9343(94)90143-0. [DOI] [PubMed] [Google Scholar]
- 14.Friedman ND, Kaye KS, Stout JE, et al. Health care–associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med. 2002;137:791–7. doi: 10.7326/0003-4819-137-10-200211190-00007. [DOI] [PubMed] [Google Scholar]
- 15.Daniel WG, Mugge A, Martin RP, et al. Improvement in the diagnosis of abscesses associated with endocarditis by transesophageal echocardiography. N Engl J Med. 1991;324:795–800. doi: 10.1056/NEJM199103213241203. [DOI] [PubMed] [Google Scholar]
- 16.Mathema B, Mediavilla J, Kreiswirth B . Sequence analysis of the variable number tandem repeat in Staphylococcus aureus protein A gene. In: Deleo F, Otto MW, editors. Methods in molecular biology: bacterial pathogenesis. Totowa, NJ: Humana Press; 2008:. xvi, 285. [DOI] [PubMed] [Google Scholar]
- 17.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–30. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci. 2002;99:7687–92. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–75. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 20.Grundmann H, Hori S, Enright MC, et al. Determining the genetic structure of the natural population of Staphylococcus aureus: a comparison of multilocus sequence typing with pulsed-field gel electrophoresis, randomly amplified polymorphic DNA analysis, and phage typing. J Clin Microbiol. 2002;40:4544–6. doi: 10.1128/JCM.40.12.4544-4546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breslow NE, Day NE. Statistical methods in cancer research. Volume I—The analysis of case-control studies. IARC Sci Publ. 1980;32:5–338. [PubMed] [Google Scholar]
- 22.Fowler VG, Jr., Sakoulas G, McIntyre LM, et al. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J Infect Dis. 2004;190:1140–9. doi: 10.1086/423145. [DOI] [PubMed] [Google Scholar]
- 23.Xiong YQ, Fowler VG Jr, Yeaman MR, Perdreau-Remington F, Kreiswirth BN, Bayer AS. Phenotypic. and genotypic characteristics of persistent methicillin-resistant Staphylococcus aureus bacteremia in vitro and in an experimental endocarditis model. J Infect Dis. 2009;199:201–8. doi: 10.1086/595738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lalani T, Federspiel JJ, Boucher HW, et al. Associations between the genotypes of Staphylococcus aureus bloodstream isolates and clinical characteristics and outcomes of bacteremic patients. J Clin Microbiol. 2008;46:2890–6. doi: 10.1128/JCM.00905-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melles DC, Gorkink RF, Boelens HA, et al. Natural population dynamics and expansion of pathogenic clones of Staphylococcus aureus. J Clin Invest. 2004;114:1732–40. doi: 10.1172/JCI23083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wertheim HF, van Leeuwen WB, Snijders S, et al. Associations between Staphylococcus aureus genotype, infection, and in-hospital mortality: a nested case-control study. J Infect Dis. 2005;192:1196–200. doi: 10.1086/444427. [DOI] [PubMed] [Google Scholar]
- 27.Rountree PM, Beard MA. Further observations on infection with phage type 80 staphylococci in Australia. Med J Aust. 1958;45:789–95. [PubMed] [Google Scholar]
- 28.Robinson DA, Kearns AM, Holmes A, et al. Re-emergence of early pandemic Staphylococcus aureus as a community-acquired methicillin-resistant clone. Lancet. 2005;365:1256–8. doi: 10.1016/S0140-6736(05)74814-5. [DOI] [PubMed] [Google Scholar]
- 29.Rindi S, Cicalini S, Pietrocola G, et al. Antibody response in patients with endocarditis caused by Staphylococcus aureus. Eur J Clin Invest. 2006;36:536–43. doi: 10.1111/j.1365-2362.2006.01675.x. [DOI] [PubMed] [Google Scholar]
- 30.Elasri MO, Thomas JR, Skinner RA, et al. Staphylococcus aureus collagen adhesin contributes to the pathogenesis of osteomyelitis. Bone. 2002;30:275–80. doi: 10.1016/s8756-3282(01)00632-9. [DOI] [PubMed] [Google Scholar]
- 31.Sinha B, Herrmann M. Mechanism and consequences of invasion of endothelial cells by Staphylococcus aureus. Thromb Haemost. 2005;94:266–77. doi: 10.1160/TH05-04-0235. [DOI] [PubMed] [Google Scholar]
- 32.Rivera J, Vannakambadi G, Hook M, Speziale P. Fibrinogen-binding proteins of gram-positive bacteria. Thromb Haemost. 2007;98:503–11. [PubMed] [Google Scholar]
- 33.O’Brien L, Kerrigan SW, Kaw G, et al. Multiple mechanisms for the activation of human platelet aggregation by Staphylococcus aureus: roles for the clumping factors ClfA and ClfB, the serine-aspartate repeat protein SdrE and protein A. Mol Microbiol. 2002;44:1033–44. doi: 10.1046/j.1365-2958.2002.02935.x. [DOI] [PubMed] [Google Scholar]
- 34.Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev. 2000;13:16–34. doi: 10.1128/cmr.13.1.16-34.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becker K, Friedrich AW, Lubritz G, Weilert M, Peters G, Von Eiff C. Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J Clin Microbiol. 2003;41:1434–9. doi: 10.1128/JCM.41.4.1434-1439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verkaik NJ, Dauwalder O, Antri K, et al. Immunogenicity of toxins during Staphylococcus aureus infection. Clin Infect Dis. 2010;50:61–8. doi: 10.1086/648673. [DOI] [PubMed] [Google Scholar]
- 37.Ellis M, Serreli A, Colque-Navarro P, et al. Role of staphylococcal enterotoxin A in a fatal case of endocarditis. J Med Microbiol. 2003;52:109–12. doi: 10.1099/jmm.0.05003-0. [DOI] [PubMed] [Google Scholar]
- 38.Feil EJ, Cooper JE, Grundmann H, et al. How clonal is Staphylococcus aureus? J Bacteriol. 2003;185:3307–16. doi: 10.1128/JB.185.11.3307-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holtfreter S, Grumann D, Schmudde M, et al. Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J Clin Microbiol. 2007;45:2669–80. doi: 10.1128/JCM.00204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price J, Baker G, Heath I, et al. Clinical and microbiological determinants of outcome in Staphylococcus aureus bacteraemia. Int J Microbiol. 2010; doi: 10.1155/2010/654858. doi: 10.1155/2010/654858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng Y, Chen CJ, Su LH, Hu S, Yu J, Chiu CH. Evolution and pathogenesis of Staphylococcus aureus: lessons learned from genotyping and comparative genomics. FEMS Microbiol Rev. 2008;32:23–37. doi: 10.1111/j.1574-6976.2007.00086.x. [DOI] [PubMed] [Google Scholar]
- 42.Harris SR, Feil EJ, Holden MT, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–74. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diep BA, Sensabaugh GF, Somboona NS, Carleton HA, Perdreau-Remington F. Widespread skin and soft-tissue infections due to two methicillin-resistant Staphylococcus aureus strains harboring the genes for Panton-Valentine leucocidin. J Clin Microbiol. 2004;42:2080–4. doi: 10.1128/JCM.42.5.2080-2084.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lina G, Piemont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–32. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 45.Ruotsalainen E, Karden-Lilja M, Kuusela P, et al. Methicillin-sensitive Staphylococcus aureus bacteraemia and endocarditis among injection drug users and nonaddicts: host factors, microbiological and serological characteristics. J Infect. 2008;56:249–56. doi: 10.1016/j.jinf.2008.01.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.