Abstract
Background. Human immunodeficiency virus type 1 (HIV-1)–specific CD8+ responses contribute to the decline in acute peak viremia following infection. However, data on the relative immunogenicity of CD8+ T-cell epitopes during and after acute viremia are lacking.
Methods. We characterized CD8+ T-cell responses in 20 acutely infected, antiretroviral-naive individuals with HIV-1 subtype C infection using the interferon-γ enzyme-linked immunosorbent spot assay. Eleven of these had not fully seroconverted at the time of analysis. Viruses from plasma were sequenced within defined cytotoxic T-lymphocyte (CTL) cell epitopes for selected subjects.
Results. At approximately 28 days after estimated initial infection, CD8+ T-cell responses were directed against an average of 3 of the 410 peptides tested (range, 0–6); 2 individuals had no detectable responses at this time. At 18 weeks, the average number of peptides targeted had increased to 5 (range 0–11). Of the 56 optimal Gag CTL epitopes sequenced, 31 were wild-type in the infecting viruses, but only 11 of 31 elicited measurable CD8+ T-cell responses.
Conclusions. These data demonstrate that the majority of CD8+ responses are not elicited during acute HIV infection despite the presence of the cognate epitope in the infecting strain. There is a need to define factors that influence lack of induction of effective immune responses and the parameters that dictate immunodominance in acute infection.
Thirty years after the first clinical cases of AIDS were identified, more than 60 million people have been infected with the human immunodeficiency virus type 1 (HIV-1) [1]. South Africa has the world’s largest population of people living with HIV, and the province of KwaZulu-Natal is experiencing the most severe HIV epidemic with nearly one-third of adults reported to be infected [2, 3]. Stemming the AIDS epidemic will require the development of an efficacious vaccine to protect people from HIV-1 infection and/or attenuate disease progression. To this end, understanding the immune correlates of protection against the virus is critical [4].
HIV-1–specific CD8+ T lymphocyte (CTL) cells have been a key focus for HIV vaccine development efforts, in part because approaches based on the development of a neutralizing antibody–based vaccine have thus far failed to induce broadly cross-reactive neutralizing antibodies [4–8] and in part because induction of T-cell responses in animal models of HIV infection have impacted viral set point and disease progression [9–11].
CD8+ T cells may constitute a critical component of controlling viral replication because their emergence has been reported to coincide with a decline in acute-phase viremia in HIV and simian immunodeficiency virus (SIV) infection [12–14]. Furthermore, it has been demonstrated that CD8+ T cells exert significant selective pressure on HIV-1 and SIV during acute and chronic infection [14–18] and that their depletion in SIV infection results in loss of control of SIV replication [19–21].
Numerous studies in clade B infection have shown that early CD8+ T-cell responses are narrowly directed, and specific immunodominant T-cell responses detected during acute HIV-1 infection are associated with the subsequent viral set point [22–25]. However, these studies have had a number of limitations. In most studies to date, although subjects have been identified prior to seroconversion, the initial assays of T-cell function have been performed following seroconversion. Studies that link CD8+ T-cell responses to seroconversion status and contemporaneous viral load are lacking, and overall there is a paucity of data on HIV-1–specific CD8+ T-cell responses for the ethnicities most ravaged by the HIV-1 pandemic and for the clades responsible in these regions.
Moreover, most studies on immune responses in acute infection have relied on identification of subjects based on symptoms of acute infection staged as days following onset of symptoms [13, 25, 26], whereas the evolution of antibody responses used in the Fiebig staging of acute infection were obtained from a cohort of asymptomatic blood donors [27] who would not have qualified for donation if they had had typical acute infection symptoms such as fever and malaise.
To address these limitations and to define the characteristics of HIV-1–specific immunity in acute HIV-1 subtype C (HIV-1C) infection, we screened persons at voluntary counseling and testing (VCT) centers testing negative by standard antibody assays for evidence of HIV RNA [28]. Subjects were enrolled after initial screening, allowing us to document the decline in viral load concurrent with the measurement of immune responses. Using the enzyme-linked immunosorbent spot (ELISPOT) assay, we evaluated the breadth and magnitude of HIV-specific CD8+ T-cell responses over the first few weeks of infection using synthetic overlapping peptides and defined HLA class I–restricted epitopes. Our data indicate that limited T-cell breadth is associated with the initial decline in viremia in acute HIV-1C infection and that the majority of epitopes known to be targeted in the context of expressed class I molecules are not immunogenic in the earliest stages of infection.
METHODS
Study Participants
The study design, recruitment strategies, and experiences are described elsewhere ([28] and Chonco et al, manuscript in preparation). In brief, individuals testing negative or discordant by dual commercial rapid HIV-1 tests (Bioline, Standard Diagnostics; and Sensa, Hitech Healthcare) were recruited for HIV-1 RNA testing. Acute HIV-1 infection was defined by a positive HIV-1 RNA test, negative HIV-1 enzyme immunoassay (SD HIV1/2 enzyme-linked immunosorbent assay [ELISA] 3.0, Standard Diagnostics), and a negative or indeterminate Western blot (Genetic Systems, Bio-Rad). Centers for Disease Control and Prevention (CDC) criteria were used for the interpretation of Western blot results such that a positive sample had at least 2 of the major bands (gp160, gp120, gp41 and p24), with either gp160 or gp120 present as well as gp41 or p24. An indeterminate result was one in which these requirements were not met but 1 or more bands were present, or where the band intensity was less than the weak positive control. Negative results exhibited no reactive bands. We estimated the infection date as occurring 14 days prior to the first positive HIV RNA and negative HIV antibody test as previously described [29].
A total of 42 subjects were identified, of whom 20 were enrolled. The study was approved by the Biomedical Research Ethics Committee (BREC) of the University of KwaZulu-Natal and the institutional review board of Massachusetts General Hospital. All study participants provided written informed consent for participation in the study.
Viral Load Quantification and CD4+ T-Cell Enumeration
HIV-1 RNA in plasma was measured using the Roche Amplicor version 1.5 or Cobas Taqman HIV-1 Test according to the manufacturer’s instructions. CD4+ T cells were enumerated using Tru-Count technology and analyzed on a 4-color flow cytometer (Becton Dickinson) according to the manufacturer’s instructions.
HLA Typing
HLA class I typing was performed by DNA polymerase chain reaction (PCR) using sequence-specific primers as previously described [30].
Synthetic HIV-1 Peptides
A panel of 410 peptides (18-mers overlapping by 10 amino acid residues), spanning the entire HIV-1 clade C consensus sequence together with optimal peptides with known HLA class I restriction patterns, were synthesized on an automated peptide synthesizer (MBS 396; Advanced ChemTech) and used in the ELISPOT assay [31].
Interferon-γ ELISPOT Assay
Assessment of HIV-1–specific CD8+ responses was performed on peripheral blood mononuclear cell (PBMC) samples collected at 2–4, 4–6, 6–8, 8–12 and 12–18 weeks (14–28, 29–56, 57–84, and 85–126 days, respectively) after initial viral load screening. PBMCs (50 000–100 000 cells/well) were plated in 96-well polyvinylidene plates (MAIP S45; Millipore), precoated with antihuman interferon (IFN)–γ monoclonal antibody (mAb) 1-D1k (0.5 μg/mL; Mabtech). Peptides were added at 2 μg/mL in a matrix [31]. Phytohemagglutinin (1 μg/mL) stimulation was included as a positive control, and medium alone as a negative control. Plates were incubated overnight at 37°C and 5% CO2, then washed with phosphate-buffered saline before addition of the biotinylated anti–IFN-mAb, 7-B6-1 biotin (Mabtech) at 0.5 μg/mL, and incubated for 90 minutes. Following washing, streptavidin-conjugated alkaline phosphatase (Mabtech) was added for 45 minutes. IFN-γ–producing cells were noted by direct visualization following development with alkaline phosphatase color reagents (Bio-Rad). A response was defined as positive when there were at least 100 spot-forming cells (SFCs)/million PBMCs and the total number of spots was 3 standard deviations above the negative control value. In addition to overlapping peptides, peptides corresponding to optimal HIV-1 cytotoxic T-cell (CTL) epitopes described for the individual’s HLA class I type were tested at a peptide concentration of 2 μg/mL.
To verify peptides identified as positive in the peptide pool matrix, PBMCs from the same time point were stimulated with individual peptides that were common to 2 peptide pools in the initial ELISPOT screen. The criteria used to define positive responses in the peptide pool matrix were also used for the verification assay. In 98% of the matrix positive responses, at least 1 single peptide positive reaction was confirmed. The sensitivity of the matrix approach for the detection of peptide-specific T-cell responses was found to be 82% for Nef and Gag. The sensitivity was calculated by dividing the number of responses detected in the peptide matrix by the number of responses detected using individual peptides [32].
All responses at the first sampling time point were evaluated using fresh PBMCs, which provide greater sensitivity in ELISPOT assays in comparison to viably frozen cells [33]. Fresh cells were also used in all but 10% of assays performed at later time points. PBMCs were frozen (90% calf serum and 10% DMSO) at −160°C until use. Cell viability was determined using a Guava automated counter (Guava Technologies), and the ELISPOT assay was performed only when PBMC viability was greater than 80%.
Gag Amplification and Sequencing
Viral RNA was extracted from plasma samples using the QIAamp Viral RNA Mini Kit from Qiagen. Gag was amplified by reverse-transcriptase polymerase chain reaction (RT-PCR) using the Superscript III One-Step RT-PCR kit (Invitrogen) followed by a second round of PCR with the Takara Ex Taq HS enzyme kit, using Gag-specific primers [34].
The PCR product was population-sequenced using the Big Dye ready reaction termination mix V3 (Applied Biosystems), as per manufacturer’s instructions. Sequences were edited in Sequencher 4.8. A modified nucleotide–amino acid alignment program algorithm [35] was used to align sequences to HXB2 (GenBank accession number K03455), and insertions with respect to HXB2 were stripped before further analysis.
Statistical Analysis
The Mann–Whitney nonparametric analysis was used to test for significant differences between HLA groups. For assessments of the relationship between immune responses and viral load, Spearman rank correlations were used. Statistical analyses were performed using GraphPad Prism software version 5.0 for Windows.
RESULTS
Viral Load and CD4 Counts in HIV-1 Clade C Acute Infection
HIV-specific CD8+ T-cell responses have been documented in primary HIV infection; however, most studies have evaluated responses well after the documented decline in viremia. To begin to address this critical phase, we recruited persons with acute HIV-1C infection before seroconversion. Twenty subjects were identified who were viremic, HIV-1 ELISA negative, and Western blot negative (Fiebig stage I or II). All subjects subsequently seroconverted.
The characteristics of the recruited subjects are described elsewhere (Chonco et al, manuscript in preparation). Viral loads at the time of diagnosis ranged from 28 720 to 33 200 000 RNA/mL (median 5 572 000). Initial screening was limited to obtaining a plasma sample; the median viral load at enrollment, when the first ELISPOT assays were performed, was 371 000 RNA/mL (range, 1090–2 840 000), and the average CD4+ T-cell counts obtained at this time were 389 cells/mm3 (range, 151–691 cells/mm3). At 12–18 weeks after the estimated date of infection, the median viral load was 127 000 RNA/mL (range, 351–430 000). Together these results indicate that a strategy based on nucleic acid testing on individuals who test negative at VCT sites can identify acute HIV-1C virus infection during times of peak viremia following acute HIV infection, and these subjects can be successfully recruited for follow-up studies.
HIV-1–Specific CD8+ T-Cell Responses Before and After Seroconversion
Although studies of acute HIV infection have assessed CD8+ T-cell responses in persons identified prior to seroconversion, there are some important caveats: most of the initial assays were performed after seroconversion, and/or with cryopreserved samples [13, 36, 37]. Moreover, few studies have assessed responses to all expressed viral proteins [25, 36–38]. To address these gaps in knowledge, we performed a detailed analysis of CD8+ T-cell responses to all expressed HIV proteins using fresh PBMCs obtained prior to antibody seroconversion. Since only plasma was obtained during initial screening, these assays were limited to the 11 subjects for whom subsequent enrollment cell samples were available prior to seroconversion (Table 1). All 11 subjects were Fiebig stage IV or earlier.
Table 1.
The Dynamics of HIV-1–Specific CD8+ T-Cell Responses to Peptides Spanning the Entire HIV Proteome and Viral Load Kinetics in Individuals Who Had Not Fully Seroconverted at Approximately 28 Days After the Estimated Time of Infection
| Participant number | Screening VL (RNA copies/mL) | Enrollment VL (days after screening) | WB bands at enrollment | Peptides targeted at enrollmenta | Dominant response (SFCs/million) | Total magnitude of responses (SFCs/million) | Peptides targeted at 12–18 weeksa | Total magnitude of responses (SFCs/million) |
| AS1-703 | 4 560 000 | 119 000 (14) | p24, p40, p55/51 | Nef-81, Nef-83 | Nef-83 (490) | 850 | #81 | 500 |
| AS1-919 | 6 280 000 | 484 000 (15) | p24, p40 | Nef-83, Nef-84, RT-202, RT203 | Nef-83 (280) | 740 | None | 0 |
| AS2-174 | 7 390 000 | 330 000 (21) | p18, p24 | gp41-366 | gp41-366 (700) | 700 | #25, #35, #36, #41, #65, #69, #231, #366 | 11240 |
| AS2-802 | 732 260 | 138 300 (15) | p18, p24, p40 | Nef-82, Nef-83, gp120-289, gp120-294 | gp120-294 (4000) | 5710 | #11, #35, #36, #44, #64, #65, #82, #83, #294 | 2740 |
| AS2-945 | 1 100 000 | 1872 (32) | p24, p40, p55/51 | None | None | 0 | #20, #22, #33, #84, #181 | 6390 |
| AS2-973 | 10 000 000 | 66 530 (15) | p24 | None | None | 0 | #275, #294, #407 | 780 |
| AS2-1037 | 12 100 000 | 708 000 (23) | p24, p40, p55/51 | Gag-41, Rev-100 | Rev-100 (580) | 720 | #40, #41, #46, #76, #100, #101, #116, #411 | 5610 |
| AS3-017 | 22 000 000 | 2 840 000 (14) | 0 | Gag-16, Gag-40, Nef-76, Tat-115 | Gag-16 (1020) | 2700 | #40 | 1320 |
| AS3-458 | 280 000 | 1 650 000 (14) | p24, p55/51 | gp120-316 | gp120-316 (420) | 420 | #42, #157, #194, #225, #252, #316 | 1990 |
| AS3-513 | 112 448 | 1 967 000 (14) | p24 | Gag-41 | Gag-41 (200) | 200 | #4, #40, #41 | 920 |
| AS3-740 | 28 716 | 3493 (22) | p24, p40, p55/51 | Gag-41, Nef-78, Nef-81, Nef-83 | Nef-78 (700) | 2280 | #41, #78, #81, #83 | 1120 |
NOTE. SFC, spot-forming cells; VL, viral load; WB, Western blot.
The sequences of tested peptides and their location in the proteome are given in Table 1 (online only).
HIV-1–specific CD8+ T-cell responses were detected in 9 of the 11 subjects but were narrowly directed in each case (Table 1). Two subjects with no detectable responses had each experienced an approximate 50- to 100-fold drop in viremia at the time of testing; both made detectable responses at later time points. Although the 2 most frequently recognized proteins at the earliest time point were Nef (peptides 69 to 84) and Gag (peptides 4 to 65), targeted by 50% and 36% of the subjects, respectively, there was great heterogeneity among subjects with respect to the specificity of the initial responses. Moreover, among those with detectable responses, there were dramatic differences in magnitude, with total detectable epitope-specific responses ranging from as high as 4000 SFCs/million PBMCs detected to as low as 100 SFCs/million PBMCs.
HIV-1–specific responses were subsequently tested longitudinally in these individuals up to 18 weeks (Table 1). At this time, responses were detectable in 10 of the 11 subjects, albeit at different magnitudes, and both subjects who initially tested negative (AS2-945 and AS2-973) now had detectable responses. Of the 9 with initially positive responses, at least 1 of these responses was still detected in 8 subjects at the later time point. There were only 2 subjects in whom the initial breadth of responses declined at the follow-up time point. Of the 9 subjects in whom an initial immunodominant response was detected, this response was still present at follow-up weeks later in 7. However, there was no consistent pattern in terms of specificity, magnitude, or evolution of responses in these individuals. These results indicate that the initial responses largely persist, and that subjects with no initial responses were able to mount immune responses subsequently.
Evolution of CD8+ T-Cell Responses Following Acute HIV Infection
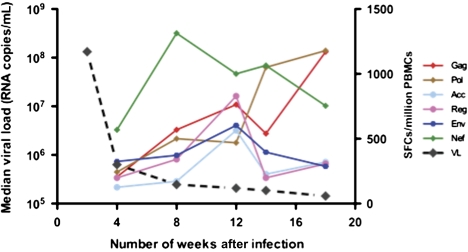
To further characterize the evolution of CD8+ T-cell responses following acute HIV-1 infection, we performed a longitudinal analysis in all 20 subjects. The magnitude of responses at approximately 4 weeks after the estimated date of infection ranged from 150 to 8770 SFCs/million PBMCs. There was a 15-fold decline in viral loads from the initial screening value, CD8+ T-cell responses were directed against an average of 3 peptides (range, 0–6), and the most targeted protein was Nef followed by Pol and Gag (Figure 1). At 18 weeks, the total magnitude of responses ranged from 500 to 11 240 SFCs/million PBMCs and the average number of peptides targeted was 5 (range 0–11).
Figure 1.
Average magnitude of HIV-1–specific CD8+ T-cell responses to human immunodeficiency virus type 1 (HIV-1) proteins in acute/early infection for all participants. The magnitude of HIV-1–specific CD8+ T-cell responses to overlapping HIV peptides spanning each of the designated proteins are shown on the y-axis as spot-forming cells (SFCs)/million peripheral blood mononuclear cells (PBMCs), and the x-axis denotes when the assays were done (number of days after the estimated time of infection). Solid lines represent interferon (IFN)–γ release in response to HIV-1 CD8+ T-cell peptides, and the dashed line represents viral load dynamics.
At 6–8 weeks after the estimated date of infection, our analyses for all 20 subjects revealed that there was a significant positive correlation between the magnitude of Nef CD8+ T-cell responses and plasma viremia (P = .003). There was no correlation observed between the magnitude of virus-specific T-cell responses targeting other HIV-1 proteins and plasma viral load.
Limited Breadth of HIV-1–Specific CD8+ T-Cell Epitopes During Primary Infection
The above analyses determined HIV-1–specific CD8+ T-cell responses using overlapping peptides and thus did not define the actual targeted epitopes within those peptides. To delineate the precise regions being targeted, a subset of 134 peptides corresponding to previously described epitopes for each subject’s respective HLA class I allotypes were used [39]. The HLA class I alleles expressed in the study cohort, as well as the number of epitopes tested for each HLA allele, are summarized in Table 2. The majority of HIV-1C epitopes described in chronic HIV infection were not targeted during these early stages, demonstrating marked differences in the inductive phase of the immune response that must depend on the epitope rather than the restricting HLA allele. In addition, although all class I alleles tested were capable of eliciting responses, many subjects did not target epitopes through these alleles (Table 2).
Table 2.
HLA Frequencies in Study Cohort and the Percentage of Subjects With Responses Against Peptides Associated With Individual HLA Expression
| HLA class I allele | HLA frequency, % | Epitopes targeted/epitopes tested (no.) at 6–8 wk and 12–18 wka | Subjects (%) with responses at 6–8 wk and 12–18 wk | ||
| A23 | 21 | 3/9 | 5/9 | 40 | 60 |
| A26 | 17 | 1/3 | 1/3 | 25 | 50 |
| A29 | 25 | 4/9 | 5/9 | 60 | 67 |
| A30 | 38 | 5/14 | 8/14 | 75 | 63 |
| B1503 | 25 | 4/6 | 6/6 | 100 | 100 |
| B1510 | 21 | 6/11 | 7/11 | 100 | 100 |
| B42 | 29 | 8/26 | 13/26 | 100 | 100 |
| B44 | 33 | 4/13 | 4/13 | 100 | 67 |
| B58 | 21 | 3/27 | 5/27 | 20 | 67 |
| Cw4 | 25 | 4/9 | 2/9 | 67 | 50 |
| Cw6 | 21 | 1/7 | 2/7 | 20 | 40 |
| Cw17 | 25 | 1/5 | 1/5 | 17 | 25 |
| Median values | 25 | 4/9 (44%) | 5/9 (56%) | 63 | 65 |
NOTE. Data are shown only for HLA class I alleles that were expressed in at least 3 individuals, and for which at least 3 HIV-1–specific optimal CD8+ T-cell epitopes had been defined.
Epitope sequences and their location in the proteome are given in Table 2 (online only).
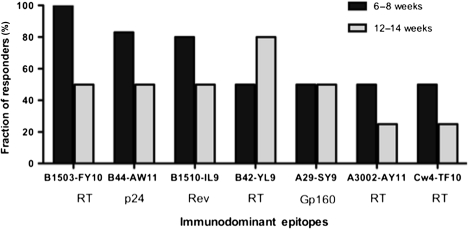
To further define these relationships, we assessed the persistence of early responses for the most frequently targeted early epitopes (Figure 2). For 5 of 7 epitopes tested, there was a decrease over time in the fraction of persons expressing the restricting HLA allele targeting these epitopes but the immunodominance patterns of CD8+ T-cells remained unchanged in the individuals with detectable responses. There was no shift in the immunodominance hierarchy in HLA-B*1503–restricted responses measured at 6–8 and 12–14 weeks against the HLA-B*1503–restricted epitopes RT(Int)-FY10 and p24-IY9, and the epitopes were found to be immunodominant and subdominant, respectively. Similarly, CD8+ T-cell responses against the HLA-B*44–restricted epitope p24-AW11 and the HLA-B*1510–restricted epitope Rev-IL9 remained immunodominant during the earliest and latest tested time points in individuals expressing these respective alleles. However, there was a shift in the hierarchy of responses in individuals expressing the HLA-A*30 where the initially immunodominant RT-AY11 epitope later became subdominant and CD8+ T-cell responses against the initially subdominant Int-KIY9 epitope became immunodominant (Figure 2).
Figure 2.
Immunodominance patterns for HIV-1–specific CD8+ T-cell responses restricted by individual HLA class I alleles at 6–8 weeks and 12–14 weeks after the estimated time of infection. The percentage of participants expressing the respective allele that had detectable peptide-specific CD8+ T-cell responses is shown on the y-axis. HLA-restricted HIV-specific CD8+ T-cell epitopes are aligned on the x-axis and their location on the major HIV-1 proteins is also indicated. The sequences of immunodominant epitopes and their location in the genome are listed in Supplementary Table 2 (available online).
Overall, these data demonstrate that the immunodominance patterns of specific epitopes during acute infection are persistent for the majority of HLA class I alleles that could be assessed, but that many class I alleles are infrequently utilized in early infection and many potentially immunogenic epitopes are not targeted.
Effect of Sequence Variation in Gag CTL Epitopes on Recognition by Specific CTL
Our previous research [40] and that of other [26] has shown that up to 30% of early responses may be missed when using peptides representing a reference strain of virus. We therefore sequenced plasma virus to determine if lack of CD8+ recognition related to lack of expression of the cognate epitope. We focused these efforts on the gag region, as targeting of Gag has been associated with lower viral loads [30]. Plasma virus sequencing revealed that of the 56 epitope sequences evaluated, 31 were wild-type virus sequences; however, these did not elicit detectable CD8+ T-cell responses in 20 of 31 (65%) of cases (Table 3). Both the subjects in whom we failed to detect any CD8+ T-cell responses prior to seroconversion (Table 1, AS2-945 and AS2-973) were infected with viruses that contained epitopes present in the reference set of peptides used in the assays. These data indicate that despite expression of the restricting HLA class I allele and the presence of cognate viral sequence, CD8+ responses to the virus were often not mounted, suggesting that the immunogenicity of these epitopes is suboptimal and that factors other than IFN-γ–expressing CD8+ T cells may be involved in the initial decline in viremia.
Table 3.
Sequence Analysis of Autologous Virus Gag Epitopes in Subjects at 6–8 Weeks After the Estimated Date of Infection
| Participant number | HLA type | HLA-restricted epitope | CD8+ T-cell response |
| EVIPMFTAL | |||
| AS2-016 | A26 | .. .. .. … | – |
| AS3-017 | .. .. .. … | + | |
| AS1-703 | . I… .. .. | – | |
| AS3-513 | .. .. .. … | – | |
| RSLYNTVATLY | |||
| AS3-268 | A29 | K. . F.. .. … | + |
| AS3-458 | .. .. .. I… . | – | |
| AS1-919 | K. . F… .. .. | – | |
| AS1-323 | K. . F… .. .. | – | |
| AS2-184 | K. . F.. .. .. C | – | |
| AS2-110 | K… .. .. V. . | – | |
| RLRPGGKKHY | |||
| AS2-802 | A30 | .. .. .. .. Q. | – |
| AS2-050 | .. .. .. .. .. | + | |
| AS2-973 | .. .. .. .. .. | – | |
| AS2-483 | .. .. .. .. Q. | – | |
| AS2-358 | .. .. .. .. R. | – | |
| AS2-016 | .. .. .. .. C. | + | |
| AS1-703 | .. .. .. .. .. | – | |
| AS2-174 | K.. .. … Q. | – | |
| RSLYNTVATLY | |||
| AS2-802 | A30 | .. .. .. .. … | + |
| AS2-050 | .. .. .. .. … | – | |
| AS2-973 | K… .. I… . | – | |
| AS2-483 | K… .. .. … | – | |
| AS2-358 | … F… .. .. | – | |
| AS2-016 | … F… .. .. | – | |
| AS1-703 | … F… .. .. | – | |
| AS2-174 | .. .. .. .. … | – | |
| VKVIEEKAF | |||
| AS2-1037 | B1503 | .. .. .. … | – |
| AS1-876 | .. .. .. … | – | |
| AS3-268 | .. .. .. … | + | |
| AS2-483 | .. .. … G. | – | |
| AS2-341 | .. .. … G. | – | |
| AS2-050 | .. .. .. … | + | |
| VHQAISPRTL | |||
| AS2-945 | B1510 | .. .. .. .. .. | – |
| AS3-369 | .. .. .. .. .. | – | |
| AS2-1037 | .. .. .. .. .. | + | |
| AS3-513 | .. .. .. .. .. | – | |
| GHQAAMQML | |||
| AS2-945 | B1510 | .. .. .. … | – |
| AS3-369 | .. .. .. … | – | |
| AS2-1037 | .. .. .. … | – | |
| AS3-513 | .. .. … I. | + | |
| EQATQDVKNW | |||
| AS1-703 | B44 | .. .. .. .. … | + |
| AS3-458 | .. .. .. .. … | + | |
| AS2-184 | .. .. .. .. … | + | |
| AS3-369 | .. .. S… .. . | + | |
| AS1-876 | … G. . E… . | – | |
| SEGATPQDL | |||
| AS1-703 | B44 | .. .. .. … | – |
| AS3-458 | .. .. .. … | – | |
| AS2-184 | .. .. .. T. . | – | |
| AS3-369 | .. .. .. … | – | |
| AS1-876 | .. .. .. … | + | |
| RDYVDRFFKTL | |||
| AS2-110 | B44 | .. .. .. .. … | – |
| AS1-703 | .. .. .. .. R. . | – | |
| AS3-458 | .. .. .. .. R. . | – | |
| AS2-184 | .. .. .. .. … | + | |
| AS3-369 | .. .. .. .. … | – | |
| AS1-876 | .. .. .. .. … | – |
DISCUSSION
Despite the dominance of HIV-1 subtype C worldwide, the evolution of virologic and immunologic parameters in acute HIV-1C infection have not been extensively characterized. By RNA screening of persons testing negative by standard antibody assays, we were able to recruit 20 subjects before seroconversion and characterize viral kinetics and evolving HIV-specific CD8+ T-cell responses during acute infection. Despite high viremia, responses were narrowly directed, and the majority of epitopes targeted in chronic infection [30] did not induce detectable responses during the rapid decline of viremia. Although the ELISPOT assay can underestimate the true magnitude of T-cell responses [14, 26, 41] and autologous virus sequences can differ from the reference strains used, HIV-1–specific CD8+ responses ultimately arose in these persons, indicating that many immunogenic epitopes are not targeted in the earliest stages of infection, at a time when viral load is rapidly declining.
Longitudinal assays for responses during and following acute infection allowed us to address not only the specificity of responses but also their persistence. In the entire cohort, responses to Nef-derived peptides were dominant in the earliest stages of infection, consistent with other data [38, 42, 43]. However, only approximately half of the individuals tested targeted Nef in the early stages, although 82% targeted this protein at some time during the average 5 months of follow-up (data not shown), again suggesting impaired induction of responses in acute infection. A trend was observed where high T-cell responses against the Nef proteins correlated positively with high viral loads, as has been reported in other studies [38, 42–45], suggesting that these responses are driven by level of antigenemia, rather than being causal in lowering viral load, again suggesting impaired functional CD8+ T-cell responses in the earliest stages of acute infection. However, there was no correlation between the magnitude and breadth of CD8+ T-cell responses for other viral proteins and the concurrent plasma viral load, consistent with other reports [17, 32, 46].
By assessing responses using peptides representing optimal epitopes, we were able to assess the hierarchy in the development of epitope-specific CD8+ T-cell responses restricted by specific HLA alleles, their immunodominance patterns, and the timing of induction of these responses. Within a month of the estimated time of infection, CD8+ T-cell responses were detected against 44% of peptide epitopes matched for each subject’s HLA and against 56% of the epitopes presented by their expressed alleles at 3 months. These data confirmed that the detectable responses in early infection are largely maintained, although immunodominance often shifts. Further investigation is required to determine whether the changes in the magnitude of responses and immunodominance are the result of sequence evolution within targeted epitopes or their flanking regions [24], immunoregulation [47–50], or other mechanisms.
In conclusion, we demonstrate that HIV-1–specific CD8+ T-cell responses can be detected before complete seroconversion, but many of the epitopes that elicit responses in chronic infection are not immunogenic during acute infection. We also confirm previous studies showing that the initial HIV-1–specific immune responses are narrowly directed, and extend these by showing the paucity of responses even when dramatic drops in viremia have occurred. Moreover, although response breadth and magnitude expand with duration of infection, a significant proportion of individuals are still unable to make responses despite the presence of cognate peptides restricted by the corresponding HLA allele. Further studies are needed to address why recognition of HIV-1 peptides appears to be selective during acute infection as the lack of recognition may contribute to the failure to control viremia to a low set point. The paucity of responses during the dramatic reduction in viremia suggests that tissue-specific responses as well as measures of CD8+ T-cell function other than IFN-γ should be examined to better define the role of HIV–specific CD8+ T-cells in the initial decline in viremia. Further studies are needed to understand the determinants and role of HIV-1–specific CD8+ T-cell responses in high-burden settings where an effective HIV-1 vaccine is urgently needed.
Funding
The HPP Acute Infection Study is funded by the National Institutes of Health (grant ROI-AI067073), the South African AIDS Vaccine Initiative, and the Collaboration for AIDS Vaccine Discovery of the Bill and Melinda Gates Foundation. Additional funding came from the Mark and Lisa Schwartz Foundation and the South African Research Chairs Initiative. M. R. and J. K. W. are funded by the National Research Foundation and the Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology and Harvard University. I. V. B. is funded by the National Institute of Allergy and Infectious Diseases (grant K23 AI 068458). T. N. holds the South African Department of Science and Technology/National Research Foundation Research Chair in Systems Biology of HIV/AIDS.
Acknowledgments
We are grateful to the staff and management at McCord Hospital, St Mary’s Hospital, Prince Mshiyeni Memorial Hospital, and all participating clinics in Durban, South Africa. We acknowledge the support and cooperation of the KwaZulu-Natal Department of Health. We thank Sisters Thandi Sikhakhane, Nonhlanhla Maphalala, Landiwe Nxele, and Nono Nkupiso for their assistance in the recruitment of study participants. We acknowledge Dr Johannes Viljoen and the Africa Center laboratory, Durban, South Africa, for providing access to the sequencing facility. Taryn Green and Lungile Maphumulo provided excellent technical assistance.
References
- 1.UNAIDS. Report on the global AIDS epidemic. Geneva, Switzerland: Joint United Nations Programme on HIV/AIDS (UNAIDS); 2009. [Google Scholar]
- 2.National Department of Health. National antenatal sentinel HIV and syphilis prevalence survey. Republic of South Africa. Pretoria: National Department of Health of South Africa; 2009. [Google Scholar]
- 3.UNAIDS. Report on the global AIDS epidemic. Geneva, Switzerland: Joint United Nations Programme on HIV/AIDS (UNAIDS); 2008. [Google Scholar]
- 4.Virgin HW, Walker BD. Immunology and the elusive AIDS vaccine. Nature. 2010;464:224–31. doi: 10.1038/nature08898. [DOI] [PubMed] [Google Scholar]
- 5.Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis. 2005;191:654–65. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 6.Pitisuttithum P, Gilbert P, Gurwith M, et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194:1661–71. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 7.Buchbinder SP, Mehrotra DV, Duerr A, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–93. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McElrath MJ, De Rosa SC, Moodie Z, et al. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet. 2008;372:1894–905. doi: 10.1016/S0140-6736(08)61592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen SG, Vieville C, Whizin N, et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med. 2009;15:293–9. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, O’Brien KL, Lynch DM, et al. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature. 2009;457:87–91. doi: 10.1038/nature07469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson NA, Keele BF, Reed JS, et al. Vaccine-induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J Virol. 2009;83:6508–21. doi: 10.1128/JVI.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–10. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koup RA, Safrit JT, Cao Y, et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–5. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goonetilleke N, Liu MK, Salazar-Gonzalez JF, et al. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med. 2009;206:1253–72. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borrow P, Lewicki H, Wei X, et al. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–11. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 16.Allen TM, O’Connor DH, Jing P, et al. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–90. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- 17.Cao J, McNevin J, Holte S, Fink L, Corey L, McElrath MJ. Comprehensive analysis of human immunodeficiency virus type 1 (HIV-1)-specific gamma interferon-secreting CD8+ T cells in primary HIV-1 infection. J Virol. 2003;77:6867–78. doi: 10.1128/JVI.77.12.6867-6878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen TM, Altfeld M, Geer SC, et al. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J Virol. 2005;79:13239–49. doi: 10.1128/JVI.79.21.13239-13249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matano T, Kobayashi M, Igarashi H, et al. Cytotoxic T lymphocyte-based control of simian immunodeficiency virus replication in a preclinical AIDS vaccine trial. J Exp Med. 2004;199:1709–18. doi: 10.1084/jem.20040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin X, Bauer DE, Tuttleton SE, et al. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–8. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitz JE, Kuroda MJ, Santra S, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–60. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 22.Yu XG, Addo MM, Rosenberg ES, et al. Consistent patterns in the development and immunodominance of human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T-cell responses following acute HIV-1 infection. J Virol. 2002;76:8690–701. doi: 10.1128/JVI.76.17.8690-8701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lichterfeld M, Yu XG, Le Gall S, Altfeld M. Immunodominance of HIV-1-specific CD8(+) T-cell responses in acute HIV-1 infection: at the crossroads of viral and host genetics. Trends Immunol. 2005;26:166–71. doi: 10.1016/j.it.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Altfeld M, Kalife ET, Qi Y, et al. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8(+) T cell response against HIV-1. PLoS Med. 2006;3:e403. doi: 10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Streeck H, Jolin JS, Qi Y, et al. Human immunodeficiency virus type 1-specific CD8+ T-cell responses during primary infection are major determinants of the viral set point and loss of CD4+ T cells. J Virol. 2009;83:7641–8. doi: 10.1128/JVI.00182-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turnbull EL, Wong M, Wang S, et al. Kinetics of expansion of epitope-specific T cell responses during primary HIV-1 infection. J Immunol. 2009;182:7131–45. doi: 10.4049/jimmunol.0803658. [DOI] [PubMed] [Google Scholar]
- 27.Fiebig EW, Wright DJ, Rawal BD, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17:1871–9. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 28.Bassett IV, Chetty S, Giddy J, et al. Screening for acute HIV infection in South Africa: finding acute and chronic disease. HIV Med. 2011;12:46–53. doi: 10.1111/j.1468-1293.2010.00850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Loggerenberg F, Mlisana K, Williamson C, et al. Establishing a cohort at high risk of HIV infection in South Africa: challenges and experiences of the CAPRISA 002 acute infection study. PLoS One. 2008;3:e1954. doi: 10.1371/journal.pone.0001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiepiela P, Ngumbela K, Thobakgale C, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 31.Thobakgale CF, Ramduth D, Reddy S, et al. Human immunodeficiency virus-specific CD8+ T-cell activity is detectable from birth in the majority of in utero-infected infants. J Virol. 2007;81:12775–84. doi: 10.1128/JVI.00624-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Addo MM, Yu XG, Rathod A, et al. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol. 2003;77:2081–92. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao J, Lou Y, Pinczewski J, et al. Boosting of SIV-specific immune responses in rhesus macaques by repeated administration of Ad5hr-SIVenv/rev and Ad5hr-SIVgag recombinants. Vaccine. 2003;21:4022–35. doi: 10.1016/s0264-410x(03)00266-4. [DOI] [PubMed] [Google Scholar]
- 34.Wright JK, Brumme ZL, Carlson JM, et al. Gag-protease-mediated replication capacity in HIV-1 subtype C chronic infection: associations with HLA type and clinical parameters. J Virol. 2010;84:10820–31. doi: 10.1128/JVI.01084-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang X, Zhang J. Methods for comparing a DNA sequence with a protein sequence. Comput Appl Biosci. 1996;12:497–506. doi: 10.1093/bioinformatics/12.6.497. [DOI] [PubMed] [Google Scholar]
- 36.Altfeld M, Rosenberg ES, Shankarappa R, et al. Cellular immune responses and viral diversity in individuals treated during acute and early HIV-1 infection. J Exp Med. 2001;193:169–80. doi: 10.1084/jem.193.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alter G, Merchant A, Tsoukas CM, et al. Human immunodeficiency virus (HIV)-specific effector CD8 T cell activity in patients with primary HIV infection. J Infect Dis. 2002;185:755–65. doi: 10.1086/339338. [DOI] [PubMed] [Google Scholar]
- 38.Gray CM, Mlotshwa M, Riou C, et al. Human immunodeficiency virus-specific gamma interferon enzyme-linked immunospot assay responses targeting specific regions of the proteome during primary subtype C infection are poor predictors of the course of viremia and set point. J Virol. 2009;83:470–8. doi: 10.1128/JVI.01678-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brander C, Goulder P. The evolving field of HIV CRL epitope mapping: new approaches for the identification of novel epitopes. In: Korber BTM, Brander C, Walker BD, et al., editors. HIV molecular database. Los Alamos, NM: Los Alamos National Laboratory; 2000. [Google Scholar]
- 40.Altfeld M, Addo MM, Shankarappa R, et al. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J Virol. 2003;77:7330–40. doi: 10.1128/JVI.77.13.7330-7340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Y, Iglesias E, Samri A, et al. A systematic comparison of methods to measure HIV-1 specific CD8 T cells. J Immunol Methods. 2003;272:23–34. doi: 10.1016/s0022-1759(02)00328-9. [DOI] [PubMed] [Google Scholar]
- 42.Lichterfeld M, Yu XG, Cohen D, et al. HIV-1 Nef is preferentially recognized by CD8 T cells in primary HIV-1 infection despite a relatively high degree of genetic diversity. AIDS. 2004;18:1383–92. doi: 10.1097/01.aids.0000131329.51633.a3. [DOI] [PubMed] [Google Scholar]
- 43.Masemola A, Mashishi T, Khoury G, et al. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J Virol. 2004;78:3233–43. doi: 10.1128/JVI.78.7.3233-3243.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novitsky V, Gilbert P, Peter T, et al. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. J Virol. 2003;77:882–90. doi: 10.1128/JVI.77.2.882-890.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masemola AM, Mashishi TN, Khoury G, et al. Novel and promiscuous CTL epitopes in conserved regions of Gag targeted by individuals with early subtype C HIV type 1 infection from southern Africa. J Immunol. 2004;173:4607–17. doi: 10.4049/jimmunol.173.7.4607. [DOI] [PubMed] [Google Scholar]
- 46.Betts MR, Ambrozak DR, Douek DC, et al. Analysis of total human immunodeficiency virus (HIV)-specific CD4(+) and CD8(+) T-cell responses: relationship to viral load in untreated HIV infection. J Virol. 2001;75:11983–91. doi: 10.1128/JVI.75.24.11983-11991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Day CL, Kaufmann DE, Kiepiela P, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–4. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 48.Trautmann L, Janbazian L, Chomont N, et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat Med. 2006;12:1198–202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- 49.Petrovas C, Casazza JP, Brenchley JM, et al. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J Exp Med. 2006;203:2281–92. doi: 10.1084/jem.20061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaufmann DE, Kavanagh DG, Pereyra F, et al. Upregulation of CTLA-4 by HIV-specific CD4+ T cells correlates with disease progression and defines a reversible immune dysfunction. Nat Immunol. 2007;8:1246–54. doi: 10.1038/ni1515. [DOI] [PubMed] [Google Scholar]