Abstract
Microbial pathogens have evolved mechanisms to overcome immune responses and successfully infect their host. Here, we studied how Listeria monocytogenes evades immune detection by peptidoglycan (PGN) modification. By analyzing L. monocytogenes muropeptides, we detected O-acetylated muramic acid residues. We identified an O-acetyltransferase gene, oatA, in the L. monocytogenes genome sequence. Comparison of PGN from parental and isogenic oatA mutant strains showed that the O-acetyltransferase OatA O-acetylates Listeria PGN. We also found that PGN O-acetylation confers resistance to different types of antimicrobial compounds targeting bacterial cell wall such as lysozyme, β-lactam antibiotics, and bacteriocins and that O-acetylation is required for Listeria growth in macrophages. Moreover, oatA mutant virulence is drastically affected in mice following intravenous or oral inoculation. In addition, the oatA mutant induced early secretion of proinflammatory cytokines and chemokines in vivo. These results suggest an important role for OatA in limiting innate immune responses and promoting bacterial survival in the infected host.
Listeria monocytogenes is a Gram-positive bacterium widespread in the environment and the etiologic agent of listeriosis, a life-threatening food-borne disease. Clinical manifestations of the disease range from gastroenteritis to septicemia, central nervous system infections, abortions, and perinatal infection [1]. After ingestion of contaminated food, L. monocytogenes can survive in the intestine, cross the intestinal barrier, disseminate to the liver and spleen, and ultimately spread to the central nervous system or fetoplacental unit. Invasive listeriosis relies on the Listeria capacity to enter and replicate in phagocytes and nonphagocytic cells such as epithelial cells, endothelial cells, and hepatocytes [2]. After internalization in a vacuole, Listeria secretes listeriolysin O (LLO), a pore-forming toxin essential for virulence that promotes bacterial escape from the vacuole [3]. In the cytosol, Listeria replicates and exploits the actin polymerization machinery to propel itself, escape control by autophagy, and spread from cell to cell, protected from extracellular immune effectors such as complement and antibodies.
Innate immune response is critical to controlling Listeria infection [4, 5]. Neutrophils are major effectors of the innate response, attracted to the liver to eliminate hepatocytes infected by Listeria [6, 7]. Activation of neutrophils relies on cytokines, notably interleukin 6 (IL-6) [8]. IL-6–deficient mice have a defective neutrophil response and are highly susceptible to listeriosis [9, 10]. Macrophages and natural killer (NK) cells are also essential for Listeria clearance, and cytokines are central to their activation. Secretion of tumor necrosis factor–α (TNF-α) and interleukin 12 (IL-12) by macrophages induces interferon-γ (IFN-γ) release by NK cells [11], which activate macrophage bactericidal activity in mice infected by Listeria [12]. TNF-α and IFN-γ are important for resistance to listeriosis, as mice that lack TNF-α receptor or IFN-γ fail to eradicate Listeria and rapidly die [13–18].
Pathogenic bacteria have a variety of mechanisms to subvert innate immunity [19]. We previously showed that Listeria evades innate defenses by peptidoglycan (PGN) N-deacetylation [20]. PGN is an essential component of bacterial cell wall and a pathogen-associated molecular pattern that activates pattern-recognition receptors (PRRs; eg, nucleotide-binding oligomerization domain [NOD] proteins), which trigger antimicrobial signaling cascades and pathogen clearance [21]. Modification of PGN N-acetylglucosamine residues by Listeria deacetylase PgdA confers resistance to lysozyme, a major antibacterial of the innate immune system [20]. Indeed lysozyme, by hydrolyzing PGN β-(1,4)-glycosidic bonds between N-acetylglucosamine and N-acetylmuramic acid residues, has a bactericidal activity and enhances presentation of pathogen-associated molecular patterns (PAMPs) to PRRs [22]. Accordingly, Listeria PGN deacetylation suppresses NOD1-dependent and toll-like receptor 2 (TLR2)–dependent IL-6 and interferon-β secretion, possibly by decreasing accessibility of cell wall components to PRRs [20]. Listeria PGN has not been fully characterized yet. Thus, it is currently unknown whether deacetylation of N-acetylglucosamine residues is the only PGN modification evolved by Listeria to evade innate immunity.
Here, we report that Listeria evades innate defenses by PGN O-acetylation. We show that L. monocytogenes PGN O-acetyltransferase OatA confers resistance to lysozyme and other antibacterials. Furthermore, we demonstrate that OatA contributes not only to intracellular survival in macrophages in vitro but also to the suppression of IL-6 secretion in vivo. Lastly, we provide evidence that immune escape by PGN O-acetylation is critical for Listeria virulence.
METHODS
Bacterial Strains, Growth Conditions, and Cell Wall Preparation
L. monocytogenes EGDe (serovar 1/2a, BUG1600) was used as the parental strain [23]. L. monocytogenes ΔpgdA (BUG2288), L. monocytogenes Δhly (BUG2133), and L. monocytogenes ΔoatA (BUG2410) were obtained by gene deletion from EGDe, and L. monocytogenes ΔoatAΔpgdA (BUG2519) was obtained by gene deletion from ΔpgdA as described [24]. L. monocytogenes ΔoatA complemented strain (BUG2520) was obtained from ΔoatA as described [25]. Oligonucleotides used for gene deletion and complementation are listed in Supplementary Table S1; online only. Listeria strains were grown in brain-heart infusion (BHI) broth (BD) at 37°C and 200 rpm. Highly purified cell wall from L. monocytogenes strains was prepared as described [20].
Cells, Culture Media, and Cell Infection
Murine macrophage-like cell line RAW264.7 (ATCC:TIB-71) was cultured in Dulbecco’s modified Eagle medium (Gibco) containing 10% decomplemented fetal bovine serum (Biowest) at 37°C in 10% CO2 atmosphere. Peritoneal-elicited macrophages (PEM) and bone marrow–derived macrophages (BMDM) isolated from 8-week-old female C57BL/6J mice (Charles River Laboratories) as described [20, 26] and human acute monocytic leukemia cell line THP-1 (ATCC:TIB-202) were cultured in RPMI-1640 (Gibco) containing 10% decomplemented fetal bovine serum (Biowest) at 37°C in 10% CO2 atmosphere. PEM and BMDM cells were infected with L. monocytogenes at multiplicity of infection (MOI) 10:1 and 20:1, respectively, centrifuged at 300g for 2 minutes, and incubated at 37°C for 15 minutes to allow bacterial phagocytosis. RAW264.7 and THP-1 cells were infected with L. monocytogenes at MOI 25:1 and 10:1, respectively, over a period of 1 hour at 37°C. The number of intracellular bacteria was assessed at various time points, and infected RAW264.7 cells were observed by electron microscopy as described [20]. Experiments were repeated 2 to 3 times independently.
Antibacterial Activity Assays
Lysozyme antibacterial activity was assessed by the disk diffusion method. Overnight cultures of Listeria strains were diluted in BHI broth to 106 colony-forming unit (CFU)/mL and spread on BHI agar plates. Disks (10 mm) were loaded with 10 μL of chicken egg white lysozyme (Sigma). Plates were incubated for 48 hours at 37°C and inhibition zone diameter was measured. Cefotaxime minimum inhibitory concentration (MIC) was determined using E-test strips (Biomérieux) on BHI agar plates that were inoculated with Listeria strains. Inhibitory activity of antimicrobial peptides was determined in 96-well plates. Overnight Listeria cultures were diluted in BHI broth to 106 CFU/mL and incubated with antimicrobial peptides at 37°C and 200 rpm overnight. CFUs in each well were assessed by plating serial dilutions on BHI agar.
Virulence Studies and Cytokine Production in Mice
Experiments were performed according to the Institut Pasteur guidelines for animal experimentation. Median lethal dose (LD50), mice survival, and quantification of bacterial multiplication were carried out in BALB/c mice following intravenous challenge or in iFABP-hEcad transgenic mice after oral infection as described [20]. Cytokine concentrations in infected mice were measured by the Mouse cytokine 20-plex bead immunoassay (Biosource) and the Mouse Interferon-Beta enzyme-linked immunosorbent assay (ELISA; PBL).
Statistical Analysis
Results are expressed as mean values ± SD of 3–8 samples. Student t test was performed to determine statistical significance (*, **, and *** indicate P < .05, P < .01, and P < .001, respectively).
RESULTS
The lmo1291 Gene Encodes a Peptidoglycan O-Acetyltransferase
The genome of L. monocytogenes EGDe encodes a putative O-acetyltransferase, Lmo1291, with a calculated molecular mass of 70423 Da and an estimated isoelectric point (pI) of 10.1. The lmo1291 gene is surrounded by lmo1290 and lmo1292, which encode the internalin-like protein InlK (27) and a putative phosphodiesterase, respectively, and are both transcribed divergently compared with lmo1291 (Figure 1A). While lmo1290 is absent from the L innocua genome, lmo1291 orthologs are present in Listeria species whose genome has been sequenced, including L. innocua, L. welshimeri, L. seeligeri, and L. grayi. The 622-amino-acid protein encoded by lmo1291 contains a signal peptide followed by an acyltransferase domain (COG1835) and a YrhL-like hydrolase domain (CD01840), which belongs to the superfamily of SGNH hydrolases (Figure 1B). Orthologs of Lmo1291 are present in Staphylococcus aureus [28] and other bacteria such as Enterococcus faecalis and Lactococcus lactis. Alignment of amino-acid sequences from Lmo1291 orthologs shows that the Ser/Asp/His catalytic triad found in SGNH hydrolases is conserved (Figure 1C). Lmo1291 and its orthologs contain the same domains and several putative transmembrane regions, suggesting that these enzymes are associated with the cell wall.
Figure 1.
Lmo1291 is a peptidoglycan O-acetyltransferase. A, Genetic organization of the L monocytogenes lmo1291 gene locus (in black) showing chromosomal coordinates (bp), putative terminators (black hairpins), and the flanking genes lmo1290 encoding a putative internalin (in white) and lmo1292 encoding a putative glycerophosphodiester phosphodiesterase (in gray). B, Map of putative domains of Lmo1291 (amino acid). C, Sequence alignment of O-acetyltransferases from Enterococcus faecalis (Efa), Lactococcus lactis (Lla), Staphylococcus aureus (Sau), and L. monocytogenes Lmo1291 (Lmo). Sequences were aligned using T-Coffee software. Amino acids in red are identical and residues in green are similar. Asterisks indicate the conserved catalytic triad of SGNH hydrolases. Efa, Lla, and Sau orthologs show 56%, 50%, and 55% identity to Lmo1291, respectively. D, Muropeptide profile of the highly purified cell walls of EGDe and its isogenic ΔoatA mutant. Peaks highlighted by an asterisk indicate muropeptides absent from the ΔoatA mutant. Major peaks were analyzed by MALDI MS/MS and their respective structure are indicated above the corresponding peak. Deacetylated muropeptides are indicated in red. Muropeptides that are O-acetylated are highlighted by the addition of the OAc moiety in blue. Dotted arrows indicate the relationship between the different O-acetylated muropeptides and their parent muropeptides that lack the OAc moiety. E, Localization of PGN modifications by OatA (O-acetylation in blue) and PgdA (N-deacetylation in red). MALDI indicates matrix-assisted laser desorption ionization; MS/MS, tandem mass spectrometry.
In order to assess the function of Lmo1291, the lmo1291 gene of L. monocytogenes EGDe was deleted by allelic exchange. Purified cell wall was prepared from EGDe and Δlmo1291 strains and hydrolyzed by mutanolysin to generate muropeptides. Muropeptides were separated by high-pressure liquid chromatography (HPLC) and their composition was determined by mass spectrometry (Figure 1D). Analysis of each peak revealed that EGDe has O-acetylated N-acetyl muramic acid residues. Approximately 23% of L. monocytogenes muropeptides contained O-acetylated residues (Supplemental Table S2; online only). The Δlmo1291 mutant lacked these peaks. Reduction of EGDe muropeptides at pH 9, a treatment that removes O-acetylation, resulted in disappearance of the muropeptides that were found only in EGDe and not in Δlmo1291 (data not shown). Thus, these muropeptides were all modified by O-acetylation. These results indicate that lmo1291, renamed oatA, encodes a PGN O-acetyltransferase (Figure 1E).
OatA Confers Resistance to Antimicrobial Compounds
As we previously demonstrated, Listeria PGN N-deacetylase was a major determinant of lysozyme resistance [20]. We then tested whether OatA could contribute to lysozyme resistance. EGDe and ΔoatA growth rates were similar in BHI (data not shown). Growth of ΔoatA was significantly inhibited by lysozyme as shown by a disk diffusion assay, in contrast to EGDe (Figure 2A). To demonstrate that lysozyme sensitivity was directly due to absence of PGN O-acetylation, ΔoatA was complemented by chromosomal insertion of a single copy of oatA and its promoter. Complementation fully restored lysozyme resistance (Figure 2A). To determine if the OatA protective role was specific to lysozyme, we tested ΔoatA susceptibility to other compounds targeting cell wall such as the β-lactam antibiotic cefotaxime and the Staphylococcus gallinarum lantibiotic gallidermin. Cefotaxime MIC for ΔoatA was 8-fold lower than that of EGDe, which was similar to that of ΔoatA+oatA (Figure 2B). Addition of increasing concentrations of gallidermin to cultures correlated with decreasing survival of ΔoatA compared with EGDe (Figure 2C). Hence, OatA protection is not specific to lysozyme and extends to other antimicrobial compounds.
Figure 2.
Inactivation of oatA increases sensitivity to antimicrobial molecules that target bacterial cell wall and impairs L monocytogenes survival in macrophages. A, Lysozyme disk-diffusion assay. Growth inhibition caused by lysozyme loaded on a paper disk (1 mg/disk) was measured on BHI agar plates inoculated with EGDe (black bars), the ΔoatA mutant (white bars), or ΔoatA+oatA−complemented strains (hatched bars). B, Cefotaxime minimum inhibitory concentration was determined using E-test strips on BHI agar plates that were inoculated with EGDe (black bars), ΔoatA mutant (white bars), or ΔoatA+oatA−complemented strains (hatched bars). C, Gallidermin inhibitory activity was determined in 96-well plates. 106 CFU/mL of EGDe (black bars) or ΔoatA (white bars) were incubated with increasing concentrations of gallidermin. The number of CFU in each well was assessed after overnight incubation at 37°C by plating serial dilutions on BHI agar plates. D, THP-1 cells (n = 4); E, peritoneal-elicited macrophages (n = 5); and F, bone marrow–derived macrophages (n = 4) were infected with EGDe (black circles) or ΔoatA (white squares). G, RAW264.7 cells (n = 5) were infected with EGDe (black circles), ΔoatA (white squares), or the ΔoatA+oatA complemented strain (gray triangles). The number of CFU was determined at different time points after cell lysis with 0.2% triton. Data are means ± SD. (H–I) Electron microscopy analysis of RAW264.7 cells infected with EGDe or ΔoatA. (H) RAW264.7 cells after 4 hours of infection with EGDe or ΔoatA. Left panel: EGDe in vacuoles and protrusions; right panel: ΔoatA free in the cytosol. Scale bars: 2 μm. I, The number of bacteria per cell was determined by counting intravacuolar and cytosolic bacteria 30 minutes and 4 hours postinfection. Data are means ± SD (n = 25). Student t test was performed to determine statistical significance (*** indicates P < .001). CFU, colony-forming unit.
OatA Contributes to Listeria Resistance to Macrophage Antimicrobial Activity
Since killing mechanisms of macrophages include production of lysozyme and other antimicrobial compounds, we assessed OatA contribution to the intracellular survival of Listeria. While EGDe could replicate in THP-1 cells from 30 minutes to 4 hours post infection, ΔoatA growth was controlled (Figure 2D). Furthermore, ΔoatA was killed faster and more efficiently than EGDe in PEM (Figure 2E) and BMDM (Figure 2F), as there were 10 times fewer ΔoatA than EGDe 24 hours post infection. The ability of ΔoatA to multiply in RAW264.7 cells was also impaired after 2 hours of infection compared with EGDe and ΔoatA+oatA complemented strain (Figure 2G). Next, we observed infected RAW264.7 cells by electron microscopy. In contrast to ΔpgdA [20], ΔoatA did not accumulate in vacuoles and was readily observed in the cytosol (Figure 2H). However, in contrast to EGDe, ΔoatA was apparently not forming protrusions (Figure 2H). While similar to the number of EGDe per cell 30 minutes post infection, the number of ΔoatA was lower than that of EGDe after 4 hours (Figure 2I). These results indicate that OatA contributes to survival within macrophages, possibly by protecting Listeria from killing mechanisms, such as lysis mediated by antimicrobial polypeptides.
OatA Is Required for Pathogenesis
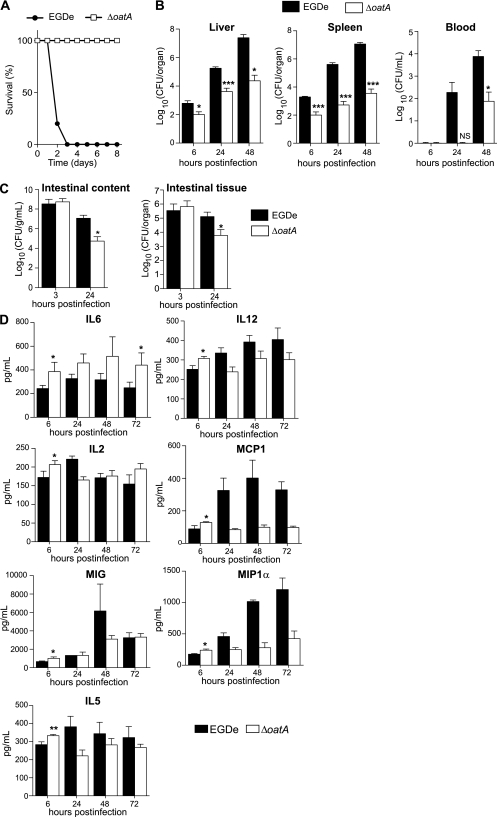
Because antimicrobial polypeptides and macrophages are major components of innate immunity, we next addressed the role of OatA in vivo. We first determined ΔoatA LD50 in BALB/c mice infected intravenously. Virulence of ΔoatA was severely attenuated, as values of LD50 were 1.1 × 107 and 0.7 × 103 bacteria for ΔoatA and EGDe, respectively. Intravenous injection of 106 EGDe resulted in 100% mortality in 3 days (Figure 3A). In contrast, mice infected with 106 ΔoatA survived (Figure 3A). Next, we analyzed the survival of ΔoatA in BALB/c mice after intravenous injection of a sublethal infectious dose. ΔoatA had a reduced capacity to replicate in liver and spleen as early as 6 hours post infection, while EGDe readily colonized both organs (Figure 3B). At 48 hours post infection, liver and spleen colonization by ΔoatA was reduced 100 times and 1000 times, respectively, compared with EGDe. Bacteremia could not be detected at 24 hours following infection with ΔoatA (Figure 3B). At 48 hours post infection, inactivation of oatA decreased bacteremia by 2 log10. Thus, OatA is a novel major virulence determinant that contributes to early stages of listeriosis in mice.
Figure 3.
The ΔoatA mutant has a strongly attenuated virulence in mice and triggers an increased cytokine response early after infection. A, BALB/c mice were challenged by intravenous injection of 106 EGDe (black circles) or ΔoatA (white squares). Survival of infected mice was determined over time (n = 5). B, BALB/c mice were challenged by intravenous injection of a sublethal dose (104 bacteria per mouse) of EGDe (black bars) or ΔoatA (white bars). Colonization of liver, spleen, and blood was followed 6, 24, and 48 hours postinfection. Data are means ± SD (n = 4). C, Transgenic human E-cadherin mice were used as a permissive model for the oral route of infection. These mice were infected with 1010 EGDe (black bars) or ΔoatA (white bars). After 3 hours and 24 hours, mice were euthanized and the number of bacteria in the intestinal lumen and intestinal tissue was determined. Data are means ± SD (n = 3–5). D, BALB/c mice were inoculated intravenously with 5 × 103 bacteria. The liver was dissected and homogenized. Homogenates were assayed using a multiplex immunoassay to determine cytokine level in response to infection with EGDe (black bars) or ΔoatA (white bars). Data are means ± SD (n = 4). Student t test was performed to determine statistical significance: *, **, and *** indicate P < .05, P < .01, and P < .001, respectively.
The oral route is the natural way of infection by Listeria and allows for the study of early stages of infection, including survival of bacteria in the intestinal lumen. Transgenic mice expressing human E-cadherin, which are permissive to Listeria oral infection, were thus infected intragastrically with EGDe or ΔoatA. In the intestinal lumen, the number of ΔoatA bacteria was strongly decreased 24 hours postinfection (Figure 3C). There were also significantly fewer mutant than wild-type bacteria in small intestine tissue (Figure 3C). Hence, ΔoatA is deficient in very early stages of infection in the intestinal lumen and at later stages of colonization in intestinal tissue.
OatA Decreases Cytokine Production in Infected Mice
We previously demonstrated that PGN N-deacetylation reduces Listeria capacity to induce secretion of inflammatory cytokines by macrophages in vitro [20]. We hypothesized that PGN O-acetylation also affects host cytokine response in vivo. We thus studied ΔoatA capacity to induce cytokine production in the liver, a major site of Listeria replication. We first infected BALB/c mice intravenously with a sublethal dose of EGDe or ΔoatA. Production of 21 cytokines was then determined by immunoassay. In mice infected with EGDe, levels of most cytokines tested increased in correlation with an increased number of bacteria (data not shown). ΔoatA induced higher levels of 7 cytokines compared with EGDe 6 hours post infection (Figure 3D). Strikingly, production of the inflammatory cytokine IL-6 in response to EGDe reached only 60% of that induced by ΔoatA both at early (6 hours) and late (72 hours) time points of infection. At 6 hours post infection, the production of T-helper 1 (Th1) differentiation cytokine IL-12, proinflammatory cytokine interleukin 2 (IL-2), chemokine monocyte chemoattractant protein 1 (MCP-1), monokine induced by IFN-γ (MIG) and macrophage inflammatory protein-1α (MIP-1α), and T-helper 2 (Th2) cytokine interleukin 5 (IL-5) increased more in response to ΔoatA compared with EGDe (Figure 3D), while liver colonization by ΔoatA was already lower than that of EGDe (Figure 3B). At later time points, the cytokine production in response to ΔoatA was similar (IL-2, MIG, IL-5) or even lower (IL-12, MCP-1, MIP-1α) compared with that induced by EGDe (Figure 3D). Interestingly, cytokine production in response to infection with the Δhly mutant was severely reduced compared with that induced by EGDe (Supplemental Figure S1A). This mutant, which does not produce LLO, was cleared from mice more rapidly than ΔoatA (Supplemental Figure S1B). Consequently, it did not activate any cytokine response in contrast to ΔoatA. Overall, OatA controls the production of inflammatory mediators at early stages of Listeria infection.
Peptidoglycan O-Acetylation and N-Deacetylation Control Different Cytokine Responses In Vivo
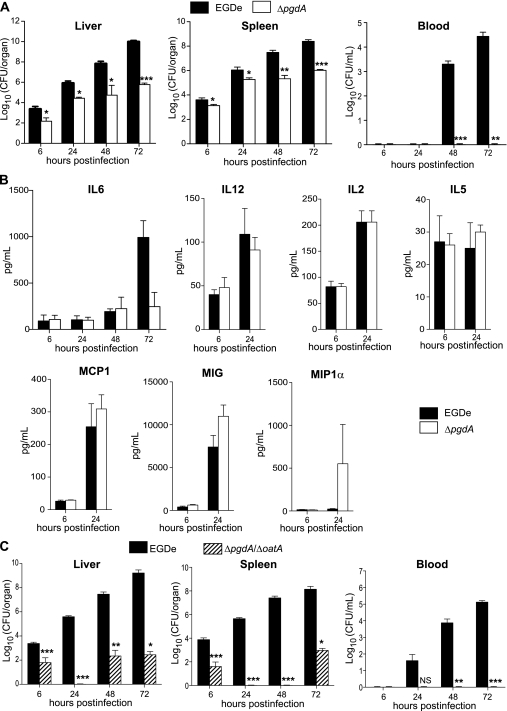
In order to test whether O-acetylation led to a specific cytokine profile or if another PGN modification could control production of the same cytokines, 21 cytokines were quantitated in the liver of mice infected with ΔpgdA, which does not produce PGN deacetylase. BALB/c mice were first injected intravenously with a sublethal dose of EGDe or ΔpgdA. Bacterial growth and cytokine production were determined in the liver at 6, 24, 48, and 72 hours after infection. Colonization of liver and spleen by ΔpgdA was strongly impaired compared with EGDe (Figure 4A). At 48 hours post infection, the number of ΔpgdA bacteria decreased by 3 and 2 log10 compared with EGDe in the liver and spleen, respectively. At 72 hours post infection with ΔpgdA, colonization decreased by 4 and 2 log10 in the liver and spleen, respectively. In contrast to EGDe, ΔpgdA could not be detected in the bloodstream (Figure 4A).
Figure 4.
The ΔpgdA mutant, while highly attenuated in virulence, does not induce IL6 and is more virulent than a ΔpgdAΔoatA double mutant. A, BALB/c mice were challenged by intravenous injection of a sublethal dose (104 bacteria per mouse) of the parental EGDe strain (black bars) and its isogenic ΔpgdA mutant (white bars). Colonization of liver, spleen, and blood was followed 6, 24, 48, and 72 hours postinfection. Data are means ± SD (n = 4). B, BALB/c mice were inoculated intravenously with 5 × 103 bacteria. The liver was dissected and homogenized. Homogenates were assayed to determine cytokine level in response to infection with the parental EGDe strain (black bars) or its isogenic ΔpgdA mutant (white bars). Data are means ± SD (n = 4). C, BALB/c mice were challenged by intravenous injection of a sublethal dose (104 bacteria per mouse) of the parental EGDe strain (black bars) and its isogenic ΔpgdAΔoatA mutant (hatched bars). Colonization of liver, spleen, and blood was followed at 6, 24, 48, and 72 hours postinfection. Data are means ± SD (n = 4). Student t test was performed to determine statistical significance: *, **, and *** indicate P < .05, P < .01, and P < .001, respectively.
Unlike ΔoatA, ΔpgdA did not induce IL-6 in the liver of infected mice (Figure 4B). Production of IL-6 in response to ΔpgdA was similar to that of EGDe until 48 hours post infection and lower at 72 hours. At 6 hours post infection, production of IL-12, IL-2, MCP-1, MIG, MIP-1α, and IL-5 was similar in the liver of mice infected by ΔpgdA and EGDe (Figure 4B). While strongly attenuated in its capacity to colonize the liver and spleen, as ΔpgdA, ΔoatA stimulates the production of a specific subset of cytokines, including IL-6. Thus, PGN O-acetylation and N-deacetylation represent 2 nonredundant modifications allowing Listeria to escape from innate immune response.
We hypothesized that the nonredundant functions of OatA and PgdA in immune escape could synergize and increase the capacity of Listeria to colonize the host. We thus created a strain of L. monocytogenes unable to produce OatA and PgdA and studied its virulence in vivo. BALB/c mice were infected intravenously with a sublethal dose of EGDe or ΔpgdAΔoatA. Growth of the double mutant was strongly impaired compared with that of EGDe in the liver and spleen (Figure 4C). Mutant bacteria could not be detected in the liver and spleen 24 hours post infection, while EGDe efficiently colonized both organs and single mutants had a defective but detectable capacity to colonize these organs (Figures 3B and 4A, 4C). At 48 hours post infection, ΔpgdAΔoatA was undetectable in the spleen and poorly colonized the liver compared with EGDe. At 72 hours post infection, there were 7−log10 and 5−log10 differences between the number of ΔpgdAΔoatA and EGDe in the liver and spleen, respectively. Furthermore, ΔpgdAΔoatA could not be detected in the bloodstream (Figure 4C). Overall, the virulence of the double mutant is more attenuated than that of single mutants. Thus, OatA and PgdA are nonredundant virulence determinants of L. monocytogenes that contribute to listeriosis in mice.
DISCUSSION
The composition of the L. monocytogenes cell wall and its role in virulence remain to be fully characterized. Here, we report that Listeria O-acetylates its PGN and that this modification is critical for virulence and escape from innate immune response. Analysis of the L. monocytogenes EGDe genome revealed a gene encoding a putative membrane-bound O-acetyltransferase encoded by the gene lmo1291. Comparison of PGN from EGDe and an lmo1291 deletion mutant confirmed that this gene, which we renamed oatA, encodes a PGN O-acetyltransferase. This is the second enzyme involved in Listeria PGN modification after PgdA, an enzyme that deacetylates N-acetylglucosamine residues [20].
Inactivation of oatA increased L. monocytogenes’ sensitivity to lysozyme, a major component of the innate defense that hydrolyzes PGN between N-acetylglucosamine and N-acetylmuramic acid residues [22]. We reported that PgdA confers resistance to lysozyme [20]. Interestingly, the ΔpgdA mutant is more sensitive to lysozyme than the ΔoatA mutant because it is killed by 10 μg/mL of lysozyme [20], a concentration that did not alter ΔoatA viability. Importantly, the OatA protective role was not specific to lysozyme because ΔoatA was more sensitive to cefotaxime and gallidermin, antibacterial compounds targeting the cell wall. Since ΔpgdA is not more sensitive to cefotaxime (data not shown), our results suggest that fine structural modifications of muropeptides change the sensitivity to different inducers of bacterial cell lysis and that the 2 modifications are not redundant.
Inactivation of oatA strongly reduced Listeria capacity to replicate in THP-1 and RAW264.7 cells and to survive in murine PEM and BMDM. In contrast, the mutant was not affected in its capacity to enter and survive in epithelial cells (data not shown). Inactivation of pgdA also severely impairs the ability of Listeria to survive and multiply in macrophages but not in epithelial cells [20]. However, while ΔpgdA accumulates in vacuoles after phagocytosis [20], ΔoatA does not. PGN modifications by OatA and PgdA thus protect Listeria from killing by phagocytes through nonredundant mechanisms, possibly by conferring different levels of resistance to cell wall–targeting molecules such as antimicrobial peptides and lysozyme or by conferring resistance to different spectra of host antimicrobial molecules.
OatA is critical for efficient host colonization by Listeria. Virulence of ΔoatA is highly attenuated in mice. The LD50 of ΔoatA was comparable to that of an hly mutant, one of the Listeria mutants whose virulence is the most attenuated [29]. OatA plays a role both at early and later stages of murine listeriosis, as indicated by host control of ΔoatA in intestinal lumen and tissue, the bloodstream, liver, and spleen. The other PGN modification, N-deacetylation, is also required for Listeria survival at early stages of infection and after crossing the intestinal barrier [20]. Thus, in Listeria, PGN modification enzymes are among the most important virulence factors.
Inactivation of OatA in L. monocytogenes amplified the magnitude of inflammatory cytokine and chemokine responses, but not interferon-β response, early after infection and, in the case of IL-6, at later stages of infection. Infection by ΔoatA and ΔpgdA led to different cytokine responses. Thus, OatA and PgdA have nonredundant roles in dampening inflammatory cytokine production. Several other L. monocytogenes virulence factors interfere with cytokine production. InlH, a cell-surface protein of the internalin family, controls IL-6 production during murine listeriosis by an unknown mechanism [30]. We recently reported that InlC, a secreted internalin, dampens production of MIP-1α and IL-8 homolog KC in infected mice by targeting the α subunit of IκB kinase [31]. These studies demonstrate that, as in other pathogenic bacteria, L. monocytogenes evades innate immune response through synthesis of multiple virulence determinants.
The oatA gene has been studied in S. aureus [28] and was shown to have a minor, although significant, role in bacterial survival in a murine model of skin infection [32]. Interestingly, S. aureus ΔoatA induces IL-1β secretion by infected macrophages in vitro and triggers an IL-1β- and IL-18-dependent skin inflammation after subcutaneous infection of mice [32]. L. monocytogenes ΔoatA did not increase IL-1β production in our murine model of infection. Thus, the magnitude of virulence defect of ΔoatA mutants and the spectrum of cytokine induced in response to infection with ΔoatA mutants vary between pathogens.
In conclusion, we have identified a new modification of L. monocytogenes PGN that is critical for infection. This modification relies on the O-acetyltransferase OatA and confers resistance to antimicrobial compounds, including lysozyme. OatA is required for intracellular survival of L. monocytogenes in macrophages and for the efficient control of cytokine response in vivo. Our study reveals that PGN O-acetylation is a novel mechanism used by Listeria to evade innate immunity and ultimately colonize the host successfully. It reinforces the notion that PGN is a pivotal component in microbe–host interactions.
Supplementary Data
Supplementary data are available at The Journal of Infectious Diseases online.
Funding
This work was supported by Institut Pasteur; Institut National de la Santé et de la Recherche Médicale; Institut National de la Recherche Agronomique; European Research Council (Advanced grant 233348 to P. C. and Starting grant 202283 to I. G. B.); Fondation Pasteur-Weizmann; Fondation Jeantet; and Fondation le Roch Les Mousquetaires. P. C. is an international research scholar of the Howard Hughes Medical Institute. C. A. is a doctoral fellow of the Ministère de l’Enseignement Supérieur et de la Recherche.
Acknowledgments
We thank Laurent Dortet for providing E-tests and for assistance with MIC studies, Marc Lecuit for providing hEcad transgenic mice, and Nadia Khelef for construction of the Listeria monocytogenes EGDe Δhly strain. We are grateful to Armelle Phalipon, Geneviève Milon, and members of the Cossart lab for insightful discussions.
References
- 1.Vazquez-Boland JA, Kuhn M, Berche P, et al. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pizarro-Cerda J, Cossart P. Listeria monocytogenes membrane trafficking and lifestyle: the exception or the rule? Annu Rev Cell Dev Biol. 2009;25:649–70. doi: 10.1146/annurev.cellbio.042308.113331. [DOI] [PubMed] [Google Scholar]
- 3.Hamon M, Bierne H, Cossart P. Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol. 2006;4:423–34. doi: 10.1038/nrmicro1413. [DOI] [PubMed] [Google Scholar]
- 4.Pamer EG. Immune responses to Listeria monocytogenes. Nat Rev Immunol. 2004;4:812–23. doi: 10.1038/nri1461. [DOI] [PubMed] [Google Scholar]
- 5.Stavru F, Archambaud C, Cossart P. Cell biology and immunology of Listeria monocytogenes infections: novel insights. Immunol Rev. 2011;240:160–84. doi: 10.1111/j.1600-065X.2010.00993.x. [DOI] [PubMed] [Google Scholar]
- 6.Conlan JW, North RJ. Neutrophils are essential for early anti-Listeria defense in the liver, but not in the spleen or peritoneal cavity, as revealed by a granulocyte-depleting monoclonal antibody. J Exp Med. 1994;179:259–68. doi: 10.1084/jem.179.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers HW, Unanue ER. Neutrophils are involved in acute, nonspecific resistance to Listeria monocytogenes in mice. Infect Immun. 1993;61:5090–6. doi: 10.1128/iai.61.12.5090-5096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mocci S, Dalrymple SA, Nishinakamura R, Murray R. The cytokine stew and innate resistance to L. monocytogenes. Immunol Rev. 1997;158:107–14. doi: 10.1111/j.1600-065x.1997.tb00996.x. [DOI] [PubMed] [Google Scholar]
- 9.Dalrymple SA, Lucian LA, Slattery R, et al. Interleukin-6-deficient mice are highly susceptible to Listeria monocytogenes infection: correlation with inefficient neutrophilia. Infect Immun. 1995;63:2262–8. doi: 10.1128/iai.63.6.2262-2268.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopf M, Baumann H, Freer G, et al. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature. 1994;368:339–42. doi: 10.1038/368339a0. [DOI] [PubMed] [Google Scholar]
- 11.Tripp CS, Wolf SF, Unanue ER. Interleukin 12 and tumor necrosis factor alpha are costimulators of interferon gamma production by natural killer cells in severe combined immunodeficiency mice with listeriosis, and interleukin 10 is a physiologic antagonist. Proc Natl Acad Sci USA. 1993;90:3725–9. doi: 10.1073/pnas.90.8.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai WJ, Bartens W, Kohler G, Hufnagel M, Kopf M, Brombacher F. Impaired macrophage listericidal and cytokine activities are responsible for the rapid death of Listeria monocytogenes–infected IFN-gamma receptor-deficient mice. J Immunol. 1997;158:5297–304. [PubMed] [Google Scholar]
- 13.Havell EA. Evidence that tumor necrosis factor has an important role in antibacterial resistance. J Immunol. 1989;143:2894–9. [PubMed] [Google Scholar]
- 14.Kato K, Nakane A, Minagawa T, et al. Human tumor necrosis factor increases the resistance against Listeria infection in mice. Med Microbiol Immunol. 1989;178:337–46. doi: 10.1007/BF00197452. [DOI] [PubMed] [Google Scholar]
- 15.Nakane A, Minagawa T, Kohanawa M, et al. Interactions between endogenous gamma interferon and tumor necrosis factor in host resistance against primary and secondary Listeria monocytogenes infections. Infect Immun. 1989;57:3331–7. doi: 10.1128/iai.57.11.3331-3337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfeffer K, Matsuyama T, Kundig TM, et al. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell. 1993;73:457–67. doi: 10.1016/0092-8674(93)90134-c. [DOI] [PubMed] [Google Scholar]
- 17.Rothe J, Lesslauer W, Lotscher H, et al. Mice lacking the tumor necrosis factor receptor 1 are resistant to TNF-mediated toxicity but highly susceptible to infection by Listeria monocytogenes. Nature. 1993;364:798–802. doi: 10.1038/364798a0. [DOI] [PubMed] [Google Scholar]
- 18.Harty JT, Bevan MJ. Specific immunity to Listeria monocytogenes in the absence of IFN gamma. Immunity. 1995;3:109–17. doi: 10.1016/1074-7613(95)90163-9. [DOI] [PubMed] [Google Scholar]
- 19.Diacovich L, Gorvel JP. Bacterial manipulation of innate immunity to promote infection. Nat Rev Microbiol. 2010;8:117–28. doi: 10.1038/nrmicro2295. [DOI] [PubMed] [Google Scholar]
- 20.Boneca IG, Dussurget O, Cabanes D, et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci USA. 2007;104:997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 22.Callewaert L, Michiels CW. Lysozymes in the animal kingdom. J Biosci. 2010;35:127–60. doi: 10.1007/s12038-010-0015-5. [DOI] [PubMed] [Google Scholar]
- 23.Glaser P, Frangeul L, Buchrieser C, et al. Comparative genomics of Listeria species. Science. 2001;294:849–52. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 24.Archambaud C, Gouin E, Pizarro-Cerda J, Cossart P, Dussurget O. Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence of Listeria monocytogenes. Mol Microbiol. 2005;56:383–96. doi: 10.1111/j.1365-2958.2005.04551.x. [DOI] [PubMed] [Google Scholar]
- 25.Archambaud C, Nahori MA, Pizarro-Cerda J, Cossart P, Dussurget O. Control of Listeria superoxide dismutase by phosphorylation. J Biol Chem. 2006;281:31812–22. doi: 10.1074/jbc.M606249200. [DOI] [PubMed] [Google Scholar]
- 26.Alford CE, King TE, Jr, Campbell PA. Role of transferrin, transferrin receptors, and iron in macrophage listericidal activity. J Exp Med. 1991;174:459–66. doi: 10.1084/jem.174.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dortet L, Mostowi S, Samba Louaka A, et al. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathogens. doi: 10.1371/journal.ppat.1002168. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bera A, Herbert S, Jakob A, Vollmer W, Gotz F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol Microbiol. 2005;55:778–87. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 29.Portnoy DA, Jacks PS, Hinrichs DJ. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med. 1988;167:1459–71. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Personnic N, Bruck S, Nahori MA, et al. The stress-induced virulence protein InlH controls interleukin-6 production during murine listeriosis. Infect Immun. 2010;78:1979–89. doi: 10.1128/IAI.01096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gouin E, Adib-Conquy M, Balestrino D, et al. The Listeria monocytogenes InlC protein interferes with innate immune responses by targeting the IκB kinase subunit IKKα. Proc Natl Acad Sci USA. 2010;107:17333–8. doi: 10.1073/pnas.1007765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimada T, Park BG, Wolf AJ, et al. Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1beta secretion. Cell Host Microbe. 2010;7:38–49. doi: 10.1016/j.chom.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]