Abstract
IsdI, a heme-degrading protein from Staphylococcus aureus, binds heme in a manner that distorts the normally planar heme prosthetic group to an extent greater than that observed so far for any other heme-binding protein. To understand better the relationship between this distinct structural characteristic and the functional properties of IsdI, spectroscopic, electrochemical, and crystallographic results are reported that provide evidence that this heme ruffling is essential to the catalytic activity of the protein and eliminates the need for the water cluster in the distal heme pocket that is essential for the activity of classical heme oxygenases. The lack of heme orientational disorder in 1H-NMR spectra of the protein argues that the catalytic formation of β- and δ-biliverdin in nearly equal yield results from the ability of the protein to attack opposite sides of the heme ring rather than from binding of the heme substrate in two alternative orientations.
Keywords: direct electrochemistry, NMR spectroscopy, X-ray crystallography
IsdG and IsdI are paralogues that catalyze protoheme IX degradation as components of the iron-regulated surface determinant (Isd) system of Staphylococcus aureus (1–4). Recently, the heme oxidation products formed by these enzymes have been shown to be a nearly equal mixture of 5-oxo-δ-bilirubin and 15-oxo-β-bilirubin (staphylobilins) (5). These unprecedented catalytic products distinguish these proteins from the widely distributed, well-studied, classic heme oxygenases (HOs) that in general catalyze formation of α-biliverdin exclusively (6, 7). The three-dimensional structure of IsdI (5) differs significantly from that of a classic HO (6, 8, 9). For IsdI, the electron density attributed to a dioxygen species bound to the active site iron is aligned with the plane of the coordinating imidazole that lies along the β, δ-axis of the porphyrin ring (5), and this orientation is restricted by steric hindrance from Phe22 and an H-bond formed with Asn6 (5). In contrast, the regiospecificity of classic HOs is achieved by steric restriction of the hydroperoxy ligand that forms during heme oxidation so that only the α-meso carbon is accessible for hydroxylation (6, 8, 9). In addition, the distal heme pockets of classic HOs also possess a group of ordered water molecules that direct protons to the catalytic center to stabilize the reactive hydroperoxy intermediate (8, 10–14). This water network is proposed to be required for catalysis by all classic HOs (11, 15–18), but it is absent from the active sites of IsdG and IsdI (5, 19). Therefore, the mechanisms of heme oxidation by the Isd enzymes and HOs must differ considerably.
The active sites of the Isd enzymes and classic HOs also differ significantly in that heme bound at the active sites of the Isd enzymes is highly ruffled (4, 5, 19). Normal-coordinate structural decomposition (NSD) analysis (20) reveals that the heme at the active sites of these enzymes is distorted 2.1 Å out of plane, far more than has been observed for any other heme-binding protein. This distinctive structural characteristic has been suggested to be a major factor in the mechanism by which these enzymes oxidize heme (4, 5, 19).
Studies of model heme derivatives reveal that the chemical and photophysical properties of heme are influenced by nonplanar structural distortion. Although heme ruffling is energetically unfavorable (21), most if not all heme proteins for which crystallographically determined structures are known exhibit some degree of heme distortion. Heme proteins or domains that exhibit significant heme distortion include the nitrophorins (approximately 0.8 Å) (22), heme nitric oxide/oxygen binding (H-NOX) domains (approximately 1 Å) (23), and c-type cytochromes (approximately 1 Å) (24), each of which exhibits distortion of the heme plane to the extent indicated. Heme structure distortion has been proposed to influence the reduction potentials (25–27), ligand binding affinities (22, 28–30), and spectroscopic characteristics (31) of these proteins, but systematic variation of the extent of heme distortion has been achieved only recently (32). Whereas out-of-plane distortion of model porphyrins by > 2 Å can be achieved by side-chain substitution or insertion of larger central metal ions, site-directed mutagenesis of proteins with distorted heme centers has so far resulted only in decreasing the extent of distortion. Through a combination of structural, spectroscopic, and functional studies of wild-type IsdI, we provide evidence that the remarkable nonplanarity of heme bound at the active site of IsdI changes the electronic structure of the bound heme in a manner that should significantly enhance susceptibility of the heme group to oxidation and formation of the unusual products that have been reported.
Results
Electronic Absorption Spectra.
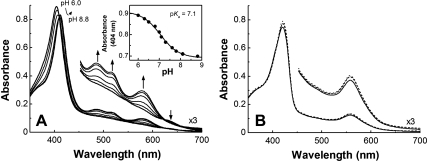
The pH-dependence of the electronic spectrum of the IsdIFe3+ (20 °C) is shown in Fig. 1A. At pH 6, the protein has a Soret maximum at 404 nm and no other apparent band in the visible region except for a weak transition at approximately 640 nm. At pH 9, this spectrum exhibits maxima at 412, 485, 520, and 578 nm, and the pKa for this acid-alkaline transition is 7.1 (Fig. 1A, Inset). The HOs, methemoglobin and metmyoglobin also exhibit pH-dependent spectra that are known to correspond to the transition between high-spin water bound form and low-spin hydroxide bound form. However, neither the spectra of low- nor high-pH forms of IsdIFe3+ resemble those of HOs or globins, except that the Soret maxima of both low- and high-pH forms of IsdI match those of other proteins (Table S1). In the absence of a relevant crystallographically defined structure of IsdIFe3+ to provide direct experimental information concerning the identity of a pH-dependent distal ligand, the simplest interpretation of the pH dependence of the visible spectrum of IsdI is the protonation/deprotonation of a distally coordinated water molecule. This hypothesis is supported by the observation of MCD spectra of IsdI at low- and high-pH that are similar to those of HO-1 (Fig. S1). The unusual absorption spectra shown in Fig. 1A could result from the ruffling of the heme that distorts the electronic structure of the heme. The absorption spectrum of the cyanide complex of IsdI is independent of pH and is similar to that of HOs and globins except for a slight red-shift in both the Soret and Q-bands (Fig. 1B) that is a common feature for ruffled model heme compounds (33).
Fig. 1.
(A) The pH dependence of absorption spectra of IsdIFe3+ in sodium phosphate buffer (20 mM, 20 °C). (Inset) The pH titration of IsdIFe3+ monitored at 404 nm. The line represents a nonlinear least-squares fit to a titration curve assuming n = 1. (B) Absorption spectra of IsdIFe3+CN at pH 6.0 (solid line), 7.0 (broken line), and 9.0 (dotted line).
NMR Spectroscopy.
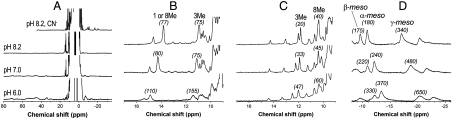
The 1H NMR spectra (30 °C) of IsdI at acidic pH [IsdIFe3+ (low-pH)], alkaline pH (IsdIFe3+(high-pH)) and with cyanide bound (IsdIFe3+CN) are shown in Fig. 2. At acidic pH, the paramagnetically shifted resonances of the heme substituents are observed in the range of 20–80 ppm, which is typical for high-spin heme and thus consistent with coordination of water to high-spin Fe3+ at the active site of IsdI.
Fig. 2.
(A) The 1H NMR spectrum of IsdIFe3+ as a function of pH (pH 6.0, 7.0, and 8.2 as indicated) and IsdIFe3+CN- (pH 8.2) in sodium phosphate buffer (20 mM, 10% D > 2O, 30 °C). (B–D) The 1H NMR spectra observed at 10 °C (Bottom), 20 °C (Middle), and 30 °C (Top). (B) Low-field region of IsdIFe3+ (high-pH). (C) Low-field region of IsdIFe3+CN. (D) High-field region of IsdIFe3+CN. Half line widths (Hz) of some signals are indicated in brackets.
At pH 8.2, the hyperfine-shifted proton resonances exhibited at acidic pH diminished in intensity and low-spin paramagnetically shifted signals appeared. Both high-spin and low-spin signals are observed at pH 7.0 (Fig. 2A) indicating that the pH-dependent transition of IsdI is slow on the NMR time scale. The two strong signals observed at 13.8 and 10.9 ppm at pH 8.2 are attributed to two heme methyl groups. 2D NMR experiments reveal that the other 2 heme methyl signals occur at 8.1 and 4.5 ppm (Fig. S2), and thus the average value of chemical shifts of four heme methyl signals was determined to be 9.3 ppm, a value considerably smaller than those (34) reported for other heme proteins (Table S2). This result indicates that the unpaired electron density on the heme methyl groups of IsdI is small. At lower temperature, all the paramagnetically shifted resonances of the heme substituents of IsdIFe3+(high-pH) exhibited considerable line broadening (Fig. 3A), consistent with chemical exchange at the active site. This line broadening was sufficiently problematic that none of heme meso-signals could be assigned for this form of the protein.
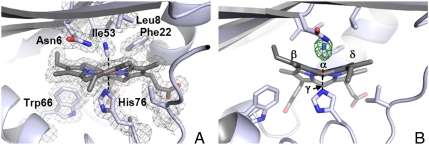
Fig. 3.
(A) Active site of IsdIFe3+CN. The 2Fo-Fc map is represented as a mesh contoured at 1 σ (gray). (B) The active site depicted as in (A) but in another orientation with the Fo-Fc omit map contoured at 3 σ (red). The heme meso-carbons are labeled.
In contrast to IsdIFe3+(high-pH), the chemical shifts for all the heme substituents of IsdIFe3+CN could be assigned (Fig. S3 and Table S3). The average chemical shift of four heme methyl groups was even smaller than that observed for IsdIFe3+(high-pH), and, surprisingly, broad meso-H signals were apparent at high field (-2–-25 ppm, Fig. 2 A and D). As well, the paramagnetically shifted resonances of IsdIFe3+CN exhibit much less line broadening at low temperature than observed for IsdIFe3+(high-pH) (Fig. 2 C and D).
Because IsdI produces a mixture of β- and δ-isomers of oxo-bilirubin in the ratio of 56.2 : 43.8 (5), it is reasonable to expect the occurrence of two heme insertion isomers that are rotated 180° about the α/γ-axis in the protein as observed in various HOs. However, only one orientation for heme binding at the active site of IsdI is apparent in these spectra. Consequently, catalysis of the heme oxygenase reaction must be possible on either side of the heme pocket.
X-Ray Crystal Structure of IsdIFe3+CN.
The X-ray crystal structure of IsdIFe3+CN was determined to 1.8 Å resolution with one homodimer in the asymmetric unit [Protein Data Bank (PDB) ID code 3QGP]. Overall, the IsdIFe3+CN structure retains the native protein fold and heme-binding mode. The homodimer forms a β-barrel at the dimeric interface, and within each subunit, heme is bound in a hydrophobic cleft such that the propionate groups are buried. The striking heme ruffling occurs to a greater extent than in the native structure with the out-of-plane rms displacement increasing from 2.1 to 2.3 Å. The heme pocket of IsdIFe3+CN with His76 as the heme-iron proximal ligand is shown in Fig. 3A. In the Fo-Fc omit map, elongated density was observed at the distal side of the heme and was assigned to a coordinated CN- with a ligand bond length of 1.9–2.0 Å in both active sites (Fig. 3B). The bound CN molecules refine with B-factors of approximately 12 Å2 similar to that of the heme iron. The CN ligand adopts a bent binding mode toward the heme with Fe-C-N angles of 171 to 158° for chains A and B, respectively. In both active sites, the bound cyanide ligand forms an H-bond to the ND1 atom of Asn6. The CN- is bound in a hydrophobic pocket and makes contacts with Leu8, Phe22, and Ile53 (Fig. 3A). Of these residues, Phe22 likely contributes to heme ruffling by contacting the γ-meso carbon. The extreme ruffling reduces the distance between N atom of bound CN to within 3.9 Å of the β and δ meso-carbons. Consistent with the NMR spectra collected at 30 °C, a single orientation for heme is observed in the crystal structure at 100 K. An alternative orientation in which the heme is rotated about the α/γ meso-carbon axis is unfavorable owing to steric interactions between the 4-vinyl group and the side-chains of Val79, Leu81, and Ile92 (Fig. S4).
Cyclic Voltammetry.
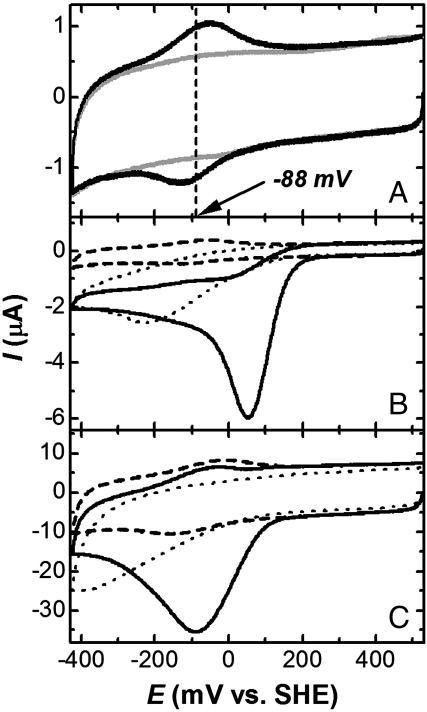
Cyclic voltammetry (CV) of IsdI (sodium phosphate buffer (20 mM), pH 7.0) under N2 purge yields a midpoint reduction potential (Em) of -88 ± mV vs. standard hydrogen electrode (SHE) (Fig. 4A) and represents a well-defined direct electrochemical response of a heme degrading enzyme. This Em value is similar to that for rat [-87 mV (35)] and human [-65 mV (36)] HO-1 measured by spectroelectrochemistry. Aerobic CV of IsdI (20 mV/ sec scan rate) in the same buffer exhibited a substantial reduction wave at approximately +50 mV (vs. SHE) (Fig. 4B) that was more intense at this lower scan rate (Fig. 4 B and C). In the absence of IsdI, a peak for direct reduction of dioxygen occurs below -200 mV, we attribute the intense reduction wave observed in the presence of IsdI and dioxygen to the catalytic reduction of O2 by IsdI. Similar catalytic reduction of dioxygen is also observed for the molten globule state myoglobin [pH 4.8 (37)] and hemoglobin and myoglobin entrapped in methyl cellulose films (38). For the globins, this catalytic peak is attributed to the following reactions:
Fig. 4.
(A) Direct electrochemistry of IsdI. Black, cyclic voltammogram of IsdI entrapped between the surface of a pyrollytic graphic electrode and a semipermeable membrane (sodium phosphate buffer (20 mM, pH 7.0, 20 °C). Scan rate, 50 mV/s. Gray, background measured with the same electrode and semipermeable membrane in the absence of IsdI. (B) CV of IsdI in the presence (black solid line) and absence (broken line) of O2 (20 mV/s). Dotted line, background obtained in the presence of O2 without IsdI. (C) Same as B but with a scan rate of 500 mV/s.
Similar reactions are expected to occur for IsdI, but it is not yet clear whether the third reaction is heme degradation rather than conversion of the oxyferrous complex to IsdIFe3+ and peroxide.
Discussion
Coordination Environment of the IsdI Heme Iron.
Electronic absorption and magnetic circular dichroism spectra (Fig 1A and Fig. S1) indicate that IsdIFe3+ possesses a distally coordinated water molecule at acidic pH that deprotonates with a pKa of 7.1 as pH is increased, a value considerably lower than that of HO-1 (pKa = 7.6(39)), HO-2 (pKa = 8.5(40)), Pseudomonas aeruginosa HO (pa-HO, pKa = 8.3(41)), and HmuO (pKa = 9.0(42)) but similar to that reported (pKa = 6.8(32)) for the H-NOX protein from Thermoanaerobacter tengcongensis. The two structural characteristics of the active site of IsdI that are most likely to account for this low pKa are (a) stabilization of the bound hydroxide through H-bond formation with the side-chain amide of Asn6 (5, 19) and (b) stabilization of the electrostatically neutral ferric-hydroxide heme iron center by the unusually hydrophobic nature of the heme-binding site. The 1H-NMR spectra obtained for IsdIFe3+ at pH 6, 7, and 8.2 (Fig. 2A) indicate that the interconversion of coordinated water and coordinated hydroxide is slow on the NMR time scale as previously reported for bovine myoglobin variants in which the distal His is replaced by Gln or Asn (43) and for two bacterial heme oxygenases (41, 44).
The red-shift of Soret and Q-band maxima observed for the IsdIFe3+CN complex (Fig. 1B and Table S1) is commonly observed for distorted model heme compounds, but attribution of this observation to heme ruffling, perturbation from side-chain substitutions or shortening of pyrrole-N to metal bond length has been debated (45–47). This latter possibility can be ruled out for the IsdIFe3+CN complex because the crystal structure indicates no change in this bond distance upon binding cyanide. Interestingly, this red-shift is greater for porphyrins with larger central metal ions (e.g., Ni2+, Zn2+) (47). The ruffling of heme bound to IsdI with neither metal substitution nor side-chain substitution may make spectroscopic characterization of IsdI reconstituted with various heme derivatives a useful approach to understanding better how structural distortion affects the spectroscopic properties of heme.
Electronic Structure of IsdI.
The chemical shifts of the heme methyl protons observed in 1H NMR spectra of IsdIFe3+CN are considerably smaller and the paramagnetic shifts observed for the meso-protons are considerably greater than those observed for other heme proteins. These observations indicate that the unpaired electron of the heme iron is more delocalized to the meso-positions than to the β-pyrrole positions. Spectroscopic analysis and density functional theory calculations of variously substituted ferriporphyrin and ferrichlorin model compounds indicate that the ruffling of a ferriheme stabilizes the dxz and dyz orbitals of the iron and changes the electron configuration of low-spin Fe3+ from the more common (dxy)2(dxz,dyz)3 to the less common (dxz,dyz)4(dxy)1 configuration (48–53). This change in configuration causes major changes in the 1H-NMR chemical shifts of the β-pyrrole substituent and meso-H signals of the heme (48–52). In ruffled heme, the pz orbital from the porphyrin 2a2u(π) orbital twists away from orthogonality with the porphyrin plane to permit interaction of the 2a2u(π) orbital with the 3dxy orbital of the iron. The 2a2u(π) orbital has large spin density at the nitrogens and meso-positions and very small spin density at the β-pyrrole positions. Thus, observation of a large paramagnetic shift for the meso-H and a small shift for heme methyl groups of IsdIFe3+CN is evidence that the (dxz,dyz)4(dxy)1 state dominates. Such a high-field shift of meso-H signals, which is the signature of the (dxz,dyz)4(dxy)1 configuration, has been reported only for model compounds and has never been observed for heme bound to a protein, presumably because the degree of heme distortion in proteins studied previously has been insufficient. As a result, IsdI provides an exceptional opportunity to explore the spectroscopic and functional consequences of heme ruffling on the properties of the active site of a heme enzyme.
The paramagnetic shifts of heme methyl signals in 1H NMR spectra of IsdIFe3+(high-pH) are greater than those observed for IsdIFe3+CN but are nevertheless considerably smaller than those of other heme proteins, indicating that IsdIFe3+(high-pH) retains a considerable (dxz,dyz)4(dxy)1 component. Detection of the chemical shift of meso-H signals of IsdIFe3+(high-pH) was prevented by exchange broadening. The EPR spectrum (X-band, 10 K) of IsdI(high-pH) (Fig. S5) exhibited g-values of 2.61, 2.24, and 1.78 that are very similar to those of HO-1 (g = 2.67, 2.21, 1.79) (39), indicating that the electron configurations of IsdIFe3+(high-pH) and HO-1 are very similar at cryogenic temperatures (i.e., (dxy)2(dxz,dyz)3). Thus, we propose that the electron configuration of IsdIFe3+ is in dynamic exchange between (dxy)2(dxz,dyz)3 and (dxz,dyz)4(dxy)1 and that this exchange accounts for the temperature-dependent signal broadening noted in the Results section (vide supra) as suggested for heme model compounds and pa-HO (54, 55). For IsdIFe3+CN, exchange broadening of heme signals is much smaller, suggesting that any exchange favors (dxz,dyz)4(dxy)1, consistent with stabilization of the (dxz,dyz)4(dxy)1 configuration by axial ligands that are strong π acceptors (51) and with the greater distortion of the heme from planarity observed in the crystal structure of the cyanide complex.
The (dxz,dyz)4(dxy)1 configuration is expected to be significant for IsdI catalysis because delocalization of the single, unpaired dxy electron has been proposed to increase susceptibility of the meso-position to oxidative attack (31, 54, 55). Although structures reported for HOs exhibit less distortion (approximately 0.5 Å) of the heme than seen for IsdI, 13C NMR analysis of pa-HO suggested that the alkaline form of this enzyme nevertheless exhibits approximately 2% of a (dxz,dyz)4(dxy)1 component (41, 54). Thus, we propose that it is the increased stabilization of the (dxz,dyz)4(dxy)1 state at the active site of IsdI that enables the oxidation of heme without assistance of an ordered water cluster as required by classical HOs.
Electrochemical Properties of IsdI.
Many structural factors have been proposed to control the Em of heme proteins (e.g., the identity of the axial ligands to the heme iron, H-bonding interactions of the axial ligands and heme propionate groups, and the dielectric of the heme binding environment) (56). The proximal His of IsdI forms an H-bond with the main chain carbonyl of Phe73, whereas proximal His residues of rat and human HO-1 form an H-bond with the side-chain of Glu29 (9, 57). Glu is a better electron donor than a main chain carbonyl, so this factor favors a more positive Em for IsdI relative to HOs. Ordered waters dominate the distal heme pockets of HOs whereas the same region of IsdI is very hydrophobic, so the dielectric environments of the hemes of these proteins favor a lower potential for HOs. The similarity of the Em we observe for IsdI with those reported for HOs, therefore, provides evidence that heme ruffling, the remaining significant structural difference between the active sites of these proteins, lowers the potential of IsdI to the range observed for HOs. This interpretation is consistent with studies of model heme compounds (33) and heme proteins (22, 27, 32).
Interestingly, the Em value of IsdI obtained under N2 purge (-88 mV) is more negative than that of ascorbate, and the potential of the catalytic reduction peak seen aerobically is similar to the Em of ascorbate. Thus, the catalytic wave observed for IsdI in the presence of O2 is consistent with the inability of ascorbate to reduce IsdI in the absence of O2 and with heme degradation following aerobic addition of ascorbate to IsdIFe3+ (1, 19).
Materials and Methods
Sample Preparation.
Recombinant IsdI was expressed, purified, and reconstituted with heme as reported (1, 19), and IsdIFe3+CN was prepared by addition of excess potassium cyanide (KCN).
Electronic Absorption Spectroscopy.
The electronic spectra of IsdI (approximately 10 μM) were recorded in sodium phosphate buffer (20 mM, 20 °C) with a Cary Model 6000i spectrophotometer equipped with a Peltier thermoregulator. Solution pH was adjusted with small volumes of NaOH or HCl (0.1–0.5 M).
The 1H NMR spectra.
1D 1H NMR spectra were collected with a Varian INOVA 500 MHz spectrometer. IsdI samples (approximately 1 mM) were prepared in sodium phosphate buffer (20 mM, 10% D2O), and presaturation was used to suppress the solvent signal. Chemical shifts are given in ppm downfield from sodium 2,2-dimethyl-2-silapentane-5-sulfonate with H2O.
X-ray Crystallography.
IsdIFe3+CN (10 mg/mL) in 10 mM NaCN, 20 mM Tris, pH 7.5, and 200 mM NaCl was mixed with an equal volume of reservoir solution (0.2 M MgCl2, 0.1 M Bis-Tris, pH 5.5, and 25% w/v PEG 3350). Crystals of the IsdIFe3+CN complex were bright red in color and formed after 3 d at room temperature. Crystals were dipped in reservoir solution supplemented with 10% ethylene glycol as a cryoprotectant and immersed in liquid nitrogen prior to data collection at 100 K. Diffraction data was collected at Stanford Synchrotron Radiation Lightsource (SSRL) on beamline 7-1 and processed with iMosflm (58). Initial phases were obtained with Molrep (59, 60) using the native IsdIFe3+ structure (PDB ID code 3LGN) as the search model. Crystallographic refinement and model building was done using the programs Refmac5 (61) and Coot (62), respectively. Data collection and structure refinement statistics are provided in Supporting Information (Table S4). The complete peptide chain was modeled, residues -1 (including an Ala-His cloning artifact) to 108. Refinement of the heme groups required the use of a modified stereochemical library for the heme as described previously (19). No angle or distance restraints were used for the iron-cyanide ligand bond. A total of 252 water molecules and 2 Mg2+ ions were modeled around the protein. The extent of heme ruffling in the final IsdIFe3+CN structure was analyzed using the NSD) program (20). Graphic renderings of the structure were created with PyMOL (Schrödinger).
Cyclic Voltammetry.
CV was performed with an Autolab PGSTAT12 potentiostat-galvanostat (Eco Chemie) using an edge-plane pyrrolytic carbon electrode (PGE) polished with alumina slurry and then sonicated in deionized water for 1 min. IsdI solution (3 μL, approximately 300 μM) was spread evenly with a microsyringe onto the surface of the PGE. The PGE surface was then covered with a semipermeable membrane and used as the working electrode. An SCE electrode and a Pt wire were employed as the reference and counter electrodes, respectively. All experiments were performed (20 °C) in sodium phosphate buffer (20 mM, pH 7.0). For anaerobic measurements, the buffer was purged (≥10 m) with N2 prior to data collection.
Supplementary Material
Acknowledgments.
We thank Professor Lawrence McIntosh for use of the NMR spectrometer and Fred Rosell for help in collection of the EPR and MCD spectra. This work was supported by a postdoctoral fellowship from the Uehara Memorial Foundation (S.J.T.), a Canada Research Chair (A.G.M.), and a Canadian Blood Services–Canadian Institutes of Health Research partnership grant (A.G.M.), CIHR operating Grant MOP-49597 (M.E.P.M.), and the Canadian Foundation for Innovation (A.G.M.). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the US Department of Energy (USDE), Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the USDE, Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3QGP).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101459108/-/DCSupplemental.
References
- 1.Skaar EP, Gaspar AH, Schneewind O. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J Biol Chem. 2004;279:436–443. doi: 10.1074/jbc.M307952200. [DOI] [PubMed] [Google Scholar]
- 2.Reniere ML, Skaar EP. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol Microbiol. 2008;69:1304–1315. doi: 10.1111/j.1365-2958.2008.06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu R, et al. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J Biol Chem. 2005;280:2840–2846. doi: 10.1074/jbc.M409526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grigg JC, Ukpabi G, Gaudin CFM, Murphy MEP. Structural biology of heme binding in theStaphylococcus aureus Isd system. J Inorg Biochem. 2010;104:341–348. doi: 10.1016/j.jinorgbio.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 5.Reniere ML, et al. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol Microbiol. 2010;75:1529–1538. doi: 10.1111/j.1365-2958.2010.07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unno M, Matsui T, Ikeda-Saito M. Structure and catalytic mechanism of heme oxygenase. Nat Prod Rep. 2007;24:553–570. doi: 10.1039/b604180a. [DOI] [PubMed] [Google Scholar]
- 7.Matsui T, Unno M, Ikeda-Saito M. Heme oxygenase reveals its strategy for catalyzing three successive oxygenation reactions. Acc Chem Res. 2010;43:240–247. doi: 10.1021/ar9001685. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Syvitski RT, Chu GC, Ikeda-Saito M, Mar GNL. Solution 1H NMR investigation of the active site molecular and electronic structures of substrate-bound, cyanide-inhibited HmuO, a bacterial heme oxygenase from Corynebacterium diphtheriae. J Biol Chem. 2003;278:6651–6663. doi: 10.1074/jbc.M211249200. [DOI] [PubMed] [Google Scholar]
- 9.Schuller DJ, Wilks A, Ortiz de Montellano PR, Poulos TL. Crystal structure of human heme oxygenase-1. Nat Struct Mol Biol. 1999;6:860–867. doi: 10.1038/12319. [DOI] [PubMed] [Google Scholar]
- 10.Friedman J, et al. Crystal structures of the NO- and CO-bound heme oxygenase from Neisseriae meningitidis. J Biol Chem. 2003;278:34654–34659. doi: 10.1074/jbc.M302985200. [DOI] [PubMed] [Google Scholar]
- 11.Lad L, et al. Crystal structures of the ferric, ferrous, and ferrous-NO forms of the Asp140Ala mutant of human heme oxygenase-1: Catalytic implications. J Mol Biol. 2003;330:527–538. doi: 10.1016/s0022-2836(03)00578-3. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Syvitski RT, Auclair K, Ortiz de Montellano P, La Mar GN. Solution 1H, 15N NMR spectroscopic characterization of substrate-bound, cyanide-inhibited human heme oxygenase: Water occupation of the distal cavity. J Am Chem Soc. 2003;125:13392–13403. doi: 10.1021/ja036176t. [DOI] [PubMed] [Google Scholar]
- 13.Gorst CM, Wilks A, Yeh DC, Ortiz de Montellano PR, La Mar GN. Solution 1H NMR investigation of the molecular and electronic structure of the active site of substrate-bound human heme oxygenase: The nature of the distal hydrogen bond donor to bound ligands. J Am Chem Soc. 1998;120:8875–8884. [Google Scholar]
- 14.Peng D, et al. Coupling of the distal hydrogen bond network to the exogenous ligand in substrate-bound, resting state human heme oxygenase. Biochemistry. 2009;48:11231–11242. doi: 10.1021/bi901216s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsui T, Furukawa M, Unno M, Tomita T, Ikeda-Saito M. Roles of distal Asp in heme oxygenase from Corynebacterium diphtheriae, HmuO. J Biol Chem. 2005;280:2981–2989. doi: 10.1074/jbc.M410263200. [DOI] [PubMed] [Google Scholar]
- 16.Schuller DJ, Zhu W, Stojiljkovic I, Wilks A, Poulos TL. Crystal structure of heme oxygenase from the Gram-negative pathogen Neisseria meningitidis and a comparison with mammalian heme oxygenase-1. Biochemistry. 2001;40:11552–11558. doi: 10.1021/bi0110239. [DOI] [PubMed] [Google Scholar]
- 17.Chen H, Moreau Y, Derat E, Shaik S. Quantum mechanical/molecular mechanical study of mechanisms of heme degradation by the enzyme heme oxygenase: The strategic function of the water cluster. J Am Chem Soc. 2008;130:1953–1965. doi: 10.1021/ja076679p. [DOI] [PubMed] [Google Scholar]
- 18.Sugishima M, et al. Crystal structure of rat heme oxygenase-1 in complex with heme bound to azide. Implication for regiospecific hydroxylation of heme at the α-meso carbon. J Biol Chem. 2002;277:45086–45090. doi: 10.1074/jbc.M207267200. [DOI] [PubMed] [Google Scholar]
- 19.Lee WC, Reniere ML, Skaar EP, Murphy MEP. Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J Biol Chem. 2008;283:30957–30963. doi: 10.1074/jbc.M709486200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jentzen W, Song X-Z, Shelnutt JA. Structural characterization of synthetic and protein-bound porphyrins in terms of the lowest-frequency normal coordinates of the macrocycle. J Phys Chem B. 1997;101:1684–1699. [Google Scholar]
- 21.Anderson KK, et al. Planar-nonplanar conformational equilibrium in metal derivatives of octaethylporphyrin and meso-nitrooctaethylporphyrin. J Am Chem Soc. 1993;115:12346–12352. [Google Scholar]
- 22.Roberts SA, et al. Ligand-induced heme ruffling and bent NO geometry in ultra-high-resolution structures of Nitrophorin 4. Biochemistry. 2001;40:11327–11337. doi: 10.1021/bi0109257. [DOI] [PubMed] [Google Scholar]
- 23.Pellicena P, Karow DS, Boon EM, Marletta MA, Kuriyan J. Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases. Proc Natl Acad Sci USA. 2004;101:12854–12859. doi: 10.1073/pnas.0405188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jentzen W, Ma J-G, Shelnutt JA. Conservation of the conformation of the porphyrin macrocycle in hemoproteins. Biophys J. 1998;74:753–763. doi: 10.1016/S0006-3495(98)74000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olea C, Boon EM, Pellicena P, Kuriyan J, Marletta MA. Probing the function of heme distortion in the H-NOX family. Chem Biol. 2008;3:703–710. doi: 10.1021/cb800185h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shokhireva TK, et al. Electrochemical and NMR spectroscopic studies of distal pocket mutants of nitrophorin 2: Stability, structure, and dynamics of axial ligand complexes. Proc Natl Acad Sci USA. 2003;100:3778–3783. doi: 10.1073/pnas.0536641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma J-G, et al. The structural origin of nonplanar heme distortions in tetraheme ferricytochromes c3. Biochemistry. 1998;37:12431–12442. doi: 10.1021/bi981189i. [DOI] [PubMed] [Google Scholar]
- 28.Benabbas A, et al. Ultrafast dynamics of diatomic ligand binding to nitrophorin 4. J Am Chem Soc. 2010;132:2811–2820. doi: 10.1021/ja910005b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shokhireva TK, Berry RE, Zhang H, Shokhirev NV, Walker FA. Assignment of ferriheme resonances for high- and low-spin forms of nitrophorin 3 by 1H and 13C NMR spectroscopy and comparison to nitrophorin 2: Heme pocket structural similarities and differences. Inorg Chim Acta. 2008;361:925–940. doi: 10.1016/j.ica.2007.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheesman MR, et al. Two enzymes with a common function but different heme ligands in the forms as isolated. Optical and magnetic properties of the heme groups in the oxidized forms of nitrite reductase, cytochrome cd1, from Pseudomonas stutzeri and Thiosphaera pantotropha. Biochemistry. 1997;36:16267–16276. doi: 10.1021/bi971677a. [DOI] [PubMed] [Google Scholar]
- 31.Walker FA. Magnetic spectroscopic (EPR, ESEEM, Mössbauer MCD and NMR) studies of low-spin ferriheme centers and their corresponding heme proteins. Coord Chem Rev. 1999;185-186:471–534. [Google Scholar]
- 32.Olea C, Kuriyan J, Marletta MA. Modulating heme redox potential through protein-induced porphyrin distortion. J Am Chem Soc. 2010;132:12794–12795. doi: 10.1021/ja106252b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shelnutt JA, et al. Nonplanar porphyrins and their significance in proteins. Chem Soc Rev. 1998;27:31–41. [Google Scholar]
- 34.Shokhirev NV, Walker FA. The effect of axial ligand plane orientation on the contact and pseudocontact shifts of low-spin ferriheme proteins. J Biol Inorg Chem. 1998;3:581–594. [Google Scholar]
- 35.Sato H, et al. Electrochemical reduction of ferrous α-verdoheme in complex with heme oxygenase-1. J Inorg Biochem. 2007;101:1394–1399. doi: 10.1016/j.jinorgbio.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, et al. Replacement of the proximal histidine iron ligand by a cysteine or tyrosine converts heme oxygenase to an oxidase. Biochemistry. 1999;38:3733–3743. doi: 10.1021/bi982707s. [DOI] [PubMed] [Google Scholar]
- 37.Onuoha AC, Zu X, Rusling JF. Electrochemical generation and reactions of ferrylmyoglobins in water and microemulsions. J Am Chem Soc. 1997;119:3979–3986. [Google Scholar]
- 38.Li Y-M, Liu H-H, Pang D-W. Direct electrochemistry and catalysis of heme-proteins entrapped in methyl cellulose films. J Electroanal Chem. 2004;574:23–31. [Google Scholar]
- 39.Takahashi S, et al. Heme–heme oxygenase complex. Structure of the catalytic site and its implication for oxygen activation. J Biol Chem. 1994;269:1010–1014. [PubMed] [Google Scholar]
- 40.Ishikawa K, et al. Heme oxygenase-2. J Biol Chem. 1995;270:6345–6350. doi: 10.1074/jbc.270.11.6345. [DOI] [PubMed] [Google Scholar]
- 41.Caignan GA, et al. The hydroxide complex of Pseudomonas aeruginosa heme oxygenase as a model of the low-spin iron(III) hydroperoxide intermediate in heme catabolism: 13C NMR spectroscopic studies suggest the active participation of the heme in macrocycle hydroxylation. J Am Chem Soc. 2003;125:11842–11852. doi: 10.1021/ja036147i. [DOI] [PubMed] [Google Scholar]
- 42.Chu GC, Tomita T, Sönnichsen FD, Yoshida T, Ikeda-Saito M. The heme complex of HmuO a bacterial heme degradation enzyme from Corynebacterium diphtheriae. J Biol Chem. 1999;274:24490–24496. doi: 10.1074/jbc.274.35.24490. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto Y, Suzuki T, Hori H. Dynamics and thermodynamics of acid-alkaline transitions in metmyoglobins lacking the usual distal histidine residue. Biochim Biophys Acta. 1993;1203:267–275. doi: 10.1016/0167-4838(93)90093-7. [DOI] [PubMed] [Google Scholar]
- 44.Ma LH, Liu Y, Zhang X, Yoshida T, La Mar GN. 1H NMR study of the magnetic properties and electronic structure of the hydroxide complex of substrate-bound heme oxygenase from Neisseria meningitidis: influence of the axial water deprotonation on the distal H-bond network. J Am Chem Soc. 2006;128:6657–6668. doi: 10.1021/ja0584626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Z, Cao C, Liu Q, Jiang R. Hybrid orbital deformation (HOD) effect and spectral red-shift property of nonplanar porphyrin. Org Lett. 2010;12:1780–1783. doi: 10.1021/ol100434w. [DOI] [PubMed] [Google Scholar]
- 46.Haddad RE, et al. Origin of the red shifts in the optical absorption bands of nonplanar tetraalkylporphyrins. J Am Chem Soc. 2003;125:1253–1268. doi: 10.1021/ja0280933. [DOI] [PubMed] [Google Scholar]
- 47.Ryeng H, Ghosh A. Do nonplanar distortions of porphyrins bring about strongly red-shifted electronic spectra? Controversy, consensus, new developments, and relevance to chelatases. J Am Chem Soc. 2002;124:8099–8103. doi: 10.1021/ja0202659. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura M. Electronic structures of highly deformed iron(III) porphyrin complexes. Coord Chem Rev. 2006;250:2271–2294. [Google Scholar]
- 49.Cai S, Shokhireva TK, Lichtenberger DL, Walker FA. NMR and EPR studies of chloroiron(III) tetraphenyl-chlorin and its complexes with imidazoles and pyridines of widely differing basicities. Inorg Chem. 2006;45:3519–3531. doi: 10.1021/ic0515352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yatsunyk LA, Shokhirev NV, Walker FA. Magnetic resonance spectroscopic investigations of the electronic ground and excited states in strongly nonplanar iron(III) dodecasubstituted porphyrins. Inorg Chem. 2005;44:2848–2866. doi: 10.1021/ic049089q. [DOI] [PubMed] [Google Scholar]
- 51.Walker FA, et al. π-Acid ligands in iron(III) porphyrinates. Characterization of low-spin bis(tert-butylisocyanide)(porphyrinato)iron(III) complexes having ((dxz,dyz)4(dxy)1 ground states. J Am Chem Soc. 1996;118:12109–12118. [Google Scholar]
- 52.Safo MK, et al. Models of the cytochromes b. Low-spin bis-ligated (porphinato)iron(III) complexes with unusual molecular structures and NMR, EPR, and Mössbauer spectra. J Am Chem Soc. 1992;114:7066–7075. [Google Scholar]
- 53.Cheesman MR, Walker FA. Low-temperature MCD studies of low-spin ferric complexes of tetramesitylporphyrinate: Evidence for the novel (dxz,dyz)4(dxy)1 ground state which models the spectroscopic properties of heme d. J Am Chem Soc. 1996;118:7373–7380. [Google Scholar]
- 54.Rivera M, Zeng Y. Heme oxygenase, steering dioxygen activation toward heme hydroxylation. J Inorg Biochem. 2005;99:337–354. doi: 10.1016/j.jinorgbio.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 55.Rivera M, et al. Models of the low-spin iron(III) hydroperoxide intermediate of heme oxygenase: Magnetic resonance evidence for thermodynamic stabilization of the dxy electronic state at ambient temperatures. J Am Chem Soc. 2002;124:6077–6089. doi: 10.1021/ja017334o. [DOI] [PubMed] [Google Scholar]
- 56.Mauk AG, Moore GR. Control of metalloprotein redox potentials: What does site-directed mutagenesis of hemoproteins tell us? J Biol Inorg Chem. 1997;2:119–125. [Google Scholar]
- 57.Sugishima M, et al. Crystal structure of rat heme oxygenase-1 in complex with heme. FEBS Lett. 2000;471:61–66. doi: 10.1016/s0014-5793(00)01353-3. [DOI] [PubMed] [Google Scholar]
- 58.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography No. 26. 1992 [Google Scholar]
- 59.Collaborative Computational Project N. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 60.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 61.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 62.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.