Abstract
Current models of developmental evolution suggest changes in gene regulation underlie the evolution of morphology. Despite the fact that protein complexes regulate gene expression, the evolution of regulatory protein complexes is rarely studied. Here, we investigate the evolution of a protein-protein interaction (PPI) between Homeobox A11 (HoxA11) and Forkhead box 01A (Foxo1a). Using extant and “resurrected” ancestral proteins, we show that the physical interaction between HoxA11 and Foxo1a originated in the mammalian stem lineage. Functional divergence tests and coimmunoprecipitation with heterologous protein pairs indicate that the evolution of interaction was attributable to changes in HoxA11, and deletion studies demonstrate that the interaction interface is located in the homeodomain region of HoxA11. However, there are no changes in amino acid sequence in the homeodomain region during this time period, indicating that the origin of the derived PPI was attributable to changes outside the binding interface. We infer that the amino acid substitutions in HoxA11 altered Foxo1a's access to the conserved binding interface at the HoxA11 homeodomain. We also found an expansion in the number of paired Hox/Fox binding sites in the genomes of mammalian lineage species suggesting the complex has a biological function. Our data indicate that the physical interaction between HoxA11 and Foxo1a evolved through noninterface changes that facilitate the PPI, which prevents inappropriate interactions, rather than through the evolution of a novel binding interface. We speculate that evolutionary changes of intramolecular regulation have limited pleiotropic effects compared with changes to interaction domains themselves.
Keywords: protein-protein interaction evolution, transcription factor evolution
Changes in gene regulation are the driving force in the origin and evolution of novel phenotypes. Gene expression is coordinated by the formation of multiprotein complexes that bind to cis-regulatory promoter and enhancer regions for target genes and activate or repress transcription in a signal-dependent fashion (1–3). There is strong evidence that changes in both cis-regulatory elements and regulatory proteins, such as transcription factors, have led to gene regulatory evolution (4–12). However, the mechanisms by which transcription factor evolution affects gene regulation are poorly understood. For example, it has been suggested that the evolution of novel protein-protein interactions (PPIs), posttranslational modifications, and DNA- and ligand-binding specificities may all contribute to the origin of regulatory activities in transcription factors; however, to date, few studies have carefully dissected potential mechanisms.
Here, we address this question by investigating the evolution of the physical interaction between two transcription factors, Homeobox A11 (HoxA11) and Forkhead box 01A (Foxo1a), which play a major role in regulating gene expression in endometrial stromal cells during pregnancy in placental mammals (8, 13). By examining the ability of a series of extant and resurrected ancestral HoxA11 and Foxo1a proteins and testing their ability to interact physically and functionally, we found that the PPI evolved before the origin of cooperative transcriptional activation. Further, we show that the derived PPI did not originate through the evolution of a new protein-protein binding interface; rather, this interaction depends on the unmasking of an ancestral interaction site within the highly conserved homeodomain of HoxA11.
These results suggest that an important mechanism of transcription factor evolution may be regulating the accessibility of existing PPI sites, potentially increasing the combinatorial complexity of transcription factor interactions while minimizing the negative pleiotropic effects of the newly allowed protein interactions.
Results
HoxA11 Evolved Rapidly in the Stem Lineage of Mammals.
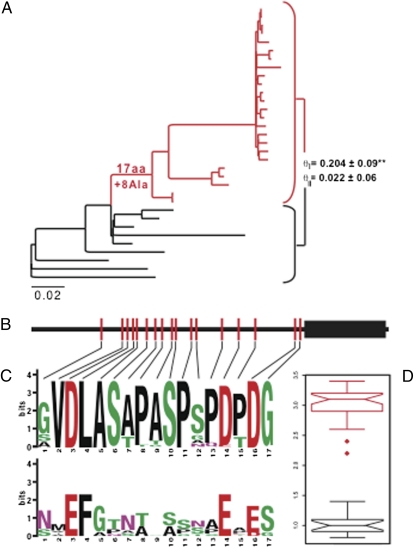
Previous studies of HoxA11 molecular evolution identified a period of rapid evolution in the stem lineage of placental mammals (Eutheria) that occurred coincident with the evolution of a novel functional interaction between HoxA11 and Foxo1A (8). To determine if HoxA11 evolved rapidly in other lineages potentially associated with the evolution of the physical interaction between these proteins, we used maximum-likelihood and parsimony to reconstruct the history of amino acid substitutions and insertion-deletion, respectively, using a larger dataset of HoxA11 genes. We found that although most lineages accumulated relatively few amino acid changes in HoxA11, 17 amino acid substitutions and eight alanine insertions occurred in the stem lineage of mammals (Fig. 1A), all of which occur outside the well-conserved DNA-binding homeodomain (Fig. 1B). The derived amino acids are well conserved within mammals but variable in nonmammals, suggesting that HoxA11 gained additional selective constraints in the stem lineage of mammals (Fig. 1 C and D). To test whether these amino acid substitutions evolve under different selection pressures in mammals and nonmammals, we estimated Gu's coefficient of functional divergence (θ) for HoxA11 across 33 jawed vertebrates (gnathostomes). Rejection of the null model (θ = 0) is evidence for the acquisition or loss of structural and/or functional constraints, such as the gain of a novel interaction partner (14, 15). Although HoxA11 generally evolves under strong constraints within vertebrates, we identified an episode of strong type I functional divergence between mammals and nonmammals (θI = 0.204 ± 0.09; P < 0.01; Fig. 1A) but found no evidence of type II functional divergence (θII = 0.022 ± 0.06; Fig. 1A). These data suggest that mammalian HoxA11 acquired additional functional constraints (type I divergence) rather than changed constraints on amino acids that were already ancestrally constrained (type II divergence).
Fig. 1.
HoxA11 evolved rapidly in the stem lineage of mammals. (A) Phylogenetic tree shows the relationships of species used in this study (species names are provided in Table S1); mammals are shown in red, and nonmammals are shown in black. Branch lengths are shown proportional to the number of nonsynonymous substitutions per codon. Seventeen amino acid substitutions and eight alanine insertions occurred in the stem lineage of mammals. Ala, alanine. Estimates of Gu's type I (θI) and type II (θII) functional divergence between mammals and nonmammals indicate there was a significant gain of constraint within mammals (θII P < 0.01) **P < 0.02. (B) Cartoon of mammalian-specific amino acid substitutions in HoxA11; note that no amino acid changes occurred in the DNA-binding homeodomain (black rectangle) or the C-terminal tail. (C) Sequence logos of the 17 amino acid sites substituted in the stem lineage of mammals from mammals (Upper) and nonmammals (Lower). Lines indicate the position of amino acid changes in HoxA11. (D) Box plots shows median and variation in bit scores of amino acids shown in C from mammals (red) and nonmammals (black), with higher bit scores indicating greater conservation. Note that the 17 amino acids substituted in the stem lineage of mammals are much more conserved within mammals than they are in other species.
Using the same tests, we found no evidence of type I or type II functional divergence in Foxo1A in the stem lineage of mammals. Together, these findings indicate that selection in the stem lineage of mammals recruited weakly constrained amino acids in HoxA11 into a novel function, rather than coopting amino acids with existing structural or functional constraints (8, 14, 15). These results also suggest that the function of HoxA11 may have been altered in the stem lineage of mammals, whereas Foxo1a has been functionally conserved.
Reconstruction and Expression of Ancestral Proteins.
To test directly whether the amino acid changes in HoxA11 enabled a novel PPI with Foxo1A, we reconstructed and synthesized ancestral HoxA11 and Foxo1A proteins and tested their ability to interact physically (16–21). We used a maximum-likelihood method that implements Bayes empirical Bayes inference of character states to reconstruct the ancestral eutherian, ancestral therian, ancestral mammalian, and ancestral amniote HoxA11 and Foxo1A sequences (22). We found that the reconstructed genes had a mean Bayesian posterior probability of >0.93 and were similar to parsimony-based reconstructions (Table S2). Ancestral genes were synthesized (GeneScript Corp.) with human optimized codon use and ligated into pcDNA3.1(+)-V5/His (HoxA11 sequences) or pcDNA3.1(+)-Flag (Foxo1a sequences).
A concern when working with resurrected proteins is that they might not function in the way a native protein does (e.g., will have no biochemical activity) because errors in the reconstruction may lead to defects in protein synthesis, folding, or function (17). We ensured that the ancestral proteins were expressed, properly localized to the nucleus, and functionally active using Western blots of nuclear lysate and luciferase reporter assays, respectively. Western blots of nuclear lysate from HeLa cells transfected with constructs expressing epitope-tagged ancestral proteins confirmed that each protein was expressed at a similar level as transfected epitope tagged native proteins, that they remained soluble, and that they localized to the nucleus (Fig. 2 and Figs. S1 and S2). In addition, the ancestral proteins functioned in luciferase assays like extant proteins, indicating that they are functionally active (Fig. S3). Together, these results show that the ancestrally reconstructed proteins, like the native proteins, are expressed, soluble, and biologically active.
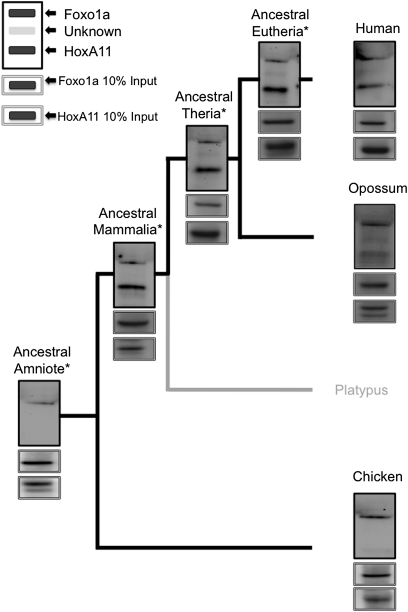
Fig. 2.
Physical interaction between HoxA11 and Foxo1a originated in the mammalian stem lineage. Note the presence of a band for human, opossum, ancestral eutherian, ancestral therian, and ancestral mammalian HoxA11-V5/His, but there is no band for chicken or ancestral amniote HoxA11-V5/His, indicating that the physical interaction arose in the mammalian stem lineage. The legend in the upper right corner of the figure indicates location/identity of bands shown in panels, including unknown peptides (observed in some panels) that cross-react with the antibody. Asterisks (*) indicate reconstructed ancestral proteins. Flag epitope-tagged Foxo1a and V5/His epitope-tagged HoxA11 mammalian expression vectors were cotransfected into HeLa cells, and nuclear lysates were incubated with anti-Flag agarose (Sigma) overnight. The following day, samples were treated with DNaseI and washed, and protein complexes were eluted with NuPage LDS sample buffer. Protein complexes were resolved by SDS/PAGE and transferred to a PDVF membrane. Blots were probed with anti-Flag (1:100,000; Sigma) and anti-V5 (1:5,000; Invitrogen) antibodies. Experiments were repeated a minimum of three times, and the results presented here are representative of typical findings.
Foxo1a-HoxA11 PPI Evolved in the Mammalian Stem Lineage.
We have previously shown that human HoxA11 and Foxo1a physically interact and that the interaction is independent of DNA binding (13) (Fig. S4). Tests with in vitro transcribed/translated proteins indicate that the interaction is direct and does not require a third “bridging” molecule (Fig. S5). To determine when the PPI between HoxA11 and Foxo1a evolved, we tested whether HoxA11 and Foxo1a from several extant species interact using coimmunoprecipitation (CoIP). We found that the human and opossum Foxo1a proteins were able to capture HoxA11, whereas the chicken Foxo1a was unable to capture chicken HoxA11. Even in chicken fibroblast cells, chicken HoxA11 and Foxo1a failed to interact (Fig. S6). We conclude that the lack of interaction between chicken HoxA11 and Foxo1a is unlikely to be an artifact of using a human cell line. These results suggest that the physical interaction between HoxA11 and Foxo1a evolved in basal mammals (i.e., before the most recent common ancestor of marsupials and placentals and after the amniote ancestor). Unfortunately, dating when in mammalian evolution the HoxA11/Foxo1a interaction originated was not possible using proteins from extant species because we were unable to identify the platypus Foxo1a homolog after extensive searches of the platypus genome or trace reads. Thus, the HoxA11/Foxo1a interaction evolved in either the stem lineage of mammals or therian mammals, or was lost during the evolution of chickens.
To define more precisely when the HoxA11/Foxo1a interaction evolved, we tested whether reconstructed ancestral eutherian, ancestral therian, ancestral mammalian, and ancestral amniote HoxA11 and Foxo1A proteins were able to interact using CoIP. Like the extant human and opossum proteins, we found that the ancestral eutherian, ancestral therian, and ancestral mammalian Foxo1as interacted with their respective HoxA11 proteins. However, as was observed with the chicken proteins (ancestral amniote), Foxo1a protein did not pull down the ancestral amniote HoxA11 protein (Fig. 2). Therefore, the PPI between HoxA11 and Foxo1a likely arose in the stem lineage of mammals, coincident with the episode of rapid change in HoxA11 described above.
Derived Interaction Between HoxA11 and Foxo1 Is Attributable to Changes in HoxA11.
Our sequence analysis indicates that HoxA11 evolved rapidly in the stem lineage of mammals and has the statistical signature of adaptive sequence change, whereas Foxo1a did not show signs of adaptive evolution. These results suggest that the new PPI likely resulted from changes in HoxA11 rather than changes in Foxo1a or changes in both HoxA11 and Foxo1a. To test this inference, we investigated whether heterologous pairs of proteins were able to interact. First, we tested the ability of human HoxA11 to interact with all possible variants of Foxo1a. As shown in Fig. S1, human HoxA11 was able to interact with all extant and ancestral Foxo1a variants, indicating that the ability of Foxo1a to interact with HoxA11 is ancestral to amniotes. Conversely, human Foxo1a was only able to interact with mammalian lineage HoxA11 proteins, similar to the results observed when species paired proteins were tested (Fig. S2, compare with Fig. 2). These results demonstrate that the evolution of the HoxA11/Foxo1a interaction results entirely from changes in HoxA11 alone, consistent with the conclusion from the evolutionary sequence analysis described above.
HoxA11 Binds to Foxo1a via the Homeodomain.
The comparative analysis of the HoxA11/Foxo1a interaction presented above demonstrates that changes in HoxA11 were responsible for the interaction with Foxo1a. To map the region of HoxA11 that mediates the interaction with Foxo1a, we constructed truncated mouse HoxA11 proteins and tested their ability to interact with Foxo1a. We found that full-length mouse HoxA11 (amino acids 1–313) and a protein fragment that contains only the homeodomain plus the C-terminal tail of 10 amino acids (amino acids 242–313, henceforth referred to as the “homeodomain region”) were both able to interact with human Foxo1a. However, despite being properly expressed and soluble, the homeodomain-deleted HoxA11 (amino acids 1–241) peptide was unable to interact with Foxo1a (Fig. 3). Thus, the evolutionarily conserved homeodomain region binds to Foxo1a. This observation is particularly unexpected, because no amino acid substitutions occurred in the homeodomain, or in the 10-aa C-terminal tail, in the stem lineage of mammals that could explain the origin of the new PPI (Fig. 1B). These results indicate that amino acid changes in HoxA11 allow an ancestral interaction potential with Foxo1a to occur rather than generating an entirely new interaction surface.
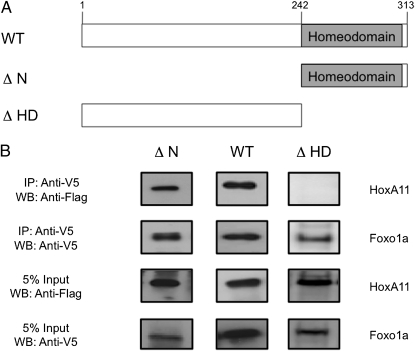
Fig. 3.
Homeodomain is sufficient to bind Foxo1a. (A) Cartoon of mouse HoxA11 deletion constructs used to determine the Foxo1a binding region. WT, full-length mouse HoxA11 (amino acids 1–313); ΔN, homeodomain plus 10 conserved amino acids (amino acids 242–313); ΔHD, homeodomain deleted (amino acids 1–241). (B) Deletion constructs were tested for their ability to coprecipitate with human Foxo1a. Flag epitope-tagged full-length HoxA11 (amino acids 1–313), homeodomain deleted (amino acids 1–241), and N-terminal deletion (amino acids 242–313) constructs were tested with V5/His-tagged Foxo1a. The presence of a WT and ΔN but no band for ΔHD after precipitation with Foxo1a, indicates that the C-terminal portion of the HoxA11 protein, which is composed of the homeodomain and 10 additional conserved amino acids, is sufficient to support the PPI with Foxo1a. Five percent input bands indicate equal expression of all constructs. Mammalian expression vectors were cotransfected into HeLa cells, and nuclear lysates were incubated with anti-V5 agarose (Sigma) overnight. The following day, samples were treated with DNaseI and washed, and protein complexes were eluted with NuPage LDS sample buffer. Protein complexes were resolved by SDS/PAGE and transferred to a PDVF membrane. Blots were probed with anti-Flag (1:100,000; Sigma) and anti-V5 (1:5,000; Invitrogen) antibodies. Experiments were repeated a minimum of three times, and results presented here are representative of typical findings. WB, Western blot.
Stepwise Evolution of the HoxA11/Foxo1a Regulatory Complex.
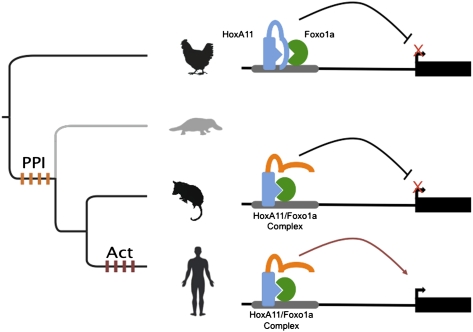
We have previously shown that HoxA11 from eutherian (placental) mammals but not other species functionally cooperates with Foxo1a to activate gene expression from the decidual prolactin enhancer (8). However, the results presented here demonstrate that the physical interaction between HoxA11 and Foxo1a originated in the mammalian stem lineage, predating the derived functional interaction in eutherian mammals and suggesting that the cooperative transactivating ability of these transcription factors evolved from an ancestral physical interaction.
To test if the origin of the HoxA11/Foxo1a interaction had consequences for their gene regulatory function, we cotransfected Hox/Fox pairs from reconstructed and extant species into HeLa cells with the luciferase reporter vector 3×IRS[luc/3×IRS]-pGL2, which contains the SV40 promoter and three repeats of the insulin response sequence (IRS) from the enhancer of insulin-like growth factor binding protein 1 (IGFBP1); this sequence drives transcription of the luciferase reporter gene luc in response to binding of Foxo1A and Abd-B type Hox genes, such as HoxA11. We found that Foxo1a proteins from all extant species and reconstructed ancestors act as transcriptional activators of the 3×IRS enhancer, indicating that the regulatory function of Foxo1a is conserved within amniotes (Fig. 4 and Fig. S3). Consistent with its function as an intrinsic repressor (23), HoxA11 repressed luciferase expression from the 3×IRS promoter, although the strength of repression varied by species (Fig. 4 and Fig. S3).
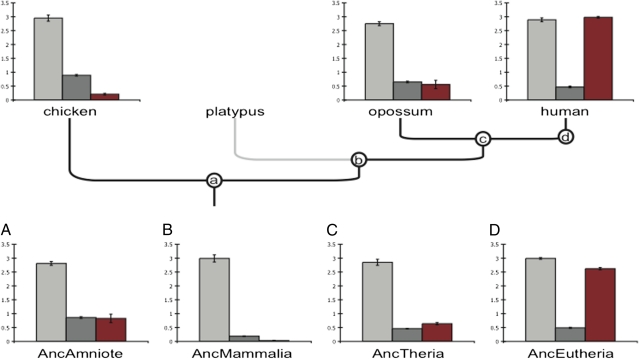
Fig. 4.
Step-wise evolution of Foxo1a/HoxA11 cooperative gene regulation. The gene regulatory function of Foxo1a and HoxA11 genes from extant and reconstructed ancestors were tested in luciferase reporter assays using a composite promoter with three pairs of Fox/Hox binding sites (3×IRS). Foxo1a genes alone activated reporter gene expression similarly across all species (light gray bars). HoxA11 genes from all species repressed reporter gene expression, but the strength of repression was variable across species (dark gray bars). Cotransfection of Foxo1a and HoxA11 (red bars) jointly repressed reporter gene expression in species other than human and the ancestral eutherian reconstructed proteins (AncEutheria), whereas cotransfection of human Foxo1a/HoxA11 and the ancestral eutherian reconstructed Foxo1a/HoxA11 genes (red bars) jointly activated reporter gene expression. Expression levels are shown as fold changes relative to luciferase expression in cells transfected with the reporter gene (3×IRS) and empty vector (y axes) (n = 4, mean ± SEM). Anc, ancestral. a, b, c, d, ancestral nodes for Amniote, Mammalia, Theria, and Eutherian, respectively. (A–D) luciferase assay results for resurrected genes corresponding to the lettered (a, b, c, d) nodes.
Coexpression of human or ancestral eutherian HoxA11/Foxo1a up-regulated luciferase expression from the 3×IRS reporter, whereas all other HoxA11/Foxo1a protein pairs repressed luciferase expression, indicating that transcriptional activation is a derived trait in eutherian mammals (Fig. 4). However, unlike the cooperative interaction observed at the decidual prolactin (PRL) enhancer (8, 13), cooperative effects were not observed on the 3×IRS enhancer. For example, the ancestral amniote Foxo1a and HoxA11 genes up-regulated luciferase expression 2.81-fold and down-regulated luciferase expression 0.86-fold, respectively, whereas coexpression down-regulated luciferase expression 0.83-fold. Similarly, the ancestral eutherian Foxo1a and HoxA11 genes up-regulated luciferase expression 2.99-fold and down-regulated luciferase expression 0.49-fold, respectively, whereas coexpression up-regulated luciferase expression 2.62-fold. Neither the repression of luciferase expression by the ancestral amniote proteins nor the activation by the ancestral eutherian proteins was greater than the repression observed for HoxA11 alone or the activation observed for Foxo1a alone. Furthermore, the activating function of Foxo1a is dominant over the repressive function of HoxA11, although when noneutherian proteins are tested, the repressive function of HoxA11 is dominant over the activating of Foxo1a. The evolution of the PPI itself does not change the functional activity of these transcription factors, at least not on the two promoters tested, decidual PRL (dPRL) and 3×IRS. These results also indicate that cooperative transcriptional activation is promoter-dependent. Finally, the results also suggest that full transcriptional activation, such as that observed at the PRL enhancer, requires the recruitment of additional factors not recruited to the 3×IRS construct.
Genome-Wide Expansion of Paired Hox-Foxo1a Binding Sites.
If the derived PPI between HoxA11 and Foxo1A is relevant for their regulatory functions, the abundance of paired Hox/Fox binding sites should increase after the PPI evolved. To test this prediction, we determined the density of paired binding sites in the genomes of 25 mammals, lineages with the HoxA11-Foxo1a physical interaction, and eight nonmammal species, lineages without the physical interaction. We found that the density of paired Hox/Fox binding sites in the genomes of nonmammalian species is 57% lower (P = 0.04, Wilcoxon rank sum test) than the density of paired sites in mammalian species (Fig. S7). This pattern, however, was not observed for a paired binding site density in permuted DNA sequences, suggesting that there was an increase in paired binding site density coincidental with the origin of the PPI. Finally, because the expansion in paired binding sites and the origin of the PPI both occurred before the origin of the functional cooperativity at the prolactin locus, it is very likely that the HoxA11-Foxo1a interaction has some other as yet unknown function that goes back to the stem lineage of mammals.
Discussion
There is an emerging consensus that gene regulation evolves through changes in cis-regulatory elements as well as in transcription factors (4–12). Although it is clear how nucleotide substitutions in cis-regulatory elements affect gene expression, it is not clear how amino acid substitutions in transcription factors influence gene regulation. Here, we explored the evolutionary history of the physical interaction between HoxA11/Foxo1a.
Intramolecular Regulatory Changes, and Not Binding Interface Changes, Lead to Derived PPI.
We observed the PPI between HoxA11 and Foxo1a in all mammalian proteins tested, including the extant human and opossum proteins and the resurrected ancestral mammalian, ancestral therian, and ancestral eutherian proteins. Conversely, we were unable to detect an interaction between the chicken and the ancestral amniote proteins. The most parsimonious interpretation of these data is that the PPI between HoxA11 and Foxo1a evolved in the stem lineage of mammals. This result implies that the derived functional cooperativity between HoxA11 and Foxo1a, which evolved in placental mammals, is not a direct consequence of a newly evolved PPI between these transcription factors. Thus, although the PPI is derived, it is many million years older than the transactivating cooperativity between HoxA11 and Foxo1a observed on the decidual enhancer of prolactin.
To determine if changes in HoxA11, Foxo1a, or both proteins were responsible for their derived physical interaction, we performed CoIP experiments that tested the ability of human HoxA11 to interact with heterologous Foxo1a proteins and found that only the mammalian HoxA11 proteins coprecipitated with Foxo1a. Thus, the physical PPI is dependent on the selectivity of HoxA11 and not on Foxo1a. These results suggest that evolutionary changes in the HoxA11 protein led to the derived PPI rather than changes in Foxo1a or in both HoxA11 and Foxo1a. HoxA11 evolved relatively rapidly in the stem lineage of mammals, accumulating 17 amino acid substitutions and eight alanine insertions; thus, there is a substantial amount of amino acid sequence evolution coincidental with the origin of the PPI, although Foxo1a remained functionally conserved. This result is reminiscent of previous findings that the evolution of functional cooperativity was attributable to changes in the HoxA11 protein rather than the Foxo1a protein (8).
Foxo1a is an important hub protein (i.e., a protein with many more interactions than other proteins), with roles in cell proliferation, differentiation, metabolic responses, and apoptosis (24–28). These functions of Foxo1a are specified by its numerous interaction partners to direct the amplitude of the transcriptional response as well as to determine which target genes will be activated/repressed (28). Hub proteins, such as Foxo1a, evolve less readily than proteins with fewer interactions (29–32). Therefore, our finding that changes in Foxo1a played no role in the evolution of the interaction with HoxA11 is consistent with the view that hub proteins tend to be evolutionarily conserved. Furthermore, our results suggest that novel biochemical functions of protein complexes can evolve through adaptation of one member of the protein complex rather than coevolution of both proteins.
One of our more surprising findings is that the PPI between HoxA11 and Foxo1a is mediated by the homeodomain of HoxA11, which is invariant among the amniotes tested in this study. This finding indicates that the derived PPI is not attributable to the evolution of a novel protein-binding interface on the HoxA11 protein. Instead, all amniote HoxA11 proteins are potentially capable of interacting with Foxo1a proteins through their homeodomain, but this inherent potential is masked by the less conserved region amino-terminal to the homeodomain in nonmammalian species. Indeed, another forkhead protein (Foxa2) has been shown to interact with the homeodomains of many transcription factors, such as Engrailed 2, HoxA5, Gsc, and Lim1 (33), through its conserved forkhead domain. These independent findings suggest that the interaction interface between forkhead and homeodomain proteins is very old. Thus, homeodomain proteins must have a regulatory mechanism for selecting if their potential to bind a given forkhead family protein is allowed or not. Within our system, all HoxA11 proteins are potentially capable of interacting with Foxo1a because the interaction occurs at the invariant homeodomain; however, because only mammalian HoxA11 sequences allow the interaction, it is likely that the interaction is prevented in HoxA11 from nonmammals by structural differences in the region N-terminal to the homeodomain (Fig. 5).
Fig. 5.
Model of the evolution of the HoxA11-Foxo1a physical interaction. In the ancestral state, HoxA11 (blue protein) was unable to interact with Foxo1a (green protein) despite the presence of an interaction interface present on each protein (blue triangle, green antitriangle). We believe the interaction interface is masked by N-terminal regions of HoxA11 (blue lines) and that selection in the mammalian stem lineage for amino acid substitutions caused a structural change (orange lines) that unmasks the binding interface.
We can envision two potential mechanisms: (i) the recruitment of a cofactor by ancestral HoxA11 that indirectly blocks the interaction with Foxo1a or (ii) masking of the homeodomain by parts of the HoxA11 peptide itself that prevents Foxo1a's access to the homeodomain (e.g., the N-terminal portion of ancestral HoxA11 is positioned over the binding interface). In either case, amino acid changes in the stem lineage of mammals unmask a preexisting ability of the homeodomain to interact with Foxo1a rather than generating a new interaction site. The evolution of a specific PPI between HoxA11 and Foxo1a is potentially another example of intramolecular regulation of transcription factor protein activities. For instance, both the DNA-binding affinity and specificity of the Ubx protein are influenced by intramolecular interactions between the homeodomain and the rest of the molecule (34, 35). It is interesting that these “regulatory” interactions are linked to intrinsically unstructured parts of the protein (34, 36, 37) in many cases, because the amino-terminal region of HoxA11 is predicted to contain two unstructured regions (38) (Fig. S8).
One of the main objections against the idea that transcription factor protein evolution may play an important role in gene regulatory network evolution is the assumption that transcription factor function is not modular, meaning that any functionally important changes to the protein will affect many, if not all, of the pleiotropic functions that transcription factors have (11, 12). For instance, it is thought that a mutation affecting DNA binding will affect its activity in all tissues and cell types in which this transcription factor is functional. Our results show that this reasoning is misleading. Specific biochemical functions of transcription factor proteins are not passively expressed but are under context-sensitive regulation. The fact that transcription factor proteins are able to show context-sensitive deployment of certain biochemical activities (39) and that evolution of transcription factor function can be achieved through the evolution of these regulatory functions (this study) shows how the extent of pleiotropy of amino acid substitutions can be limited to certain cellular and developmental contexts. Intramolecular regulation of transcription factor activity may limit the pleiotropic effects of protein coding changes, and thus greatly enhance the evolvability of transcription factors.
PPI Evolution Can Lead to Transcriptional Rewiring.
There is a long-standing debate in evolutionary and developmental biology about the gene regulatory mechanisms of acquiring novelty. One idea is that new cis-regulatory regions are created, which serve to rewire transcription (10, 40). Alternatively, it has been proposed that the acquisition of novel PPI can lead to novelty via recruitment of multiple target genes into regulatory networks (9, 41). This suggests that a transcription factor (TFa) evolves a PPI with a second transcription factor (TFb). Later, secondary mutations to the DNA create a paired binding site in the DNA sequence for TFb (41). Consistent with this idea, we found the coincidental origin of a PPI between two transcription factors and an expansion in the frequency of their paired binding sites. It is currently not known if the origin of the PPI drove selection for the evolution of paired binding sites, or vice versa. Together, the origin of a physical interaction between two transcription factors and the increase in the number of their binding sites may provide a mechanism for system-wide transcriptional rewiring.
Materials and Methods
Cell Lines and Culture.
Human endometrial stromal cells, immortalized with telomerase (catalog no. CRL-4003; American Type Culture Collection) were grown in steroid-depleted DMEM supplemented with 5% (vol/vol) charcoal-stripped FBS and 1% antibiotic/antimycotic (ABAM). ES cells were decidualized by treatment with 0.5 mM 8-BrcAMP (Sigma) and 1 mM medroxyprogesterone acetate. HeLa cells were cultured in DMEM with 4.5 g/L glucose and l-glutamine supplemented with 5% (vol/vol) FBS and 1% ABAM. Chicken fibroblast cells, (UMNSAH/DF-1, catalog no. CRL-12203; American Type Culture Collection) were cultured in DMEM with 4.5 g/L glucose and l-glutamine supplemented with 10% (vol/vol) FBS and 1% ABAM.
Reporter Constructs and Expression Vectors.
Full-length human, opossum, and chicken HoxA11 was cloned into the pcDNA3.1/V5-His Topo TA expression vector (Invitrogen) to create a HoxA11-V5/His fusion protein. Mouse deletion constructs described previously (23) were cloned into pcDNA 3 (Invitrogen) to create an N-terminal Flag epitope-tagged fusion protein via BamHI and XbaI cut sites. N-terminal Flag-tagged human Foxo1a was purchased from Addgene (plasmid 13507). N-terminal Flag-tagged opossum and chicken Foxo1a was cloned into pcDNA3.1. V5/His-tagged ancestral HoxA11 and Flag-tagged ancestral Foxo1a proteins were synthesized by GeneScript Corp. based on reconstructed ancestral sequences.
Molecular Evolutionary Analysis of HoxA11 and Foxo1a Ancestral Sequence Reconstruction.
HoxA11 and Foxo1A genes were identified from BLAST/BLAST-like alignment tool (BLAT) searches of whole-genome databases at the National Center for Biotechnology Information, University of California, Santa Cruz genome browser, and ENSEMBL (included species are provided in Table S1). Functional divergence was tested with DIVERGE 2.0 (42).
Ancestral sequences were inferred with CODEML in PAML4 (43), which uses maximum likelihood and an empirical Bayes approach to estimate ancestral character states. The Bayesian posterior probabilities of the reconstructed sequences were 0.93/0.95 for the ancestral amniote genes (shown as HoxA11/Foxo1A), 0.98/0.97 for ancestral mammalian genes, 0.96/0.97 for ancestral therian genes, and 1.0/0.97 for ancestral eutherian genes (Table S2). Ancestral genes were synthesized by GeneScript Corp. with human optimized codon use and ligated into pcDNA3.1(+)-V5/His.
CoIP Assay.
In vivo coIP assays were conducted as described previously (13). Briefly, HeLa cells were transfected with HoxA11-V5/His and either empty pcDNA3.1(+) vector or Flag-Foxo1A using Lipofectamine 2000 (Invitrogen). UMNSAH/DF-1 cells were transfected using TransIT-LT1 (Mirus). Two days after transfection, cells were harvested and cleared nuclear lysate was prepared. Nuclear lysate for each sample was mixed with 40 mL of M2 anti-FLAG agarose beads (Sigma) prewashed in TNT buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.05% Triton X-100] and rotated overnight at 4 °C. Samples were treated with 50 U DNaseI (Roche) and 2.5 μg RNase (Roche) for 60 min to remove confounding DNA. After washing to remove nonspecific binding proteins, protein complexes were eluted in NuPage LDS sample buffer (Invitrogen) and resolved on SDS/PAGE gels, transferred to a PVDF membrane using standard methods, and probed for HoxA11 with anti–V5-HRP antibody (1:5,000; Invitrogen) and Foxo1a with anti–FLAG-HRP antibody (1:100,000; Sigma). Bands were visualized using an Immun-Star WesternC chemiluminescence kit (Bio-Rad). Each experiment was repeated a minimum of three times.
In vitro coIP assays were conducted similarly using the TNT T7/T3-coupled wheat germ extract system (Promega) from the same vectors described above following the manufacturer's suggested protocol. In vitro translated protein reactions were divided between single-protein (e.g., HoxA11 only) and double-protein (e.g., HoxA11, Foxo1a) reactions and diluted to 500 μL using modified BF1 buffer (20 mM Tris HCl (pH 7.4), 100 mM NaCl, 1 mM EDTA, 10% glycerol, 0.01% Triton X-100, 0.5 mM DTT) (33). Samples were incubated overnight with anti-Flag beads; they were then washed and visualized as described above.
Luciferase Assays.
HeLa cells were grown in 96-well opaque culture plates in DMEM supplemented with 5% (vol/vol) FBS. Cells were transiently transfected with TransIT-LT1 according to the manufacturer's protocol with 2 ng of the Renilla control vector (pGL4.71) and 50 ng of the luciferase reporter construct 3×IRS-pGL2, which contains three copies of the paired Foxo1A/Hox site (13511; Addgene). Depending on treatment, cells were cotransfected with either 200 ng of empty pcDNA3.1(+)-V5/His, 100 ng of the Foxo1A expression vector, or 100 ng of one of the HoxA11 expression vectors. In addition, the effect of Foxo1A alone and HoxA11 alone on reporter gene expression was assayed by transfection with 2 ng of the Renilla control vector (pGL4.71), 50 ng of the 3×IRS-pGL2 reporter plasmid, 100 ng of empty pcDNA3.1(+)-V5/His, and either 100 ng of Foxo1A or 100 ng of HoxA11. Luciferase expression was assayed 48 h after transfection using the Dual Luciferase Reporter System (Promega). Each experiment was repeated four times, with eight replicates per experiment.
Finding Paired Hox-Fox Binding Sites.
Paired Hox-Fox binding sites were identified in the genomes of 22 mammals and 9 nonmammals (a list of genomes tested is provided in Table S3). Sites were identified using custom PERL scripts and the regular expression TTT[AT] ([AT]CAAA[AT] in both + and − configurations).
Supplementary Material
Acknowledgments
We thank J. J. Roth for providing mouse HoxA11 deletion expression vectors. This work was supported by a grant from the John Templeton Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 12977.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100990108/-/DCSupplemental.
References
- 1.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 2.Wilkins AS. The Evolution of Developmental Pathways. Sunderland, MA: Sinauer; 2002. [Google Scholar]
- 3.Smith CL, O'Malley BW. Coregulator function: A key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 4.Ronshaugen M, McGinnis N, McGinnis W. Hox protein mutation and macroevolution of the insect body plan. Nature. 2002;415:914–917. doi: 10.1038/nature716. [DOI] [PubMed] [Google Scholar]
- 5.Galant R, Carroll SB. Evolution of a transcriptional repression domain in an insect Hox protein. Nature. 2002;415:910–913. doi: 10.1038/nature717. [DOI] [PubMed] [Google Scholar]
- 6.Löhr U, Yussa M, Pick L. Drosophila fushi tarazu. A gene on the border of homeotic function. Curr Biol. 2001;11:1403–1412. doi: 10.1016/s0960-9822(01)00443-2. [DOI] [PubMed] [Google Scholar]
- 7.Lynch VJ, Wagner GP. Revisiting a classic example of transcription factor functional equivalence: Are Eyeless and Pax6 functionally equivalent or divergent? J Exp Zool B Mol Dev Evol. 2010;316B:93–98. doi: 10.1002/jez.b.21373. [DOI] [PubMed] [Google Scholar]
- 8.Lynch VJ, et al. Adaptive changes in the transcription factor HoxA-11 are essential for the evolution of pregnancy in mammals. Proc Natl Acad Sci USA. 2008;105:14928–14933. doi: 10.1073/pnas.0802355105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch VJ, Wagner GP. Resurrecting the role of transcription factor change in developmental evolution. Evolution. 2008;62:2131–2154. doi: 10.1111/j.1558-5646.2008.00440.x. [DOI] [PubMed] [Google Scholar]
- 10.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 11.Carroll SB. Evolution at two levels: On genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prud'homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch VJ, Brayer K, Gellersen B, Wagner GP. HoxA-11 and FOXO1A cooperate to regulate decidual prolactin expression: Towards inferring the core transcriptional regulators of decidual genes. PLoS ONE. 2009;4:e6845. doi: 10.1371/journal.pone.0006845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu X. Functional divergence in protein (family) sequence evolution. Genetica. 2003;118:133–141. [PubMed] [Google Scholar]
- 15.Gu X. A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Mol Biol Evol. 2006;23:1937–1945. doi: 10.1093/molbev/msl056. [DOI] [PubMed] [Google Scholar]
- 16.Jermann TM, Opitz JG, Stackhouse J, Benner SA. Reconstructing the evolutionary history of the artiodactyl ribonuclease superfamily. Nature. 1995;374:57–59. doi: 10.1038/374057a0. [DOI] [PubMed] [Google Scholar]
- 17.Thornton JW. Resurrecting ancient genes: Experimental analysis of extinct molecules. Nat Rev Genet. 2004;5:366–375. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- 18.Shi YS, Yokoyama S. Molecular analysis of the evolutionary significance of ultraviolet vision in vertebrates. Proc Natl Acad Sci USA. 2003;100:8308–8313. doi: 10.1073/pnas.1532535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: The functional synthesis. Nat Rev Genet. 2007;8:675–688. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liberles DA, editor. Ancestral Sequence Reconstruction. New York, NY: Oxford University Press; 2007. [Google Scholar]
- 21.Yokoyama S. Evolution of dim-light and color vision pigments. Annu Rev Genomics Hum Genet. 2008;9:259–282. doi: 10.1146/annurev.genom.9.081307.164228. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 23.Roth JJ, Breitenbach M, Wagner GP. Repressor domain and nuclear localization signal of the murine Hoxa-11 protein are located in the homeodomain: No evidence for role of poly alanine stretches in transcriptional repression. J Exp Zoolog B Mol Dev Evol. 2005;304:468–475. doi: 10.1002/jez.b.21061. [DOI] [PubMed] [Google Scholar]
- 24.Carlsson P, Mahlapuu M. Forkhead transcription factors: Key players in development and metabolism. Dev Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- 25.Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS. Fox's in development and disease. Trends Genet. 2003;19:339–344. doi: 10.1016/S0168-9525(03)00111-2. [DOI] [PubMed] [Google Scholar]
- 26.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 27.van der Horst A, Burgering BMT. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8:440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- 28.van der Vos KE, Coffer PJ. FOXO-binding partners: It takes two to tango. Oncogene. 2008;27:2289–2299. doi: 10.1038/onc.2008.22. [DOI] [PubMed] [Google Scholar]
- 29.Fraser HB. Modularity and evolutionary constraint on proteins. Nat Genet. 2005;37:351–352. doi: 10.1038/ng1530. [DOI] [PubMed] [Google Scholar]
- 30.Choi YS, Yang JS, Choi Y, Ryu SH, Kim S. Evolutionary conservation in multiple faces of protein interaction. Proteins. 2009;77:14–25. doi: 10.1002/prot.22410. [DOI] [PubMed] [Google Scholar]
- 31.Luo F, Li B, Wan XF, Scheuermann RH. Core and periphery structures in protein interaction networks. BMC Bioinformatics. 2009;10:1–11. doi: 10.1186/1471-2105-10-S4-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong P, et al. An evolutionary and structural characterization of mammalian protein complex organization. BMC Genomics. 2008;9:629–645. doi: 10.1186/1471-2164-9-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foucher I, Montesinos ML, Volovitch M, Prochiantz A, Trembleau A. Joint regulation of the MAP1B promoter by HNF3beta/Foxa2 and Engrailed is the result of a highly conserved mechanism for direct interaction of homeoproteins and Fox transcription factors. Development. 2003;130:1867–1876. doi: 10.1242/dev.00414. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Matthews KS, Bondos SE. Multiple intrinsically disordered sequences alter DNA binding by the homeodomain of the Drosophila hox protein ultrabithorax. J Biol Chem. 2008;283:20874–20887. doi: 10.1074/jbc.M800375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Matthews KS, Bondos SE. Internal regulatory interactions determine DNA binding specificity by a Hox transcription factor. J Mol Biol. 2009;390:760–774. doi: 10.1016/j.jmb.2009.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 37.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 38.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Joshi R, Sun L, Mann R. Dissecting the functional specificities of two Hox proteins. Genes Dev. 2010;24:1533–1545. doi: 10.1101/gad.1936910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 41.Tuch BB, Li H, Johnson AD. Evolution of eukaryotic transcription circuits. Science. 2008;319:1797–1799. doi: 10.1126/science.1152398. [DOI] [PubMed] [Google Scholar]
- 42.Gu X, Vander Velden K. DIVERGE: phylogeny-based analysis for functional-structural divergence of a protein family. Bioinformatics. 2002;18:500–501. doi: 10.1093/bioinformatics/18.3.500. [DOI] [PubMed] [Google Scholar]
- 43.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]