Abstract
Cells transformed by the p110α-H1047R mutant of PI3K show increased tyrosine phosphorylation of Stat3. This activation of Stat3 is important for the transformation process, because a dominant-negative mutant of Stat3 interferes with PI3K-induced oncogenesis. GDC-0941, a specific inhibitor of PI3K reduces the level of Stat3 phosphorylation. The effect of PI3K on Stat3 appears to be mediated by a member of the Tec kinase family. The Tec kinase inhibitor LFM-A13 blocks Stat3 phosphorylation in H1047R-transformed cells. The Janus kinase inhibitor AG490 and the Src kinase inhibitor Src-1, as well as rapamycin, have no effect on Stat3 phosphorylation in H1047R-transformed cells. The H1047R-transformed cells also release a factor that induces Stat3 phosphorylation in normal cells with possible effects on the cellular microenvironment. In some human tumor cell lines, the enhanced phosphorylation of Stat3 is inhibited by both PI3K and by Tec kinase inhibitors, suggesting that the link between PI3K and Stat3 is significant in human cancer.
Keywords: oncogenic signaling, tumor stroma, tyrosine kinase
Stat3 is a member of a transcription factor family that was discovered during the analysis of IFN-induced transcription (1–6). Stats are transcriptional regulators controlled by a pathway that can be activated by growth factor as well as cytokine receptors (7). Stat3 is activated in response to the epidermal growth factor and to IL-6 (2). Activation by IL-6 is primarily mediated by receptor-associated kinases of the Janus kinase (Jak) family, which phosphorylate cytoplasmic Stats at tyrosine, effecting dimerization, translocation into the nucleus, sequence-specific DNA binding, and transcriptional activation (8).
The role of Stats, however, goes far beyond the IFN response. Stat3 is an important and often essential factor in oncogenic cellular transformation and in cancer. It is a required target of the Src oncoprotein (9). Expression of dominant-negative Stat3 blocks Src-induced cellular transformation. Stat3 is frequently and persistently activated in a wide variety of cancers (10). Murine cells in which Stat3 has been genetically inactivated are resistant to oncogenic transformation (11, 12). Constitutively active mutants of Stat3 are sufficient to convert normal cells into cancer cells (13).
The canonical kinases of Stat are members of the Jak family. However, several other tyrosine kinases can phosphorylate and activate Stats, including both receptor tyrosine kinases, such as Egfr (14), Fgfr (15), Met (16), and Erbb2 (17), and nonreceptor tyrosine kinases, such as the Src and Fak kinase families. Such noncanonical Stat kinases are activated either through mutation or through the aberrant expression of cytokines (18–20). In chronic myelogenous leukemia, the BCR-ABL fusion kinase also mediates Stat3 phosphorylation, and the leukemic cells are dependent upon this activity for sustained proliferation (21).
Activation of Stat transcription factors induces a variety of proliferative and prosurvival proteins as they suppress immune responses (22). Stat proteins enhance the expression of the antiapoptotic Bcl2 and Bcl-XL (13) and repress the expression of proapoptotic proteins, such as p53 (23). Many growth factors are under Stat3 transcriptional control, including VEGF (24) and HGF (25). Constitutive expression of these growth factors can generate a positive autocrine feedback leading to activation of Stat3 (16).
The PI3K signaling pathway is part of the core regulatory networks in the cell and affects virtually all cellular activities, including growth, replication, movement, differentiation, and metabolism. PI3K signaling is elevated in most human cancers. This aberrant activity can result from differential regulation of PI3K itself or members of the pathway; it can also be caused by gain-of-function mutations in the pathway or loss-of-function mutations in the PI3K antagonist PTEN. The PI3K pathway connects to numerous other signaling nodes including Ras, p53, Hif1α, and Lkb1. However, crosstalk between PI3K and Stat signaling has not been reported. Here we describe a unique link between Stat3 and PI3K. In PI3K-transformed cells, Stat3 is activated. This activation is essential for the process of transformation. Inhibition of PI3K prevents Stat3 phosphorylation, and dominant-negative Stat3 interferes with PI3K-induced oncogenic transformation.
Results
Stat3 Is Activated in C3H 10T1/2 Mouse Fibroblasts Transformed by the PI3K Mutant p110α-H1047R.
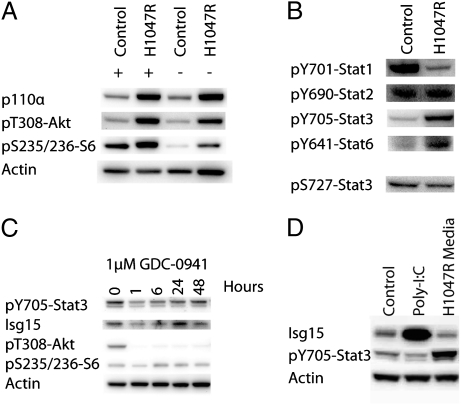
We have used stable isotope labeling with amino acids in cell culture (SILAC) in conjunction with tandem mass spectrometry to analyze the changes to the global proteome induced by the expression of the oncogenic H1047R mutant of p110α in C3H 10T1/2 cells (26). The up-regulated PI3K signaling in the H1047R-transformed cells is documented in Fig. 1A. Several of the proteins up-regulated by p110α-H1047R are known targets of Stats (Table 1) (26–32). The corresponding genes contain IFN-stimulated response elements or IFN-γ activation sites (GAS). These binding sites interact with the IFN-stimulated gene factor 3 complex which contains both IFN response factor and Stat proteins. We therefore investigated possible activation of Stat proteins by phosphorylation (Fig. 1B). The activation of Stat proteins by phosphorylation was evaluated by Western blotting. The p110α-H1047R-transformed 10T1/2 cells show enhanced phosphorylation of Stat3 and Stat6 and a decrease of phosphorylation in Stat1 (Fig. 1B). Because of the prominent role of Stat3 in cancer (9–13), we decided to investigate its significance in PI3K-induced oncogenic transformation. There is no tyrosine kinase in the canonical PI3K signaling pathway. However, activated TOR (target of rapamycin) can phosphorylate S727 of Stat3 (33, 34) and this phosphorylation enhances the activity of Stat3. However, in the two 10T1/2 cell lines, S727 is constitutively phosphorylated, regardless of the expression of p110α-H1047R (Fig. 1B). It is therefore unlikely that the enhanced expression of Stat targets in PI3K-transformed cells is mediated by TOR. In the context of the following experiments, we will use “phosphorylation of Stat3” to refer specifically to the phosphorylation of residue Y705, not of S727.
Fig. 1.
(A) PI3K signaling in cells transformed by the H1047R mutant of p110α. The Western blot shows expression of p110α and phosphorylation of Akt and of S6 in the presence and in the absence of serum. H1047R-transformed cells show constitutive activation of PI3K signaling in the absence of serum. (B) Phosphorylation of Stat proteins. 10T1/2 and 10T1/2-H1047R cells were lysed and probed for the indicated tyrosine phosphorylations of Stat proteins. Transformation by p110α-H1047R induces an increase in Stat3 and Stat6 phosphorylation and a decrease in Stat1 phosphorylation. S727 of Stat3, a known TOR phosphorylation site, is unaffected. (C) The PI3K inhibitor GDC-0941 blocks tyrosine phosphorylation of Stat3. The 10T1/2-H1047R cells were treated with 20 μM GDC-0941 for 0, 1, 6, 24, or 48 h as indicated and then analyzed by Western blot. (D) IFN stimulation. 10T1/2 cells were exposed to conditioned medium from either 10T1/2-H1047R or from normal 10T1/2 cells transfected with poly-I:C. The IFN produced by normal 10T1/2 cells in response to poly-I:C causes a sharp rise in the expression of Isg15 but does not induce phosphorylation of Stat3. In contrast, H1047R-conditioned medium leads to Stat3 phosphorylation but does not change Isg15 expression.
Table 1.
Enhanced expression of Stat3-regulated proteins in 10T1/2 cells transformed by PI3K (p110α-H1047R)*
| Name | Description | Fold overexpressed | Confidence interval | |
| Isg15 | ISG15 ubiquitin-like modifier | 13.54 | 7.08 | 25.87 |
| Oas2 | 2'-5′ oligoadenylate synthetase 2 | 8.95 | 7.80 | 10.27 |
| Ifit1 | IFN-induced protein with tetratricopeptide repeats 1 | 5.19 | 4.42 | 6.10 |
| Igtp | IFN-γ induced GTPase | 3.73 | 2.89 | 4.81 |
| Ddx58 | DEAD/H box polypeptide RIG-I | 2.44 | 1.96 | 3.05 |
| Ccnd1 | G1/S-specific cyclin-D1 | 2.17 | ||
| Eif2ak2 | IFN-induced, double-stranded RNA-activated protein kinase | 1.79 | 1.64 | 1.95 |
*As determined by SILAC analysis (26).
Inhibition of PI3K Reduces Phosphorylation of Stat3.
The p110α-H1047R protein is expressed in the 10T1/2 cells with the retroviral vector RCAS (35). It is therefore conceivable that the viral sequences of the vector could elicit an IFN response through recognition of double stranded RNA sequences of RCAS LTR by TLR3 (Toll-like receptor 3) or by Ddx58 (DEAD/H box polypeptide RIG-I) (36). In this case, PI3K activity would be irrelevant for the phosphorylation of Stat3. To test this possibility, we treated 10T1/2-H1047R cells with 1 μM GDC-0941, a potent PI3K inhibitor (37). Phosphorylation in the PI3K pathway including T308 and S473 of Akt and S235/236 of S6 was inhibited (Fig. 1C). The PI3K inhibitor GDC-0941 also interfered with the phosphorylation of Y705 on Stat3 indicating that PI3K activity is essential for this phosphorylation event.
Cells Expressing p110α-H1047R Release a Stat3-Activating Factor.
The reduction of Stat3 phosphorylation by a PI3K inhibitor does not rule out an IFN response to the retroviral vector. To explore a potential role of IFN further, we transfected 10T1/2 cells with the TLR3/RIG-I agonist poly-I:C to trigger an authentic IFN response. Cell-free conditioned medium from these cells and from nontransfected H1047R-expressing cells was then used to stimulate 10T1/2 cells. The cells were lysed 2 h after exposure to the conditioned medium and analyzed by Western blot. IFN produced in response to poly-I:C transfection triggered a large increase in the expression of Isg15, but did not affect phosphorylation of Stat3 (Fig. 1D). In contrast, medium conditioned by H1047R-transformed cells strongly stimulated the phosphorylation of Stat3, but had no effect on the production of Isg15. These data suggest that H1047R-transformed cells do not release detectable quantities of IFN, but release a factor that can stimulate the phosphorylation of Stat3. The nature of this factor is not known but is currently under investigation.
Dominant-Negative Mutant of Stat3 Interferes with PI3K-Induced Oncogenic Transformation.
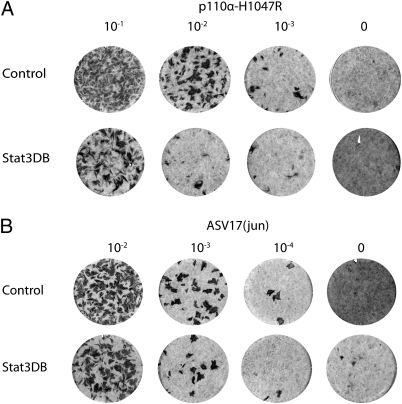
To determine the functional significance of Stat3 activation in H1047R-induced transformation, we examined a dominant negative, DNA binding-defective form of Stat3 for its ability to affect the oncogenic potency of p110α-H1047R. Chicken embryonic fibroblasts were transfected with RCAS(B)-Stat3DB (9) or RCAS(B) as a vector control. After 6 d to allow full expression of the dominant-negative Stat3, the cultures were superinfected with serial dilutions of RCAS(A) expressing the oncogenic H1047R mutant of p110α or with ASV17 expressing v-Jun, a presumably Stat3-independent oncoprotein. Potencies of oncogenic transformation were determined by focus counts 10 d after challenge infection. Stat3DB inhibited transformation induced by p110α-H1047R, but not v-Jun induced transformation (Fig. 2). Under these conditions, the efficiency of transformation by H1047R was reduced ∼10-fold, whereas the oncogenic activity of v-Jun remained at control levels. In an expansion of this experiment, we tested the ability of Stat3DB to interfere with oncogenic transformation induced by the E545K mutant of p110α and by the wild-type proteins of p110β, p110γ, and p110δ. The transforming activity of all these oncogenic PI3K constructs were reduced by Stat3DB as summarized in Table 2.
Fig. 2.
Dominant-negative Stat3 inhibits transformation by p110α-H1047R. Chicken embryonic fibroblasts were transfected with RCAS(B)-Stat3DB, a dominant-negative, DNA-binding defective form of Stat3 or RCAS(B) as an empty vector control. These cells were then superinfected with dilutions of Avian sarcoma virus 17, expressing (A) v-jun or (B) RCAS(A)-p110α-H1047R. Focus formation induced by p110α-H1047R was strongly inhibited by Stat3DB.
Table 2.
Dominant-negative Stat3 (Stat3DB) reduces the efficiency of oncogenic transformation by all class I PI3K isoforms
| Oncoprotein | EOT* |
| p110α-H1047R | 0.16 ± 0.05 |
| p110α-E545K | 0.17 ± 0.05 |
| p110β | 0.33 ± 0.10 |
| p110γ | 0.12 ± 0.03 |
| p110δ | 0.05 ± 0.01 |
| v-Src | 0.19 ± 0.06 |
*EOT, efficiency of transformation = focus count in Stat3DB-transfected CEF over focus count in CEF transfected with empty vector.
Inhibition of Stat3 Phosphorylation Induces Resistance to PI3K-Induced Transformation.
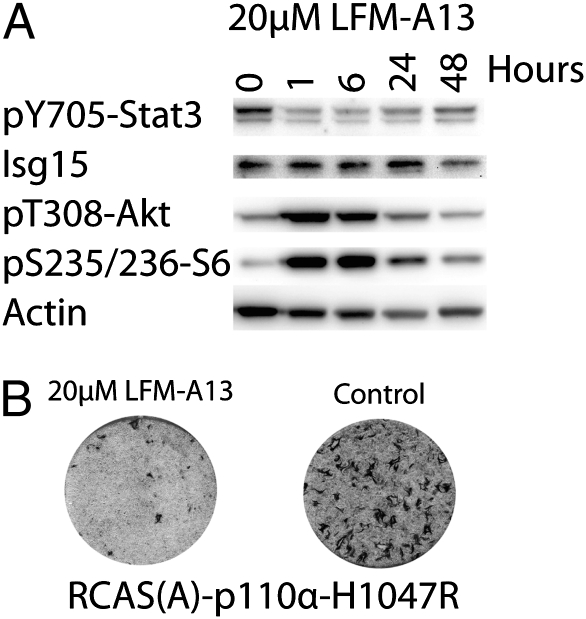
The signaling chain leading to Stat3 phosphorylation in H1047R-transformed cells has not been identified. Our observations with conditioned medium suggest that activation of PI3K can lead to the release of a cytokine or growth factor that interacts with a receptor linked to Stat3. The activation of Stat3 could also involve a tyrosine kinase that is controlled by PI3K. Candidates for such PI3K-stimulated activity are the members of the Tec kinase family (38). The Tec family tyrosine kinases (Btk, Itk, Txk, Bmx, and Tec) contain a PH domain that is selective for binding to PIP3, their activity is consequently stimulated by PI3K (39), and they are known to phosphorylate Stat3 (40). Btk, Itk, and Txk are restricted to hematopoietic cells. In the 10T1/2 mouse embryonic fibroblasts used in this study, we detected the expression of Bmx and Tec. Of these expressions, only Bmx was phosphorylated on tyrosine and, hence, activated (Fig. S1). LFM-A13 is a selective Tec family kinase inhibitor (41, 42). We treated 10T1/2-H1047R cells with 20 μM LFM-A13 and analyzed critical proteins by Western blot (Fig. 3A). LFM-A13 reduced the phosphorylation of Stat3 as effectively as did the PI3K inhibitor GDC-0941. In contrast, the Janus kinase inhibitor AG490 and the Src kinase inhibitor Src-1 failed to affect Stat3 phosphorylation in these cells. Rapamycin also did not affect the phosphorylation of Stat3 in H1047R-transformed 10T1/2 cells. The reduction in Stat3 phosphorylation by LFM-A13 is accompanied by a stimulation of Akt and S6 phosphorylation. However, despite the increased phosphorylation of PI3K signaling proteins, p110α-H1047R-induced oncogenic transformation was inhibited by LFM-A13 (Fig. 3B). These observations suggest a role of a Tec family kinase in the PI3K-induced phosphorylation of Stat3. The results obtained with the Tec family kinase inhibitor and with conditioned medium support the conclusion that both intracellular and intercellular mechanisms are responsible for the activation of Stat3 in H1047R-transformed cells.
Fig. 3.
(A) LFM-A13, a Tec kinase inhibitor, blocks Stat3 phosphorylation. 10T1/2-H1047R cells were treated with 20 μM LFM-A13 for 0, 1, 6, 24, or 48 h as indicated. Cells were lysed and analyzed by Western blotting using the indicated antibodies. (B) LFM-A13 inhibits transformation by RCAS(A)-p110α-H1047R. CEF were infected with RCAS(A)-p110α-H1047R and either not treated or treated with varying concentrations of LFM-A13. Treatment with 20 μM LFM-A13 is shown.
Stat3-PI3K Connection in Human Cancer Cell Lines.
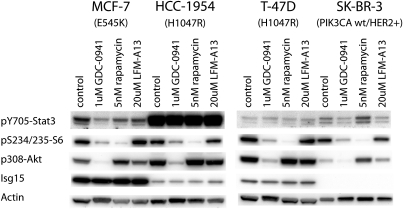
The 10T1/2 cells transformed by p110α-H1047R can be regarded as a model for human PI3K-driven cancer. We tested four human cancer cell lines for their responses to the inhibition of Tec family kinases and of PI3K signaling (Fig. 4). Three of the cell lines carry gain-of-function mutations in p110α (T-47D and HCC1954 carry the H1047R mutation, and MCF7 carries the E545K mutation). The fourth cell line, SK-BR-3, shows amplified HER2 with up-regulated signaling by wild-type PI3K. In two of these cell lines, SK-BR-3 and MCF-7, the phosphorylation of STAT3 is sensitive to the Tec family kinase inhibitor as well as the PI3K inhibitor. These results suggest that in some human cancers the activation of STAT3 is PI3K-dependent and appears to be mediated by a Tec kinase.
Fig. 4.
Phosphorylation of Stat3 in human cell lines. MCF-7, HCC-1954, T-47D, and SK-BR-3 cells were treated with 1 μM GDC-0941, 5 nM rapamycin, or 20 μM LFM-A13 for 24 h, as indicated. Cells were then lysed and analyzed by Western blotting. MCF-7 and SK-BR-3 show significant reduction in Stat3 phosphorylation in response to both PI3K and to Tec kinase inhibition, similar to the sensitivity of 10T1/2-H1047R cells.
Discussion
PI3K and Stat proteins represent two distinct cellular regulatory systems that are not known to share components. An unexpected link between PI3K and Stat-dependent transcription was recently revealed by a SILAC analysis of PI3K-transformed cells (26). The current series of experiments confirms this unique connection. Dominant-negative Stat3 interferes with PI3K-induced oncogenic transformation but does not affect transformation initiated by the Jun oncoprotein. The finding suggests a role of Stat3 in the PI3K-dependent transformation process but does not rule out the involvement of other Stats. Stat3 does not strictly homodimerize (43), and the overexpression of dominant-negative Stat3 could therefore affect other proteins. Oncogenic PI3K stimulates phosphorylation of Stat3 and of Stat6; the possible significance of the latter still remains to be explored. Stat6 is activated in certain cancers (44–46), and its increased phosphorylation in PI3K-transformed cells could be important for the oncogenic phenotype.
Could the activation of Stat3 in the 10T1/2-H1047R cells be an artifact related to an IFN response that resulted from the use of a retroviral vector? Although this possibility cannot be ruled out with absolute certainty, there are two sets of data that argue strongly against it. First, inhibiting PI3K activity with a small-molecule inhibitor blocks the phosphorylation of Stat3. This result clearly shows that PI3K is required for the activation of Stat3. Second, IFN produced in response to poly-I:C fails to induce phosphorylation of Stat3 in 10T1/2 cells when up-regulating the expression of Isg15. Testing a possible effect of the empty vector on Stat3 phosphorylation directly was not possible because cells transfected with the empty RCAS vector cannot be selected for.
A small molecule inhibitor of PI3K blocks the enhanced phosphorylation of Stat3, but there is no tyrosine kinase in the canonical PI3K pathway that could accomplish Stat3 activation. However, the Tec tyrosine kinases can link to PI3K through their PIP3-specific PH domain and are candidates for such a function. The ability of the selective Tec family kinase inhibitor LFM-A13 to interfere with PI3K-induced phosphorylation of Stat3 supports this possibility. LFM-A13 also interferes with PI3K-induced oncogenic transformation, possibly by blocking the PI3K-dependent activation of Stat3. LFM-A13 inhibits Tec family kinases with an IC50 of 2.7 μM (42), but it also inhibits Polo-like kinases with an IC50 of 60 μM (41). The concentrations of LFM-A13 used in our experiments are insufficient to affect Polo-like kinases, which therefore are unlikely to play a role in the PI3K-dependent phosphorylation of Stat3. The phosphorylation of Stat3 in PI3K-transformed cells is also unresponsive to the Janus kinase inhibitor AG490 and the Src kinase inhibitor Src-1, suggesting that these canonical activators of Stat3 are not involved. Furthermore, rapamycin fails to interfere with Stat3 phosphorylation in H1047R-transformed cells, placing the critical signaling component for the PI3K-dependent Stat3 activation upstream of the TOR kinase. Of the five members of the Tec kinase family—Btk, Itk, Txk, Bmx, and Tec—only Bmx and Tec are expressed in the fibroblast cell line studied, and only Bmx is phosphorylated on tyrosine and, hence, activated. Bmx is therefore the likely mediator between PI3K and Stat3. The molecular details of the connection between Bmx and Stat3 still remain to be explored, in particular the question whether Bmx acts on Stat3 directly or indirectly. The role Tec family kinases as link between PI3K and Stat3 identifies these kinases as complementary targets in PI3K-driven cancer and suggests the possibility of including Tec kinase inhibitors in combination therapy for such cancers.
A surprising finding is the stimulating effect of the Tec kinase inhibitor on PI3K signaling. Phosphorylation of Akt and of S6 are significantly increased in the presence of this inhibitor. This observation suggests a possible negative regulatory effect of Tec kinases on PI3K signaling. The apparent up-regulation of PI3K signaling is paradoxical in view of the fact that the Tec family kinase inhibitor interferes with PI3K-induced transformation. The phosphorylation of Akt is not always correlated with PI3K oncogenic activity (47, 48), and the result with the Tec family kinase inhibitor suggests that phosphorylation of Akt and of S6 are not sufficient for PI3K-induced oncogenic transformation. Critical factors in the transformation process clearly remain to be identified.
In human cancer cell lines, the link between PI3K and Stat3 is indicated by a sensitivity of Stat3 tyrosine phosphorylation to two defining inhibitors: inhibitors of PI3K and inhibitors of Tec kinases. This criterion of dual sensitivity can now be applied to a larger number of cancer cell lines to evaluate the incidence of the PI3K-Stat3 connection in human tumors of different tissue origin and genetic make-up. Of particular interest will be cancers that show enhanced PI3K signaling, either because of a gain-of-function in a PI3K pathway component or a loss-of-function in PTEN. The importance of the Tec kinase Bmx in bridging PI3K signaling to Stat3 has also recently been convincingly documented for the tumor-propagating stem cells of glioblastoma (49).
H1047R-transformed 10T1/2 cells release a factor that activates Stat3 without up-regulating Isg15 in normal 10T1/2 cells. This factor does not appear to be IFN because IFN produced in response to poly-I:C by 10T1/2 cells causes up-regulation of Isg15 but does not lead to enhanced Stat3 phosphorylation. The nature of this factor is currently unknown but is likely a growth factor or cytokine. One possibility is IL-1. A limiting determinant in the release of this proinflammatory cytokine is caspase-1 which processes and thereby activates the IL-1 precursor (50). Our SILAC study of H1047R-transformed 10T1/2 cells revealed a large increase in caspase-1 levels, which could enhance the generation of IL-1 (26). Another likely candidate is IL-6. Although antibody to IL-6 fails to inactivate the factor, we continue to pursue the possibility of IL-6 involvement using different approaches. The release of a Stat3-activating factor by transformed cells is of relevance for an understanding of the tumor microenvironment. The identification of this factor and studies of its possible occurrence in human cancer are clearly important topics for future work.
Materials and Methods
Cell Lines, Cell Culture, and Transformation Assays.
C3H10T1/2 cell lines were established as previously described (26). C3H10T1/2 cells were cultured in 4.5 g/L glucose DMEM (Invitrogen) supplemented with 10% FBS, 2 mM l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin (Sigma), and 200 μg/mL G418 (Invitrogen). HCC-1954, MCF-7, SK-BR-3, and T-47D were obtained from the ATCC and cultured in 4.5 g/L glucose DMEM (Invitrogen) supplemented with 10% FBS, 2 mM l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin (Sigma), and 0.1 U/mL insulin (Sigma). Cell lines were used at low passage numbers not exceeding 30 passages. Chicken embryonic fibroblasts (CEF) were prepared from pathogen free White Leghorn embryos (Charles River). For transformation assays, CEF were seeded into six-well plates and infected with RCAS(A) expressing the H1047R mutant of p110α, ASV17, or PRA as indicated and as previously described (51–53). In focus assays, inhibitors are added at the specified concentration in the first nutrient agar overlay and in all subsequent feeding overlays. For interference assays, CEF were transfected with either RCAS(B) or RCAS(B)-Stat3DB by DMSO shock. Cells were passed three times, during which the transfected retroviral construct spread throughout the cell culture. The cells were then seeded into six-well plates and superinfected with the indicated tumor viruses. All transformation assays were performed in media containing: 0.715× F-10, 0.20× Earle’s balanced salt solution, 0.6% SeaPlaque Agarose, 3% FBS, 1% heat-inactivated chicken serum, 9% tryptose phosphate broth, 1.8 mM glutamine, 89 U/mL penicillin, 89 μg/mL streptomycin, 1.1% DMSO. This mixture was applied every 2 to 3 d for 10 d at which point the overlay was removed and the cell layer stained with crystal violet.
RCAS(B)-Stat3DB. Stat3DB was cloned from RcCMV-Stat3DB (kindly provided by Jacqueline Bromberg, Memorial Sloan-Kettering Cancer Center, New York) to pBSfi by BamHI restriction, gel purification, and ligation to form pBSfi-Stat3DB. Stat3DB was cloned from pBSfi-Stat3DB to RCAS(B).Sfi by restriction with SfiI followed by gel purification and ligation to form RCAS(B)-Stat3DB. Orientation was confirmed by AccI digestion.
Western Blotting.
Cells were cultured in the presence of 10% FBS unless otherwise indicated. Cells were starved by culturing in 4.5 g/L glucose DMEM containing 0.5% FBS for 22 h followed by 4.5 g/L glucose DMEM without serum for 2 h. Inhibitors, LFM-A13 (Tocris), GDC-0941 (Toronto Research Chemicals), AG490 (Cayman), and Src-I (Sigma), were introduced into the medium 24 h before lysis unless otherwise indicated. Conditioned medium was prepared by transfection of 10T1/2 wild-type cells with 1 μg Poly-I:C and 60 μL lipofectamine or the medium from confluent 10T1/2-H1047R cells. Of these media, 500 μL were added to 60-mm dishes of wild-type 10T1/2 cells for 2 h. Cells were lysed using RIPA buffer (50 mM Tris pH 8.0, 150 mM sodium chloride, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) containing Complete Protease Inhibitor mixture (Roche), 1 mM PMSF, 1 mM Na3VO4. Protein concentrations were determined by the BCA method (Pierce) and concentrations normalized before separation with 4% to 12% gradient SDS/PAGE using Bis-Tris/MOPS buffer system under reducing conditions (Invitrogen). Separated proteins were blotted onto Immobilon-P PVDF membranes (Millipore). Membranes were blocked using 5% BSA in TBS-T, incubated with primary antibodies, washed three times with TBS-T, incubated with secondary antibody-HRP (Pierce) in 5% dry milk TBS-T, washed three times with TBS-T and developed using Superpico West ECL reagent (Pierce), and imaged using a Biorad Chemidoc XRS. Primary antibodies include PY-Stat1 (#9171), PY-Stat2 (#4441), PY-Stat3 (#9132), PY-Stat6 (#9361), PS-Stat3 (#9134), Stat3 (#9132), ISG15 (#2743), actin (#4967), PS-S6 (#2211), PT308-Akt (#9275), PS473-Akt (#9271), and p110α (#4255) (Cell Signaling).
Supplementary Material
Acknowledgments
The C3H 10T1/2 tva cells were generated in our laboratory by Masa Aoki, who kindly shared this resource with the authors of this article. RCAS(B)-Stat3DB was made by Qing Sun using materials from Jacqueline Bromberg. This work was supported by National Institutes of Health Grants R01 CA078230 and P01 CA078045 (to J.R.H. and P.K.V.), and P30 NS057096 and RR011823 (to L.L. and J.R.Y.). This is manuscript 21144 of The Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110486108/-/DCSupplemental.
References
- 1.Shuai K, Stark GR, Kerr IM, Darnell JE., Jr A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science. 1993;261:1744–1746. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- 2.Zhong Z, Wen Z, Darnell JE., Jr Stat3: A STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 3.Khan KD, et al. Induction of the Ly-6A/E gene by interferon alpha/beta and gamma requires a DNA element to which a tyrosine-phosphorylated 91-kDa protein binds. Proc Natl Acad Sci USA. 1993;90:6806–6810. doi: 10.1073/pnas.90.14.6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 5.Shuai K, Schindler C, Prezioso VR, Darnell JE., Jr Activation of transcription by IFN-gamma: Tyrosine phosphorylation of a 91-kD DNA binding protein. Science. 1992;258:1808–1812. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- 6.Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE., Jr Proteins of transcription factor ISGF-3: One gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sadowski HB, Shuai K, Darnell JE, Jr, Gilman MZ. A common nuclear signal transduction pathway activated by growth factor and cytokine receptors. Science. 1993;261:1739–1744. doi: 10.1126/science.8397445. [DOI] [PubMed] [Google Scholar]
- 8.Schindler C, Darnell JE., Jr Transcriptional responses to polypeptide ligands: The JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 9.Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JE., Jr Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu H, Jove R. The STATs of cancer—New molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 11.Chiarle R, et al. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med. 2005;11:623–629. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- 12.Poget M, Meyer R. Modification of a technic of reconstruction after total or subtotal resection of the lower lip (Translated from French) ORL J Otorhinolaryngol Relat Spec. 1976;38(Suppl 1):127–132. doi: 10.1159/000275326. [DOI] [PubMed] [Google Scholar]
- 13.Bromberg JF, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 14.David M, et al. STAT activation by epidermal growth factor (EGF) and amphiregulin. Requirement for the EGF receptor kinase but not for tyrosine phosphorylation sites or JAK1. J Biol Chem. 1996;271:9185–9188. doi: 10.1074/jbc.271.16.9185. [DOI] [PubMed] [Google Scholar]
- 15.Dudka AA, Sweet SM, Heath JK. Signal transducers and activators of transcription-3 binding to the fibroblast growth factor receptor is activated by receptor amplification. Cancer Res. 2010;70:3391–3401. doi: 10.1158/0008-5472.CAN-09-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang YW, Wang LM, Jove R, Vande Woude GF. Requirement of Stat3 signaling for HGF/SF-Met mediated tumorigenesis. Oncogene. 2002;21:217–226. doi: 10.1038/sj.onc.1205004. [DOI] [PubMed] [Google Scholar]
- 17.Ren Z, Schaefer TS. ErbB-2 activates Stat3 alpha in a Src- and JAK2-dependent manner. J Biol Chem. 2002;277:38486–38493. doi: 10.1074/jbc.M112438200. [DOI] [PubMed] [Google Scholar]
- 18.Gao SP, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwamoto T, et al. The JAK-inhibitor, JAB/SOCS-1 selectively inhibits cytokine-induced, but not v-Src induced JAK-STAT activation. Oncogene. 2000;19:4795–4801. doi: 10.1038/sj.onc.1203829. [DOI] [PubMed] [Google Scholar]
- 21.Spiekermann K, et al. Constitutive activation of STAT3 and STAT5 is induced by leukemic fusion proteins with protein tyrosine kinase activity and is sufficient for transformation of hematopoietic precursor cells. Exp Hematol. 2002;30:262–271. doi: 10.1016/s0301-472x(01)00787-1. [DOI] [PubMed] [Google Scholar]
- 22.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: Role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 23.Niu G, et al. Role of Stat3 in regulating p53 expression and function. Mol Cell Biol. 2005;25:7432–7440. doi: 10.1128/MCB.25.17.7432-7440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu G, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 25.Wojcik EJ, et al. A novel activating function of c-Src and Stat3 on HGF transcription in mammary carcinoma cells. Oncogene. 2006;25:2773–2784. doi: 10.1038/sj.onc.1209306. [DOI] [PubMed] [Google Scholar]
- 26.Hart JR, Liao L, Ueno L, Yates, JR, 3rd, Vogt PK. Protein expression profiles of C3H 10T1/2 murine fibroblasts and of isogenic cells transformed by the H1047R mutant of phosphoinositide 3-kinase (PI3K) Cell Cycle. 2011;10:971–976. doi: 10.4161/cc.10.6.15077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su ZZ, Sarkar D, Emdad L, Barral PM, Fisher PB. Central role of interferon regulatory factor-1 (IRF-1) in controlling retinoic acid inducible gene-I (RIG-I) expression. J Cell Physiol. 2007;213:502–510. doi: 10.1002/jcp.21128. [DOI] [PubMed] [Google Scholar]
- 28.Dupuis S, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33:388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 29.Nakaya T, et al. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem Biophys Res Commun. 2001;283:1150–1156. doi: 10.1006/bbrc.2001.4913. [DOI] [PubMed] [Google Scholar]
- 30.Sinibaldi D, et al. Induction of p21WAF1/CIP1 and cyclin D1 expression by the Src oncoprotein in mouse fibroblasts: Role of activated STAT3 signaling. Oncogene. 2000;19:5419–5427. doi: 10.1038/sj.onc.1203947. [DOI] [PubMed] [Google Scholar]
- 31.Kuhen KL, Samuel CE. Isolation of the interferon-inducible RNA-dependent protein kinase Pkr promoter and identification of a novel DNA element within the 5′-flanking region of human and mouse Pkr genes. Virology. 1997;227:119–130. doi: 10.1006/viro.1996.8306. [DOI] [PubMed] [Google Scholar]
- 32.Shenoy AR, et al. Emerging themes in IFN-gamma-induced macrophage immunity by the p47 and p65 GTPase families. Immunobiology. 2007;212:771–784. doi: 10.1016/j.imbio.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, et al. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci USA. 2007;104:16158–16163. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokogami K, Wakisaka S, Avruch J, Reeves SA. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr Biol. 2000;10:47–50. doi: 10.1016/s0960-9822(99)00268-7. [DOI] [PubMed] [Google Scholar]
- 35.Oh J, Julias JG, Ferris AL, Hughes SH. Construction and characterization of a replication-competent retroviral shuttle vector plasmid. J Virol. 2002;76:1762–1768. doi: 10.1128/JVI.76.4.1762-1768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abujamra AL, et al. Leukemia virus long terminal repeat activates NFkappaB pathway by a TLR3-dependent mechanism. Virology. 2006;345:390–403. doi: 10.1016/j.virol.2005.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raynaud FI, et al. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: from PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Mol Cancer Ther. 2009;8:1725–1738. doi: 10.1158/1535-7163.MCT-08-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu Y, Kung HJ. Signaling network of the Btk family kinases. Oncogene. 2000;19:5651–5661. doi: 10.1038/sj.onc.1203958. [DOI] [PubMed] [Google Scholar]
- 39.Qiu Y, Robinson D, Pretlow TG, Kung HJ. Etk/Bmx, a tyrosine kinase with a pleckstrin-homology domain, is an effector of phosphatidylinositol 3′-kinase and is involved in interleukin 6-induced neuroendocrine differentiation of prostate cancer cells. Proc Natl Acad Sci USA. 1998;95:3644–3649. doi: 10.1073/pnas.95.7.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai YT, et al. Etk, a Btk family tyrosine kinase, mediates cellular transformation by linking Src to STAT3 activation. Mol Cell Biol. 2000;20:2043–2054. doi: 10.1128/mcb.20.6.2043-2054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uckun FM, et al. Anti-breast cancer activity of LFM-A13, a potent inhibitor of Polo-like kinase (PLK) Bioorg Med Chem. 2007;15:800–814. doi: 10.1016/j.bmc.2006.10.050. [DOI] [PubMed] [Google Scholar]
- 42.Mahajan S, et al. Rational design and synthesis of a novel anti-leukemic agent targeting Bruton’s tyrosine kinase (BTK), LFM-A13 [alpha-cyano-beta-hydroxy-beta-methyl-N-(2, 5-dibromophenyl)propenamide] J Biol Chem. 1999;274:9587–9599. doi: 10.1074/jbc.274.14.9587. [DOI] [PubMed] [Google Scholar]
- 43.Wehinger J, et al. IL-10 induces DNA binding activity of three STAT proteins (Stat1, Stat3, and Stat5) and their distinct combinatorial assembly in the promoters of selected genes. FEBS Lett. 1996;394:365–370. doi: 10.1016/0014-5793(96)00990-8. [DOI] [PubMed] [Google Scholar]
- 44.Wang CG, et al. EZH2 and STAT6 expression profiles are correlated with colorectal cancer stage and prognosis. World J Gastroenterol. 2010;16:2421–2427. doi: 10.3748/wjg.v16.i19.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang WJ, et al. IL-4-induced Stat6 activities affect apoptosis and gene expression in breast cancer cells. Cytokine. 2008;42:39–47. doi: 10.1016/j.cyto.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 46.Das S, Roth CP, Wasson LM, Vishwanatha JK. Signal transducer and activator of transcription-6 (STAT6) is a constitutively expressed survival factor in human prostate cancer. Prostate. 2007;67:1550–1564. doi: 10.1002/pros.20640. [DOI] [PubMed] [Google Scholar]
- 47.Vasudevan KM, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang S, Denley A, Vanhaesebroeck B, Vogt PK. Oncogenic transformation induced by the p110beta, -gamma, and -delta isoforms of class I phosphoinositide 3-kinase. Proc Natl Acad Sci USA. 2006;103:1289–1294. doi: 10.1073/pnas.0510772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guryanova OA, et al. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer Cell. 2011;19:498–511. doi: 10.1016/j.ccr.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thornberry NA, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 51.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bos TJ, et al. Efficient transformation of chicken embryo fibroblasts by c-Jun requires structural modification in coding and noncoding sequences. Genes Dev. 1990;4:1677–1687. doi: 10.1101/gad.4.10.1677. [DOI] [PubMed] [Google Scholar]
- 53.Duff RG, Vogt PK. Characteristics of two new avian tumor virus subgroups. Virology. 1969;39:18–30. doi: 10.1016/0042-6822(69)90344-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.