Abstract
How drugs bind to their receptors—from initial association, through drug entry into the binding pocket, to adoption of the final bound conformation, or “pose”—has remained unknown, even for G-protein-coupled receptor modulators, which constitute one-third of all marketed drugs. We captured this pharmaceutically critical process in atomic detail using the first unbiased molecular dynamics simulations in which drug molecules spontaneously associate with G-protein-coupled receptors to achieve final poses matching those determined crystallographically. We found that several beta blockers and a beta agonist all traverse the same well-defined, dominant pathway as they bind to the β1- and β2-adrenergic receptors, initially making contact with a vestibule on each receptor’s extracellular surface. Surprisingly, association with this vestibule, at a distance of 15 Å from the binding pocket, often presents the largest energetic barrier to binding, despite the fact that subsequent entry into the binding pocket requires the receptor to deform and the drug to squeeze through a narrow passage. The early barrier appears to reflect the substantial dehydration that takes place as the drug associates with the vestibule. Our atomic-level description of the binding process suggests opportunities for allosteric modulation and provides a structural foundation for future optimization of drug–receptor binding and unbinding rates.
Keywords: dewetting, kinetics, ligands, drug target, alprenolol
Systematic determination of drug efficacy dates to Ehrlich’s experiments that culminated in the discovery of the first chemotherapeutic “magic bullet,” Salvarsan. In the ensuing century, such measurements of drug efficacy and—once the molecular targets of drugs, or “receptors,” were identified—of drug–receptor affinity have become de rigueur (1, 2). Affinity measurements are increasingly augmented by characterization of the kinetics of drug–receptor interaction, for several reasons: fast drug binding increases opportunities to capture transiently accessible receptor conformations; fast unbinding may confer safety advantages; and slow unbinding leads to long drug–receptor residence times that can dramatically enhance therapeutic efficacy at equivalent affinity (3, 4).
In stark contrast, essentially nothing is known about the process by which drugs bind to their receptors. By what pathway, or pathways, do drugs enter and exit the receptor binding pocket? Must drugs first associate with regions outside the binding pocket to bind? Might drugs sometimes bind (perhaps transiently) in more than one orientation or conformation within the binding pocket? Additionally, what factors determine binding and unbinding rates? Despite the innovative variety of experimental and computational techniques that have been applied to study binding mechanisms (5–16), these questions have proven extremely difficult to address, in part because they involve unstable structural states that are occupied only transiently.
G-protein-coupled receptors (GCPRs) represent the largest class of drug targets, and one-third of all drugs act by binding to GCPRs. Recent advances have yielded crystal structures that reveal the dominant GPCR-bound conformations, or “poses,” of several drugs and related ligands (17). These ligands all lie buried within a deep binding pocket—the canonical “orthosteric” site that is cradled within a characteristic bundle of seven transmembrane helices—that is typically accessed from the extracellular side of the receptor. These advances have left unresolved, however, both the process by which drugs bind to GPCRs and the precise location and nature of alternative, allosteric binding sites (18).
Here, we used unbiased molecular dynamics (MD) simulations to capture the entire process by which several drugs bind to two archetypal GPCRs. Despite the fact that our simulations lacked any “knowledge” of the binding site, and thus required the drugs to spontaneously “discover” the binding pocket, they resulted in bound conformations that recapitulate crystallographic poses to subangstrom accuracy. These binding events occur on microsecond timescales that, until very recently, were beyond the reach of atomistic MD simulations.
Our results reveal not only the predominant pathway into the binding site, but also the energetic barriers that govern drug binding and unbinding kinetics. We found that drugs encounter two major barriers on the binding pathway: the “expected” barrier suggested by receptor geometry, proximal to the binding pocket, and also an unexpected, earlier barrier at the receptor surface. This early barrier coincides with, and appears to be tied to, substantial drug–receptor dehydration that occurs far from the binding pocket.
Results
We performed all-atom MD simulations in which four distinct ligands bound spontaneously to the β2-adrenergic receptor (β2AR): three antagonists (the “beta blocker” drugs propranolol and alprenolol, used to treat hypertension and angina pectoris, and dihydroalprenolol, a key experimental tool) and one agonist (isoproterenol, used to treat bradycardia and heart block). We also simulated the binding of dihydroalprenolol to the β1-adrenergic receptor (β1AR). We performed 82 simulations lasting from 1 to 19 μs each, resulting in a total of 21 spontaneous binding events (Tables S1 and S2).
All simulations were completely blind to the location of the binding site, and did not incorporate any artificial guiding or biasing forces. Ligands were initially positioned at least 30 Å from the binding pocket and 12 Å from the receptor surface. They diffused extensively about the receptor before entering the binding pocket (Fig. 1A); once bound, ligands remained bound for the remainder of the simulation. Binding rates in simulation were comparable with experimentally observed rates: our simulations suggest an on-rate of 3.1 × 107 M-1 s-1 for alprenolol and dihydroalprenolol binding to β2AR at 37 °C, close to the experimentally derived value of approximately 1.0 × 107 M-1 s-1 (19; see SI Text). Likewise, the dihydroalprenolol–β2AR binding energy determined by free-energy perturbation calculations, -13.4 ± 1.6 kcal/mol, is within error of the experimentally derived value of -12.2 kcal/mol (20; see SI Text).
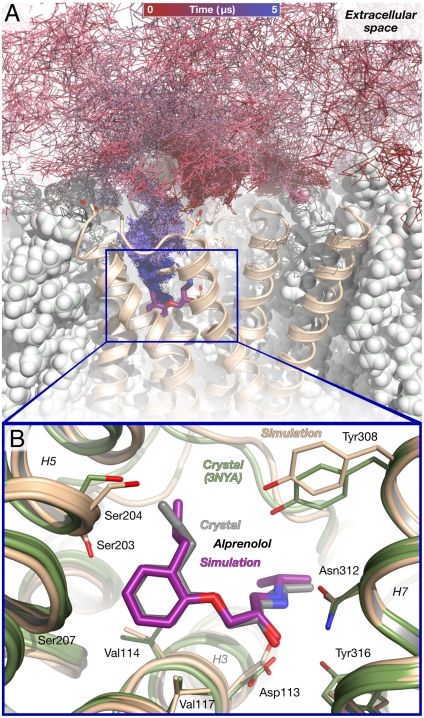
Fig. 1.
Alprenolol binds spontaneously to β2AR in unbiased molecular dynamics simulations, achieving the crystallographic pose. (A) The path taken by an alprenolol molecule as it diffuses about the receptor and then binds. The final, bound alprenolol is shown as a stick figure (purple carbon atoms); the protein as a tan cartoon; and the lipid bilayer as white spheres. (B) Close-up view of the simulated alprenolol pose shown in (A) superimposed on the alprenolol–β2AR crystal structure (gray ligand, green cartoon; PDB entry 3NYA). Data from simulation 1 (Table S2).
We describe first the binding of (S)-alprenolol and (S)-dihydroalprenolol to β2AR (Fig. 2A). These ligands interact essentially identically with β-adrenergic receptors both in our simulations and in biochemical studies (20, 21); we refer to both ligands simply as “alprenolol” except where otherwise indicated. We then turn to the other ligands and β1AR, noting extensive similarities in the binding mechanism.
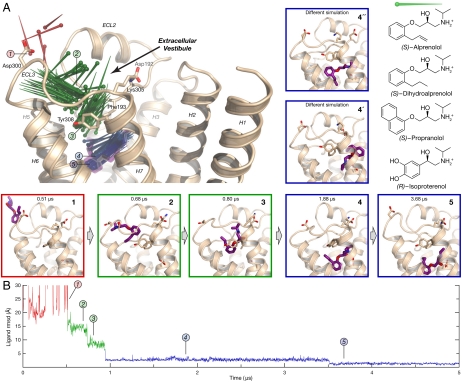
Fig. 2.
The alprenolol–β2AR binding pathway passes through several metastable states. (A) Pins indicate successive positions of an alprenolol molecule as it binds to β2AR (the pin point is at the nitrogen atom position, and the round end is at the benzene ring center). Alprenolol moves from bulk solvent (red, pose 1), into the extracellular vestibule (green, poses 2 and 3), and finally into the binding pocket (blue, poses 4 and 5). Pose 5 matches the crystallographic pose (Fig. 1B), whereas pose 4 and two poses observed in a different simulation (4′ and 4′′) represent alternative, metastable poses in the binding pocket. The structures of ligands used in this study are shown at right. (B) Rmsd of alprenolol in simulation from the alprenolol-β2AR crystal structure, calculated after aligning on protein binding pocket Cα atoms (SI Text). Poses 4′ and 4′′ from simulation 3; remaining data from simulation 1.
Bound Pose Matches Crystal Structure.
In 6 of the 12 simulations in which alprenolol bound to β2AR, it adopted a pose matching the alprenolol-bound β2AR crystal structure (Fig. 1B), with a 0.8-Å root-mean-squared deviation (rmsd) between the average simulation pose and the crystallographic pose (22; SI Text). Once adopted, this pose remained stable for the duration of these 6 simulations; more simulations ended with alprenolol in the crystallographic pose than in any other. The other 6 simulations were terminated with alprenolol in the binding pocket in one of two alternative, metastable poses (Fig. 2, poses 4′ and 4′′). These alternative poses, which were also observed transiently in several simulations that ended in the crystallographic pose, are likely less energetically favorable binding poses that convert slowly (relative to the timescale of our simulations) to the crystallographic pose.
A Dominant Binding Pathway.
Alprenolol bound to β2AR along a single dominant pathway. In 11 of the 12 simulations in which alprenolol bound to β2AR, it entered via a strikingly similar pathway, not only following the same spatial route but also pausing at common, metastable intermediate conformations. Alprenolol passed between extracellular loops 2 and 3 (ECL2/ECL3) and then through the crevice between ECL2 and transmembrane helices 5, 6, and 7 to reach the binding pocket (Fig. 2A and Movie S1). In the single remaining simulation, alprenolol instead entered between ECL2 and helices 2 and 7. In our simulations, as in experiments (23), the alprenolol hydrophobic group (i.e., the 2-allyl[propyl]-benzene) tended to partition into the lipid bilayer, remaining there for a majority of the simulation; despite this, alprenolol never entered the binding pocket from the lipid bilayer.
The dominant alprenolol binding pathway comprises two major steps. First, alprenolol associated with a surface region we term the “extracellular vestibule,” which is enclosed by ECL2, ECL3, and helices 5–7. The alprenolol hydrophobic group bound to the extracellular vestibule surface, contacting hydrophobic surfaces of residues such as Tyr3087.35, Phe193ECL2, Ala2005.39, His2966.58, and Val2976.59. [Superscripts refer to Ballesteros–Weinstein residue numbering (24)]. Alprenolol typically spent several hundred nanoseconds in the vestibule, where it assumed several distinct poses (Fig. 2A). In the most common (Fig. 2, pose 2), the alprenolol ammonium group formed a salt bridge with Asp300ECL3. This pose was often followed, temporally, by a pose in which the ammonium bound instead to the backbone carbonyl oxygen atom of Asp192ECL2 (Fig. 2, pose 3).
Second, alprenolol moved from the extracellular vestibule into the binding pocket by squeezing through a narrow passage between ECL2 and helices 5, 6, and 7. In some binding events, alprenolol immediately adopted the crystallographically observed pose upon entering the binding pocket. In others, it transiently adopted one of several alternative poses (Fig. 2, poses 4, 4′, and 4′′)—all of which possessed the alprenolol ammonium–Asp1133.32 carboxylate salt bridge—before relaxing into the crystallographic pose (Fig. 2, pose 5). In the simulation trajectory illustrated in Fig. 2, for example, alprenolol initially bound with its aliphatic chain twisted near the beta-hydroxyl group (Fig. 2, pose 4; see also Fig. S1); after 2.6 μs in this pose, it relaxed into the crystallographic pose, allowing the beta-hydroxyl to form a hydrogen bond with Asn2936.55.
The Primary Barrier to Binding Lies Far Outside the Binding Pocket.
Our simulations indicate the presence of two major energetic barriers along the binding pathway, corresponding to the two steps described above (Fig. 3). As might be expected, one of these barriers coincides with the passage leading from the vestibule to the binding pocket, by far the narrowest point alprenolol encounters along the binding pathway (Fig. 3 A and C).
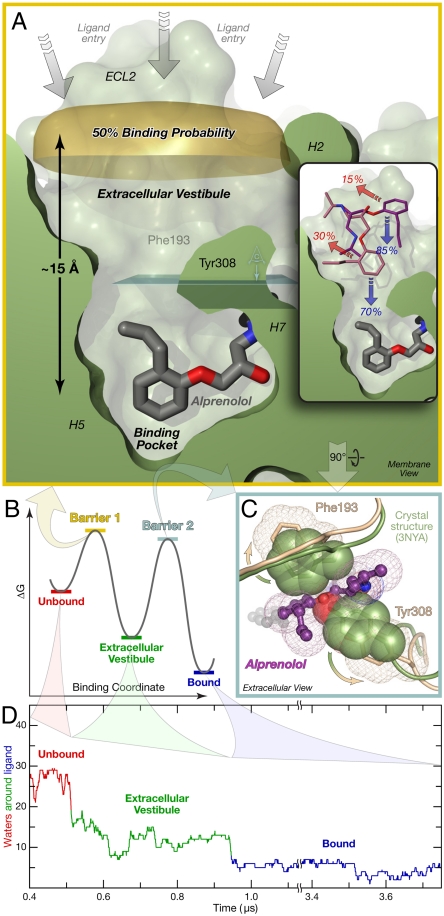
Fig. 3.
Alprenolol must traverse two primary energetic barriers to bind to β2AR. (A) The first and largest barrier precedes association with the extracellular vestibule. Alprenolol molecules for which the ring center has crossed the “50% binding probability” surface (from bulk solvent into the extracellular vestibule; above to below in this image) bind more often than they escape back into the bulk. (Inset) In repeated simulations initialized with alprenolol already in the vestibule, the ligand usually proceeded into the binding pocket. Binding/escape percentages are shown. (B) Schematic free-energy landscape for binding. (C) Phe193ECL2 and Tyr3087.35 must separate for alprenolol to move from the extracellular vestibule into the binding pocket. View plane is indicated in light blue in (A). (D) Alprenolol loses more than half of its hydration shell as it enters the extracellular vestibule, and most of the rest as it enters the binding pocket (SI Text). Data from simulation 1.
Surprisingly, however, we found that alprenolol traverses the highest barrier prior even to its entry into the extracellular vestibule. Data obtained from two distinct simulation approaches support this conclusion. First, analysis of the unbiased binding simulations described above, all of which were initiated with alprenolol far from the receptor surface, indicated that once the hydrophobic group of alprenolol moved from bulk solvent into the vicinity of the extracellular vestibule—that is, it moved across the gold surface shown in Fig. 3A—the alprenolol molecule was more likely to bind than to escape back into bulk solvent. It is notable that this “50% binding probability” surface lies, for the most part, more than 15 Å away from the position of the alprenolol hydrophobic group in its (final) bound pose. Second, for each of four vestibule-bound alprenolol positions, two of which are shown in Fig. 3A, Inset, we initiated ten to twenty additional simulation trajectories, with random initial velocities. In each case, the ligand entered the binding pocket more frequently than it returned to bulk solvent. The relative frequency with which alprenolol molecules found in the extracellular vestibule proceed to bind or instead escape back into bulk solvent suggests that the energy difference between the two barriers is small, on the order of 1 kcal/mol. Both of these analyses independently support the conclusion that ligands in the metastable vestibule-bound state have already surmounted the highest barrier on the binding pathway.
Substantial Dehydration Accompanies Drug Entry into the Extracellular Vestibule.
What accounts for the large barrier to alprenolol entry into the extracellular vestibule? Ligand entry into the vestibule does not require any noteworthy structural changes in the receptor or alprenolol itself (Fig. S1), and we found no electrostatic barrier to vestibule entry (Fig. S2). Yet experimental measurements have demonstrated that ligand–adrenergic receptor binding rates are well below diffusion-controlled rates, with correspondingly higher (enthalpic) barriers to binding (SI Text).
Our simulations suggest that this first barrier may be due instead to dehydration of both the drug and the extracellular vestibule as the drug enters the vestibule. High-affinity drug–receptor interactions are frequently stabilized by hydrophobic contacts that require substantial drug–receptor dehydration (25), and alprenolol–β2AR is no exception. Alprenolol loses about 80% of its hydration shell as it binds to β2AR. Significantly, the majority of ligand dehydration (63%; Fig. 3D) occurs as alprenolol enters the extracellular vestibule, far from the binding pocket. Ligand entry into the vestibule, which leads to substantial hydrophobic contacts and the burial of approximately 500 Å2 of hydrophobic surface area (Table S3), coincides with the rapid evacuation of approximately 15 water molecules from the vestibule within 500–1000 ps (Fig. S3), a signature of a dewetting transition (14, 15, 26). This concerted motion of the ligand and multiple water molecules appears to represent a kinetic bottleneck to ligand entry into the extracellular vestibule.
The second energetic barrier, which alprenolol crosses as it moves from the extracellular vestibule into the binding pocket, involves structural changes from the receptor’s crystallographic conformation (Fig. 3C), but also dehydration. As alprenolol passes into the binding pocket, Tyr3087.35 and Phe193ECL2 separate enough for the ligand’s hydrophobic group to pass between them, and the salt bridge between Asp192ECL2 and Lys3057.32 breaks. These events, however, do not appear to be rate-limiting: while the ligand waits in the extracellular vestibule to enter the binding pocket, Tyr3087.35 and Phe193ECL2 sometimes remain separated for over 200 ns, with the Asp192ECL2–Lys3057.32 salt bridge breaking and reforming hundreds of times during that period (Fig. S4). The rate may instead be limited, at least in part, by dehydration events: as the ligand enters the binding pocket, it loses the majority of its remaining hydration shell, and 50–60% of the water molecules in the binding pocket must also escape.
Similar Binding Pathways and Barriers for Other Drug–Receptor Pairs.
We also simulated the binding of the beta blocker (S)-propranolol and the agonist (R)-isoproterenol to β2AR. Propranolol adopted the expected pose, given those determined crystallographically for several structurally analogous antagonists (Fig. 4A) (22, 27). Isoproterenol entered the binding pocket and formed the expected salt bridge to Asp1133.32, but it exhibited much more mobility than the antagonists when bound, likely reflecting the fact that the receptor remained in its inactive conformation, which has low agonist affinity. (Receptor activation takes place on millisecond timescales (28), several orders of magnitude longer than our simulations.)
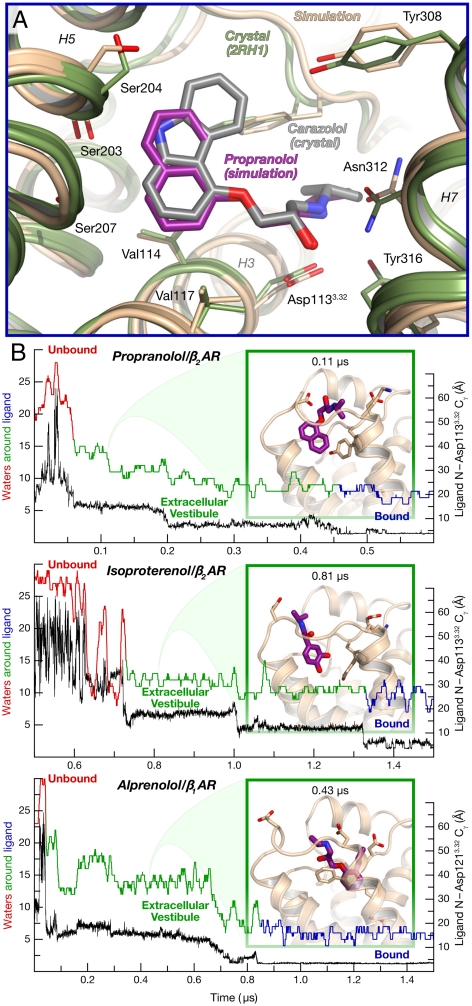
Fig. 4.
Other drug–receptor pairs follow the same binding pathway. (A) Bound pose of propranolol (purple), superimposed on the carazolol–β2AR crystal structure (PDB entry 2RH1, gray) (data from simulation 15). (B) Distance to binding pocket (ligand nitrogen atom to Asp3.32 Cγ) and ligand hydration for propranolol and isoproterenol binding to β2AR, and for alprenolol binding to β1AR (data from simulations 13, 16, and 21). The insets illustrate a typical extracellular-vestibule-bound state for each simulation.
Our simulations of propranolol and isoproterenol binding largely recapitulated our findings with alprenolol. These drugs generally followed the predominant alprenolol binding pathway, pausing in the extracellular vestibule before entering the binding pocket (Fig. 4B); one propranolol molecule followed the alternative alprenolol pathway. Although the smaller number of simulations and binding events (three events for propranolol and one for isoproterenol) prevents an accurate assessment of the relative barrier heights, the observed balance between escape from the vestibule into bulk solvent and progression to the binding pocket suggests that the two barriers are again of similar height. Like alprenolol, propranolol and isoproterenol lost approximately 60% of their hydration shells upon association with the extracellular vestibule; dewetting likely thus contributes to the vestibule entry barrier for these drugs as well.
Finally, we simulated the binding of dihydroalprenolol to β1AR, achieving a bound pose that matched crystallographic data (Fig. S5A and refs. 22 and 29). The binding pathway was similar to that observed for β2AR, with a metastable state in the extracellular vestibule and with substantial dewetting taking place upon ligand association with the vestibule (Fig. 4B). β1AR lacks the extracellular salt bridge corresponding to Asp192ECL2–Lys3057.32 in β2AR, however, allowing the β1AR vestibule to extend further toward helix 2. In addition, one of the alternative binding poses, in which the dihydroalprenolol hydrophobic group interacts with helix 2 (Fig. S5B; see also Fig. 2, pose 4′), was observed more frequently with β1AR than with β2AR. Although this alternative pose is likely transient, a mutagenesis study has suggested that a pose with this orientation may represent an important binding mode for β1AR antagonists under certain conditions (30).
Discussion
In the simulations of GPCR–drug binding we report here, diverse drugs bound to two β-adrenergic receptors and adopted crystallographically validated binding poses. Unlike standard docking approaches (31), which search for the best pose within a predefined binding site, our unbiased, atomistic MD simulations reproducibly identified and maintained the correct pose without user intervention or incorporation of any prior knowledge of the binding site. Furthermore, our approach naturally revealed the binding pathways and the energetic barriers encountered along those pathways, while taking into account the significant flexibility of both drugs and receptors. Our simulations also allowed determination of previously unknown binding sites, including transiently populated sites that might be targeted by allosteric modulators (Fig. 2A and Fig. S6). Direct simulation of the binding process thus provides access to information that is particularly difficult to obtain experimentally.
The Binding Pathway: Barriers, Kinetics, and Dewetting.
The three beta blockers and the beta agonist we studied all bound to the receptors along a predominant pathway involving entry into the extracellular vestibule, drug residence in the vestibule, and finally drug entry into the binding pocket, where the drug often took on one of several metastable poses before adopting the crystallographically observed conformation. Unexpectedly, the barrier to entry into the extracellular vestibule was comparable to—and, at least in the case of alprenolol binding to β2AR, higher than—the barrier for entry into the binding pocket, even though the latter required traversal of a narrow passage with accompanying deformation of the receptor structure. The barrier to entry into the extracellular vestibule is associated with surprisingly large dehydration of both drug and receptor that takes place as the drug associates with the vestibule.
Our simulations of drug binding also illuminate the process of unbinding: in the absence of external driving forces, the unbinding process is the reverse of the binding process, following the same pathway and traversing the same barriers in the opposite order. Thus, as alprenolol unbinds, it first moves from the binding pocket into the extracellular vestibule, passing between Tyr3087.35 and Phe193ECL2 (Fig. 3B, “Barrier 2”), and then exits from the vestibule into the bulk solvent as water enters the vestibule in a concerted fashion (Fig. 3B, “Barrier 1”). In a recent computational study, carazolol was observed to exit the β2AR binding pocket along many routes, with the extracellular vestibule route ranking second (8); this difference from our results might be due to the choice of ligand, but more likely it reflects the forces applied in that study to evict carazolol from the binding pocket within a few nanoseconds, a time period about 14 orders of magnitude shorter than its experimental dissociation half-life (32).
By characterizing the binding pathway and the major energetic barriers along it, our results provide a foundation for the rational optimization of drug binding and unbinding kinetics, which are now recognized to play a critical role in drug efficacy, selectivity, and safety (3, 4). Because the two barriers are of similar height, changes to either can substantially affect binding and unbinding rates. Geometric factors, such as ligand size and flexibility, will likely affect the barrier to binding pocket entry more than the barrier to vestibule entry. Ligand hydrophobicity and other factors associated with dehydration may, however, contribute as much to both barriers—particularly the vestibule entry barrier—as geometry and electrostatics.
The dehydration observed early on the binding pathway, and its connection to a major kinetic barrier, is particularly intriguing given the widely recognized importance of ligand and binding-site dehydration in drug–receptor affinity. Greater drug hydrophobicity usually leads to greater affinity, because changes in water structure upon drug binding—in particular, the increased disorder of water molecules released from the binding pocket—often dominate the binding free energy and thus make the largest contribution to affinity (25). Our results suggest that this dehydration, and the points on the binding pathway at which it takes place, play an important role in determining binding kinetics as well as affinity.
Beyond providing insight into elements of the binding process that have proven difficult to probe experimentally, our simulation results also suggest avenues for future experimental investigation. Free-energy perturbation calculations indicate that dihydroalprenolol binds to the β2AR extracellular vestibule—the main intermediate we observe prior to ligand entry into the orthosteric binding pocket—with a KD of about 5 μM (SI Text). Crystal structures determined with modified beta blockers, in which the ether oxygen atom is substituted by selenium, may thus reveal anomalous scattering density peaks in the vestibule. Similarly, the [19F]-NMR chemical shift of a semisynthetic β2AR in which a vestibule hydrophobic residue is substituted by Cys-SCH2CF3 should be very sensitive to the binding there of aromatic ligands. Blockade of the orthosteric site, by mutation or by presaturation with a slowly dissociating beta blocker, may even render ligand binding in the vestibule detectable by conventional means such as a radioligand binding assay. By contrast, incorporation of sufficiently large residues in the extracellular vestibule could block the dominant binding pathway, slowing ligand association with and dissociation from the orthosteric binding pocket.
Further investigation, perhaps using computational techniques similar to those demonstrated here, will be necessary to determine the similarities and differences in the pathways, energetics, and kinetics of ligand binding to different receptors. Detailed binding pathways likely vary among GPCRs, and perhaps even for a given GPCR in different conformational states. Several studies have also suggested that highly hydrophobic ligands may enter and leave certain GPCRs directly through the lipid bilayer (9, 10). Nevertheless, the conserved architecture of the GPCR family, with the binding pocket buried deep within a seven transmembrane helix bundle, suggests that many GPCRs likely share similar binding pathways. Moreover, characteristics of the binding process we have observed here for β-adrenergic receptors, including the presence of a major energetic barrier at the point where the drug first associates with the receptor surface and its connection to early drug–receptor dehydration, may even apply beyond the GPCR family, particularly to other receptors with deep, well-defined binding pockets.
Implications for Allosteric Modulation.
Allosteric GPCR modulators, which bind at sites distinct from the classical orthosteric binding pocket, hold the promise of enhanced selectivity compared to orthosteric GPCR drugs (33). The design of allosteric drugs has been hindered, however, by a lack of structural information. Although the orthosteric site was the dominant binding site observed in our simulations—and the only site to which ligands remained stably bound—we also observed several previously unknown, metastable binding sites (Fig. 2A and Fig. S6). Unbiased simulations of ligand binding, such as those presented here, may thus provide a means to identify the binding sites of known allosteric modulators and to discover potential binding sites for novel allosteric modulators.
Metastable drug binding to the extracellular vestibule (Fig. 2A) is of particular interest in light of experimental data for the binding of allosteric modulators to muscarinic acetylcholine receptors. Binding of gallamine and related compounds to these receptors dramatically slows the association and dissociation of orthosteric ligands (34). Mutagenesis data indicate that residues in ECL2 and near the ECL3/helix 7 junction are involved in gallamine binding (18). We speculate that gallamine binds in the extracellular vestibule. By “getting stuck” at this intermediate point on the dominant binding pathway, gallamine would directly block access to, or egress from, the orthosteric binding pocket. Recent data suggests that orthosteric ligands can bind weakly in the same position as gallamine (35), as would be expected if the gallamine binding site represents a metastable point on the binding pathway of orthosteric ligands.
As additional GPCR structures become available and as advances in computer technology enable ever longer simulations, the approach presented here will become applicable to more receptors and ligands. Such simulations promise to complement traditional experimental measurements by providing an atomic-level description of the binding process for orthosteric and allosteric GPCR modulators, including the binding pathway and the energetic barriers that control binding kinetics, both when the binding site is known in advance and when it is not.
Methods
We performed molecular dynamics simulations of β1AR and β2AR, with lipids and water represented explicitly, using the CHARMM force field (36) on Anton (37), a special-purpose machine that accelerates such simulations by orders of magnitude. Each simulation began with the receptor in the crystallographic conformation [Protein Data Bank (PDB) ID entries 2VT4 for β1AR (29) and 2RH1 for β2AR (27)], but with the cocrystallized antagonist (cyanopindolol or carazolol) removed. Ten identical ligand molecules were placed in the bulk solvent surrounding the receptor. No artificial forces were applied to the ligands, which diffused throughout the simulated water and lipids. Further details are provided in SI Text.
Supplementary Material
Acknowledgments.
We thank Brian Kobilka and Michael Bokoch for questions and comments that inspired this work; Woody Sherman for suggesting the study of binding kinetics; Kresten Lindorff-Larsen, Stefano Piana, Kim Palmo, and Morten Jensen for helpful advice; Ansgar Philippsen for creating the movie; Bruce Berne for comments on the manuscript; and Rebecca Kastleman and Mollie Kirk for editorial assistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104614108/-/DCSupplemental.
References
- 1.Weiland GA, Minneman KP, Molinoff PB. Fundamental difference between the molecular interactions of agonists and antagonists with the β-adrenergic receptor. Nature. 1979;281:114–117. doi: 10.1038/281114a0. [DOI] [PubMed] [Google Scholar]
- 2.Ward WH, Holdgate GA. Isothermal titration calorimetry in drug discovery. Prog Med Chem. 2001;38:309–376. doi: 10.1016/s0079-6468(08)70097-3. [DOI] [PubMed] [Google Scholar]
- 3.Lu H, Tonge PJ. Drug-target residence time: Critical information for lead optimization. Curr Opin Struct Biol. 2010;14:467–474. doi: 10.1016/j.cbpa.2010.06.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swinney DC. Applications of binding kinetics to drug discovery: Translation of binding mechanisms to clinically differentiated therapeutic responses. Pharmaceut Med. 2008;22:23–34. [Google Scholar]
- 5.Baldwin AJ, Kay LE. NMR spectroscopy brings invisible protein states into focus. Nat Chem Biol. 2009;5:808–814. doi: 10.1038/nchembio.238. [DOI] [PubMed] [Google Scholar]
- 6.Hargrove MS. Ligand binding with stopped-flow rapid mixing. Methods Mol Biol. 2005;305:323–342. doi: 10.1385/1-59259-912-5:323. [DOI] [PubMed] [Google Scholar]
- 7.Westenhoff S, et al. Time-resolved structural studies of protein reaction dynamics: A smorgasbord of X-ray approaches. Acta Crystallogr A. 2010;66:207–219. doi: 10.1107/S0108767309054361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang T, Duan Y. Ligand entry and exit pathways in the β2-adrenergic receptor. J Mol Biol. 2009;392:1102–1115. doi: 10.1016/j.jmb.2009.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hildebrand PW, et al. A ligand channel through the G protein coupled receptor opsin. PLoS One. 2009;4:e4382. doi: 10.1371/journal.pone.0004382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurst DP, et al. A lipid pathway for ligand binding is necessary for a cannabinoid G-protein-coupled receptor. J Biol Chem. 2010;285:17954–17964. doi: 10.1074/jbc.M109.041590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Tajkhorshid E. Electrostatic funneling of substrate in mitochondrial inner membrane carriers. Proc Natl Acad Sci USA. 2008;105:9598–9603. doi: 10.1073/pnas.0801786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharp K, Fine R, Honig B. Computer simulations of the diffusion of a substrate to an active site of an enzyme. Science. 1987;236:1460–1463. doi: 10.1126/science.3589666. [DOI] [PubMed] [Google Scholar]
- 13.Grubmüller H, Heymann B, Tavan P. Ligand binding: molecular mechanics calculation of the streptavidin-biotin rupture force. Science. 1996;271:997–999. doi: 10.1126/science.271.5251.997. [DOI] [PubMed] [Google Scholar]
- 14.Setny P, et al. Dewetting-controlled binding of ligands to hydrophobic pockets. Phys Rev Lett. 2009;103:187801. doi: 10.1103/PhysRevLett.103.187801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad M, Gu W, Helms V. Mechanism of fast peptide recognition by SH3 domains. Angew Chem Int Ed Engl. 2008;47:7626–7630. doi: 10.1002/anie.200801856. [DOI] [PubMed] [Google Scholar]
- 16.Shan Y, et al. How does a drug molecule find its target binding site? J Am Chem Soc. 2011;133:9181–9183. doi: 10.1021/ja202726y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.May LT, Leach K, Sexton PM, Christopoulos A. Allosteric modulation of G-protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2007;47:1–51. doi: 10.1146/annurev.pharmtox.47.120505.105159. [DOI] [PubMed] [Google Scholar]
- 19.Limbird LE, Lefkowitz RJ. Negative cooperativity among β-adrenergic receptors. J Biol Chem. 1976;251:5007–5014. [PubMed] [Google Scholar]
- 20.Caron MG, Lefkowitz RJ. Solubilization and characterization of the β-adrenergic receptor binding sites of frog erythrocytes. J Biol Chem. 1976;251:2374–2384. [PubMed] [Google Scholar]
- 21.Alexander RW, Williams LT, Lefkowitz RJ. Identification of cardiac β-adrenergic receptors by (−) [3H]alprenolol binding. Proc Natl Acad Sci USA. 1975;72:1564–1568. doi: 10.1073/pnas.72.4.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wacker D, et al. Conserved binding mode of human β2 adrenergic receptor inverse agonists and antagonist revealed by X-ray crystallography. J Am Chem Soc. 2010;132:11443–11445. doi: 10.1021/ja105108q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dax EM, Partilla JS. Adrenergic ligand liposolubility in membranes: Direct assessment in a beta-adrenergic binding system. Mol Pharmacol. 1982;22:5–7. [PubMed] [Google Scholar]
- 24.Ballesteros JA, Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure function relations in G-protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 25.Böhm H-J, Klebe G. What can we learn from molecular recognition in protein–ligand complexes for the design of new drugs? Angew Chem Int Ed Engl. 1996;35:2588–2614. [Google Scholar]
- 26.Liu P, Huang X, Zhou R, Berne BJ. Observation of a dewetting transition in the collapse of the melittin tetramer. Nature. 2005;437:159–162. doi: 10.1038/nature03926. [DOI] [PubMed] [Google Scholar]
- 27.Cherezov V, et al. High-resolution crystal structure of an engineered human β2-adrenergic G-protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vilardaga J-P, Bünemann M, Krasel C, Castro M, Lohse MJ. Measurement of the millisecond activation switch of G-protein-coupled receptors in living cells. Nat Biotechnol. 2003;21:807–812. doi: 10.1038/nbt838. [DOI] [PubMed] [Google Scholar]
- 29.Warne T, et al. Structure of a β1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rezmann-Vitti LA, et al. Site-directed mutagenesis of the rat β1-adrenoceptor. Involvement of Tyr356(7.43) in (+/−)cyanopindolol but not (+/−)[125Iodo]cyanopindolol binding. Eur J Med Chem. 2004;39:625–631. doi: 10.1016/j.ejmech.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Kolb P, et al. Structure-based discovery of β2-adrenergic receptor ligands. Proc Natl Acad Sci USA. 2009;106:6843–6848. doi: 10.1073/pnas.0812657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 33.Conn PJ, Christopoulos A, Lindsley CW. Allosteric modulators of GPCRs: A novel approach for the treatment of CNS disorders. Nat Rev Drug Discov. 2009;8:41–54. doi: 10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stockton JM, Birdsall NJM, Burgen ASV, Hulme EC. Modification of the binding properties of muscarinic receptors by gallamine. Mol Pharmacol. 1983;23:551–557. [PubMed] [Google Scholar]
- 35.Redka DS, Pisterzi LF, Wells JW. Binding of orthosteric ligands to the allosteric site of the M2 muscarinic cholinergic receptor. Mol Pharmacol. 2008;74:834–843. doi: 10.1124/mol.108.048074. [DOI] [PubMed] [Google Scholar]
- 36.MacKerell AD, Jr, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 37.Shaw DE, et al. Millisecond-scale molecular dynamics simulations on Anton; Proceedings of the Conference on High Performance Computing, Networking, Storage and Analysis (SC09); New York: ACM; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.