Abstract
CD1e is the only human CD1 protein existing in soluble form in the late endosomes of dendritic cells, where it facilitates the processing of glycolipid antigens that are ultimately recognized by CD1b-restricted T cells. The precise function of CD1e remains undefined, thus impeding efforts to predict the participation of this protein in the presentation of other antigens. To gain insight into its function, we determined the crystal structure of recombinant CD1e expressed in human cells at 2.90-Å resolution. The structure revealed a groove less intricate than in other CD1 proteins, with a significantly wider portal characterized by a 2 Å-larger spacing between the α1 and α2 helices. No electron density corresponding to endogenous ligands was detected within the groove, despite the presence of ligands unequivocally established by native mass spectrometry in recombinant CD1e. Our structural data indicate that the water-exposed CD1e groove could ensure the establishment of loose contacts with lipids. In agreement with this possibility, lipid association and dissociation processes were found to be considerably faster with CD1e than with CD1b. Moreover, CD1e was found to mediate in vitro the transfer of lipids to CD1b and the displacement of lipids from stable CD1b–antigen complexes. Altogether, these data support that CD1e could have evolved to mediate lipid-exchange/editing processes with CD1b and point to a pathway whereby the repertoire of lipid antigens presented by human dendritic cells might be expanded.
Keywords: 3D structure, glycolipid antigen presentation, human CD1b, lipid antigen editing, lipid transfer protein
Four transmembrane CD1 molecules (CD1a, -b, -c, and -d) are expressed in different cell-specific combinations by human immune cells and, among these, in dendritic cells (DCs), the professional antigen-presenting cells (APCs). These proteins present self or microbial lipid antigens to T cells, thus participating in innate and adaptive immunity (1, 2). Myeloid DCs also express a fifth isoform, CD1e, which indirectly participates in glycolipid antigen presentation. This protein has been found to facilitate the processing of complex mycobacterial hexamannosylated phosphatidylinositol (PIM6) by lysosomal α-mannosidase into dimannosylated forms (PIM2) that activate CD1b-restricted T-cell clones (3).
In several respects, CD1e behaves differently from the other human CD1 family members. After biosynthesis, all membrane-anchored CD1 molecules reach the Golgi compartments. Apart from CD1d, which is also delivered directly to endosomes (4), CD1a–d molecules are then transported to the plasma membrane, where they have been shown to bind some antigens. Subsequently, CD1 molecules are constitutively internalized into the endocytic network, where they capture antigenic ligands (4). Finally, the CD1–antigen complexes cycle back to the plasma membrane to activate specific T lymphocytes. In contrast, CD1e remains exclusively intracellular. After reaching the Golgi compartments, CD1e is addressed to sorting endosomes and from there to CD1b+ lysosomes. During its progression through the endosomal network, the CD1e α1–α3 soluble domain is released from the transmembrane domain and the propeptide, consisting of the 12 N-terminal residues, is removed by the action of undefined proteases. Hence, lysosomal CD1e proteins are soluble (sCD1e). The formation of sCD1e in late endosomes/lysosomes has been shown to be necessary for the efficient presentation of PIM6 to CD1b-restricted T cells (3, 5). Intriguingly, CD1e is detected mainly as a membrane-associated form in the Golgi compartments of immature DCs, and DC maturation results in its transfer to CD1b+ compartments and a progressive down-regulation of CD1e biosynthesis (6, 7).
Antigen presentation by CD1 molecules requires the prior transfer of lipid antigens from biological membranes to CD1-binding grooves, a key step mediated by lipid transfer proteins (LTPs) (8–11). The presence of sCD1e molecules in CD1b+ compartments raises the question of whether sCD1e also functions as an LTP. To better define its function, we determined the crystal structure of sCD1e. This structure revealed an exposed lipid-binding groove that appears well suited to mediate lipid transfer processes. This possibility was subsequently confirmed by in vitro experiments that showed that sCD1e interacts with lipids with fast rates and could mediate ligand exchange with CD1b.
Results
Di- and Triacylated Lipids Bind to CD1e.
With the intention of selecting the best CD1e recombinant proteins for crystallization, we compared the isoelectric focusing (IEF) profile and lipid-binding properties of two recombinant (r)sCD1e forms. The first one is a heterodimer of the CD1e α-chain (allele 2) and human β2-microglobulin (β2m) expressed in Drosophila S2 cells (rsCD1e-2); the second form, produced in human cells, is a single-chain CD1e (scCD1e) with β2m covalently linked to the same α-chain. RsCD1e proteins were shown to bind in vitro diacylated PIM6 and PIM2 glycolipids (3). We then enlarged the panel of lipids tested to molecules composed of two or three fatty acid chains (Fig. S1). These studies showed that rsCD1e-2 forms stable complexes with a wide range of two-tailed lipids, e.g., phosphatidylinositol (PI), sulfatides (SLF), bis-(monoacylglycero)phosphate (BMP), or diacylated sulfoglycolipids (Ac2SGL) from Mycobacterium tuberculosis (Mtb) (Fig. S2). Triacylated lipids like hemi-BMP or the Pam3CSK4 lipopeptide also associated with rsCD1e-2. No band shifts were observed with zwitterionic neutral phosphatidylcholine (PC) or sphingomyelin (SM), suggesting that endogenous CD1e ligands could be neutral or absent. Fully deglycosylated scCD1e, prepared by sequential treatment with exo- and endoglycosidases, was also found to interact with di- and triacylated lipids (Fig. S2), demonstrating that rsCD1e-2 and scCD1e display similar lipid-binding properties.
Determination of the Crystal Structure of Human CD1e.
To gain insight into the function of CD1e, we determined its 3D structure. After many unproductive crystallization trials with rsCD1e molecules, we turned our attention to the scCD1e construct, which displayed similar lipid-binding (see above) and lipid-exchange properties (see below) than rsCD1e-2. Fully deglycosylated scCD1e could be crystallized and its structure solved to a resolution of 2.90 Å (Fig. S3A and Table S1). Consistent with the sequence homologies, the backbone structure of scCD1e was similar to those of other human CD1 isoforms. Root-mean-square deviations (rmsd) of 1.10, 1.18, 1.19, and 1.15 Å were observed after superimposition of the backbone atoms of scCD1e on those of human CD1a (PDB ID 1ONQ), CD1b (2H26), CD1c (3OV6), or CD1d (1ZT4), respectively. Differences were most pronounced between residues of the α1–α2 superdomain (e.g., 1.24 Å rmsd with human CD1b). Only the first residue of the linker engineered to connect the β2m and the CD1e heavy chain could be resolved in the electron density. The loop that connects the last two β-sheets of the α2 domain of scCD1e was also poorly defined in electron density, and as a result residues Q120–I122 were excluded from the final model.
Structure of the CD1e Lipid-Binding Groove.
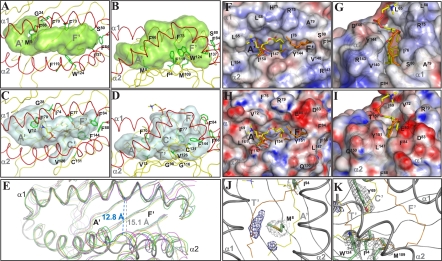
The crystal structure of scCD1e revealed a binding groove comprising two main pockets, which, according to the terminology for mouse CD1d, will be called the A′ and F′ pockets (Fig. 1 A and B and Fig. S3 B and C). As in other CD1 molecules, the binding pockets are bordered mainly by hydrophobic residues. The A′ pocket is the most conserved structural feature among CD1 grooves. Access to this pocket is gained through the main portal at the protein surface. A major portion of the A′ pocket in scCD1e is shielded from the aqueous milieu by a cover of well-conserved hydrophobic residues (Phe54, Leu62, Leu65, Leu154, and Thr158). Below this cover, the pocket follows a toroidal trace around a central pole defined by residues Met8 and Phe66. Similarly sized hydrophobic residues occupy the corresponding positions in other human CD1 isoforms, e.g., Val12 and Phe70 in CD1b (Fig. 1 C and D). The remaining residues that border the A′ pocket of CD1e or lie at the interface with the F′ compartment are comparable in size to those found in other CD1 molecules.A noteworthy difference is Ile94, which replaces the Gly98 of human CD1b and prevents the occurrence of a similar T′ tunnel (Fig. 1 B vs. D). A comparison with the superimposed structure of CD1b in complex with PC and the unknown ligand (UL) spacer indicates that the A′ pocket of human CD1e provides enough space to accommodate up to 30-carbon-long (C30) hydrophobic chains.
Fig. 1.
Structure of the scCD1e α1–α2 superdomain showing the groove architecture. Top (A) and side (B) views of the α1–α2 domain of scCD1e, with the transparent molecular surface of the groove rendered in light green. (C and D) Similar views of the human CD1b groove (2H26, gray surface). (A–D) The protein backbones are colored in yellow with the exception of the α-helical portions (red). PC associated with CD1b is shown as sticks with the carbon atoms in yellow, oxygens in red, nitrogens in blue, and phosphates in orange. All UL atoms are colored in orange. Side-chain atoms of residues differing among CD1 isoforms and/or causing notable changes in scCD1e groove architecture are shown as green sticks, with the corresponding Cα atoms depicted as balls. (E) Aperture of the CD1e groove portal. Comparison of the α1–α2 domain of scCD1e (thick gray tubes) with those of human CD1a (yellow, 1ONQ), CD1b (blue, 2H26), CD1c (magenta, 3OV6), and CD1d (green, 1ZT4). The image was generated by superimposition of residues 57–79 of the α1 helix and residues 93–101 of the β-sheet platform of scCD1e on the corresponding residues of the other human CD1 molecules. The distance between the Cα atoms of Phe73 and Tyr144 of scCD1e is compared with the distance between the corresponding Phe77 and Tyr151 of CD1b. (F–I) Comparison of the groove portals of scCD1e (F and G) and human CD1b (H and I). The molecular surfaces are presented with electrostatic potentials calculated at neutral pH (red, electronegative; blue, electropositive; −30 to +30 kT/e) in the top (F and H) and front views (G and I). PC and UL ligands are shown as sticks with atoms colored as in C and D. The position of the CD1b-bound ligands is shown for comparison in F and G after superimposition on the scCD1e structure. (J–K) Weak electron density from ligands in the scCD1e lipid-binding groove. The final FoFc map was contoured at 3.0σ (blue mesh) around the position of atoms of groove-bound ligands from superimposed CD1 structures (PC and UL of human CD1b, αGalCer of human CD1d, SLF of human CD1a, and mannosyl-β1-phosphomycoketide (MPM) and C12 of human CD1c). For ease of comparison, only the CD1b ligands are shown with lines colored as noted above. The electron densities (2FoFc, contoured at 1.0 σ) around the side-chain atoms of Met8 and Ile94 (J) or Tyr69, Ile94, Met109, and Trp124 (K) are also shown in gray. The positions of the A′, C′, F′, and T′ pockets/tunnel of human CD1b are indicated for comparison.
The most remarkable structural differences between human CD1e and other CD1 proteins lie in the F′ pocket (Fig. S3C). This structure combines the F′ and the uppermost portion of the C′ pocket of human CD1b, as a result of the wider separation between the helices of the α1 and α2 domains (see below) and the presence of the Phe73/Ile137 residues in CD1e replacing the Phe77/Phe144 pole of CD1b (Fig. 1 A–D). Merged C′ and F′ pockets also occur in human CD1a (Ser77/Phe144), CD1c (Leu77/Val144), and CD1d (Phe77/Ala144) (Fig. S4). Also in common with CD1a, CD1c, and CD1d, the T′ tunnel and the deepest part of the C′ pocket of human CD1b are blocked in CD1e by the side chains of Met109 and Trp124, respectively, at positions equivalent to the smaller Gly116 and Cys131 of CD1b. Otherwise, the remaining hydrophobic residues lining the CD1e F′ pocket resemble in size and hydrophobicity the corresponding residues of other human CD1 molecules.
Wide Open Portal Above the CD1e F′ Pocket.
The relative position of the α1 and α2 helical segments of CD1e differs considerably from what is observed in other CD1 structures. The clefts formed between the α1 and α2 domains of other CD1 molecules are about 14 Å wide and of nearly constant width. In CD1e, the distance between the two helices is ∼2 Å greater at the center of the superdomain, resulting in a wider groove entrance above the F′ pocket (Fig. 1E). Indeed, after superimposition of backbone atoms of the α1 helix and the central β-sheet platform, an rmsd of 3.2, 3.4, 3.4, and 3.10 Å was calculated between the 142–146 α2 interhelical segment of CD1e and the corresponding 149–153 residues of human CD1a, CD1b, CD1c, and CD1d, respectively. The 16-Å-wide aperture in CD1e is halfway between those in CD1 and MHC class I and class II molecules (18–20 Å in the middle of the cleft) (12).
The portal dimensions of the CD1e groove also increase in the direction running parallel to the two helices, compared with other CD1 molecules (Fig. 1F and Fig. S4). The maximal opening is dictated by the side-chain atoms of Leu65 and Phe84, which are 23.7 Å apart. In comparison, the longest aperture drops to 18.6 Å in human CD1b (Fig. 1H and Fig. S4), mostly as a consequence of the replacement of Ser80 in CD1e by the bulkier Phe84. Furthermore, the Ser80 of CD1e induces a marked wall depression on one side of the groove cleft, between the α1 helix C terminus and the α2 helix N terminus (Fig. 1G). This feature is most evident in comparison with CD1b (Fig. 1I). Interestingly, a similar lateral aperture of the F′ pocket has been recently observed in the crystal structure of hCD1c (13).
Overall, the calculated surface area of the CD1e portal (130 Å2) is significantly greater than those of human CD1a (98 Å2), CD1b (89 Å2), and CD1d (84 Å2), but much smaller than that of hCD1c (240 Å2). A wide and laterally exposed CD1e cleft might affect the way in which ligands will be anchored within the groove and how lipid polar heads will be held in place by surrounding residues. A spatially less constrained CD1e groove might also enhance the rates of lipid exchange.
No Endogenous Ligands Are Observed in the Groove of Crystallized scCD1e.
Weak and discontinuous electron density was detected within the A′ and F′ pockets of the scCD1e-binding groove (Fig. 1 J–K), suggesting that endogenous lipids are absent in crystallized scCD1e. To confirm that CD1e is secreted from human cells in association with endogenous ligands, we characterized scCD1e by native mass spectrometry (14). Electrospray ionization (ESI)-MS experiments in positive and negative modes and tandem MS-MS spectra of selected precursors demonstrated that fully deglycosylated scCD1e is associated with either PC or SM (Fig. S5). ESI-MS data also suggested the simultaneous association of scCD1e–PC and scCD1e–SM complexes with a second ligand [CD1e-associated unknown ligand (ULe)] of about 260–280 Da, reminiscent of the spacer ligands found in human CD1b (15) or mouse CD1d (16). This ULe ligand was detected at marginal intensities as a dissociation product in negative-mode tandem MS experiments (Fig. S5D, Inset). Its m/z values might correspond to those of unsaturated C17 to C20 fatty acids.
The weak electron density within the scCD1e groove therefore indicates either that endogenous ligands were lost in the course of crystallization or that ligands adopt variable conformations in the groove. Unfortunately, all our efforts to characterize crystallized scCD1e molecules by native MS proved unsuccessful, probably as a consequence of the impossibility of sufficiently removing salts and precipitants required for crystallization.
Lipid-Exchange Processes Are Faster with CD1e Than with CD1b.
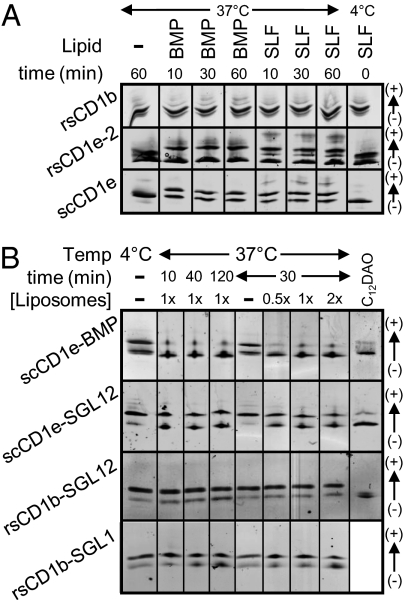
The wide and water-exposed groove cleft of CD1e suggests that lipids could associate with and/or dissociate from CD1e at faster rates than with other CD1 isoforms. To investigate this possibility, we compared the rate of lipid binding to rsCD1e-2 or scCD1e with rsCD1b. In vitro, incubation of rsCD1e-2 or scCD1e with BMP or SLF at pH 5.0 resulted in 40–60% lipid incorporation in less than 10 min (Fig. 2A). In contrast, no significant binding to rsCD1b was detected even when incubation was extended for 1 h. Only lipids with exceedingly large polar heads, e.g., GM1 or GD3 gangliosides, associated rapidly with rsCD1b. With all other lipids tested, complex formation required either the presence of detergents or longer incubation at pH ≤4.0, which is the reported optimal pH for lipid loading by CD1b (17–19). In contrast, scCD1e–lipid complex formation was complete within the first 10 min of incubation at both neutral and acid pH (Fig. S6A), in agreement with published data obtained for rsCD1e forms with fluorescent phosphatidylserine-nitro-benzoxadiazol (20).
Fig. 2.
Lipid binding to and dissociation from CD1e is rapid. (A) CD1 molecules were incubated at pH 5.0 with a 10-fold molar excess of either BMP or SLF for the time and at the temperature indicated, and the products were analyzed by IEF. The binding occurring during the time of IEF deposition and separation was controlled by mixing CD1 proteins and SLF at 4 °C immediately before deposition (lane 8). (B) CD1–lipid complexes were purified by chromatofocusing and subsequently incubated at pH 5.0 for the time and at the temperature indicated in the presence or absence of liposomes composed of PC/PE/SM/Chol (1× represents 500/200/200/250 μM final concentrations). The last lane of each gel corresponds to the given complex after incubation with 2.5 mM C12DAO at room temperature for 3–5 min, a treatment that induces ligand dissociation.
The stability of CD1e–lipid and CD1b–lipid complexes was also compared. Complexes between scCD1e and either BMP or sulfoglycolipid (SGL) 12, as well as between rsCD1b and either SGL12 or SGL1, were prepared and purified by chromatofocusing with 30–50% yields. The two synthetic SGL molecules share the same polar head and differ only by the length and the presence of methyl ramifications in one of the two hydrophobic tails (Fig. S1) (21). In the case of scCD1e–BMP, an ∼1:1 mixture of loaded and unloaded species was observed immediately after purification. This was probably due to insufficient chromatographic resolution rather than to poor stability because scCD1e–lipid complexes (like rsCD1b–lipid complexes) were stable when incubated in the absence of external lipids (Fig. S6B).
Purified complexes were next incubated at pH 5.0 in the presence of liposomes composed of mixtures of neutral lipids representing biological membranes. The persistence of CD1-bound anionic lipids was monitored by IEF. These experiments showed that anionic ligands dissociated from scCD1e considerably faster than from rsCD1b (Fig. 2B). The proportion of scCD1e–BMP and scCD1e–SGL12 complexes dropped to 10 and 50%, respectively, after the first 10 min of incubation. In contrast, dissociation of rsCD1b–SGL complexes occurred slowly, reaching 10–30% yields after 2 h. Interestingly, ligand displacement from scCD1e occurred within the first minutes of incubation, but then rapidly stagnated and was barely affected by the concentration of neutral liposomes, pointing to the presence of complexes of different stability. The dissociation behavior was dependent on the identity of the ligand, suggesting that this property is not intrinsic to the recombinant protein.
CD1e Exchanges Lipids with CD1b.
Our structural findings and the rapid kinetics of lipid association and dissociation support the view that CD1e might behave as an LTP in late endosomes/lysosomes. To further explore this hypothesis, we tested (i) whether CD1e modifies the stability of CD1b–lipid complexes and (ii) whether CD1e mediates the transfer of lipids to CD1b.
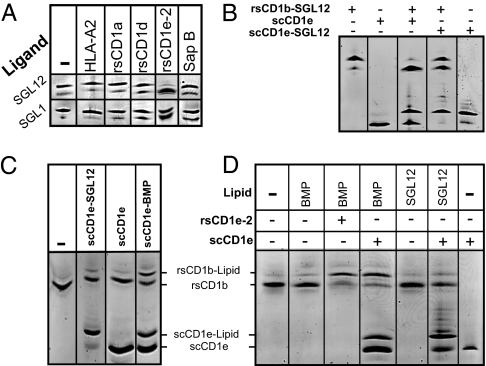
Despite the high stability of purified rsCD1b–SGL12 (see above) and its demonstrated strong T-cell–stimulatory capacity (21), incubation of the complex with rsCD1e-2 caused almost complete dissociation of SGL12 from rsCD1b within 30 min (Fig. 3A). Little or no such effect was observed in control experiments using a recombinant soluble HLA class I protein; human CD1a or CD1d; human β2m; BSA; or the LTP saposins A, B, or C (Fig. S6C). The unloading of rsCD1b–SGL12 by rsCD1e-2 was too fast to permit accurate kinetic analyses, sample loading and separation on IEF gels requiring a minimum of 10 min.
Fig. 3.
CD1e mediates the exchange of lipids with CD1b. (A) rsCD1e-2 induces the dissociation of ligands from CD1b. Purified rsCD1b-SGL12 or rsCD1b-SGL1 was incubated for 30 min at room temperature in the presence or absence of a 3-fold molar excess of HLA-A2, human rsCD1a, or rsCD1d; a 1.5-fold excess of rsCD1e-2; or a 10-fold excess of saposin B, before IEF separation. The incubation mixtures included 0.5× lipid vesicles (Fig. 2). (B) Single-chain CD1e efficiently unloads CD1b-bound lipids. Purified rsCD1b-SGL12 (8 μM) was incubated for 30 min with or without an equimolar amount of scCD1e or scCD1e-SGL12 in the presence of 0.5× lipid vesicles, before IEF separation. (C) Direct transfer of scCD1e-bound lipids to CD1b. Mixtures containing rsCD1b and the indicated scCD1e–lipid complex or unloaded scCD1e were incubated for 10 min at pH 5.0 and 37 °C, before analysis by IEF. (D) CD1e transfers lipids from vesicles to CD1b. BMP or SGL12 vesicles were incubated for 30 min at pH 5.0 with rsCD1b in the presence or absence of rsCD1e-2 or scCD1e, before analysis by IEF. To achieve higher magnification, bands from sCD1e-2 observed at lower pI values (Fig. S6E) are not presented here. Data are representative of at least three independent experiments.
RsCD1e-2 unloaded the rsCD1b–SGL12 complexes more efficiently than the rsCD1b–SGL1 complexes (Fig. 3A and Fig. S6D). The displacement of SGL12 from rsCD1b was also efficiently elicited by scCD1e (Fig. 3B and Fig. S6C). Use of scCD1e permitted us to visualize the appearance of a molecular species identical in pI to scCD1e–SGL12, demonstrating the occurrence of ligand transfer from CD1b to CD1e (lane 3, Fig. 3B). A groove-to-groove mechanism of ligand transfer was further supported by the fact that rsCD1b–SGL12 unloading was considerably impeded in incubations with scCD1e–SGL12, compared with similar incubations with scCD1e (compare lanes 3 and 4, Fig. 3B). Altogether, these experiments show that CD1b-bound ligands can be removed by and transferred to sCD1e.
When the reverse process was investigated, lipid transfer to rsCD1b occurred efficiently from scCD1e–BMP complexes, despite the presence of an ∼1:1 mixture of loaded and unloaded species, but was faint from the scCD1e–SGL12 complexes (Fig. 3C). These findings support the idea that CD1e could sustain lipid transfer to CD1b and suggest that this process displays ligand selectivity.
CD1b–Lipid Complexes Form Faster in the Presence of CD1e.
We finally investigated whether sCD1e might mediate the transfer of lipids from vesicles to CD1b. Addition of rsCD1e-2 or scCD1e to reaction mixtures containing rsCD1b and anionic lipid vesicles resulted in fast formation of rsCD1b–lipid complexes (Fig. 3D and Fig. S6E). The yields were influenced by the nature of the lipid in the following order: BMP ∼ PI > SLF > PS > SGL1 > SGL12. RsCD1e-2 molecules still transferred to rsCD1b faint and moderate quantities of triacylated hemi-BMP and Pam3CSK4, respectively. Lipid transfer was fast, the reaction with BMP being almost complete within 10 min. Moreover, substoichiometric amounts of rsCD1e-2 promoted efficient loading of rsCD1b. Thus, an eightfold lower concentration of CD1e than CD1b sufficed to induce the formation of about 40% rsCD1b–BMP after 1 h of incubation (Fig. S6F). Remarkably, BMP transfer occurred equally well under neutral and acidic conditions.
Discussion
CD1 proteins present self and nonself lipid antigens to helper and cytotoxic T cells, thereby alerting the immune system to infectious attacks and other pathological disorders of the host. The stimulation and expansion of CD1-restricted T cells in vivo depends on the nature and abundance of the lipid antigens occupying the grooves of CD1 molecules present on the APC plasma membrane, which in turn will be determined by the cellular localization and concentration of CD1 molecules, antigens, and competing binders and by their binding affinities. Lipids bind to the deep pockets of CD1 molecules through their long hydrophobic tails, and accordingly their binding affinity is assumed to be dominated by unspecific hydrophobic interactions. Not surprisingly, CD1 proteins have been described as interacting with many lipids in vitro and even each CD1 isoform as presenting diverse antigens (1, 2). It is therefore important to define how the selectivity of antigen presentation is achieved.
Two factors contribute to minimize lipid-exchange processes in the course of CD1 recycling between the plasma membrane and the endosomal network (4). First, lipids are integral components of biological membranes and need to be transferred from membranes to the CD1 molecules. Second, the endogenous lipids and spacer molecules that occupy the CD1 grooves directly after biosynthesis may be expected to hamper spontaneous lipid binding (15, 22–24). These observations suggest that additional actors must exist to optimize lipid antigen presentation. Previous studies have shown that lysosomal LTPs play an important role. Saposin C facilitates the presentation of mycobacterial lipids to CD1b-restricted T cells and was proposed to act by extracting these antigens from membranes and interacting directly with CD1b (10). The presentation of self-antigens by CD1d and the development of type 1 NKT cells also rely on saposins and the Niemann-Pick type C2 protein (NPC2) (8, 9, 11, 25, 26). Saposins, NPC2, and the GM2 activator protein were shown to mediate the exchange of CD1d ligands in vitro and the editing of CD1d–lipid complexes.
Our data support this notion and strongly suggest that CD1e behaves as an efficient CD1-related LTP. Several observations already pointed to this possibility: (i) CD1e colocalizes with CD1b in the late endosomes and lysosomes of mature DCs (7), (ii) the arrival of CD1e in these compartments is synchronized with proteolytic events that release the luminal domain as a soluble form (27), and (iii) the generation of soluble CD1e is necessary for the presentation by CD1b of CD1e-dependent antigens (3). Our in vitro data confirm that CD1e might transfer lipids to human CD1b and exercise quality control, as indicated by its ability to disrupt stable CD1b–SGL12 complexes known to stimulate T cells in CD1b-Ag–coated plate assays (21). Lipid loading onto and unloading from CD1b was found to be rapid in the presence of CD1e. Notably, CD1b–lipid complexes formed equally well at neutral and acid pH, suggesting that CD1e might enhance lipid availability/accessibility or facilitate prior endogenous lipid unloading from CD1b. Alternatively, CD1e might also disrupt CD1b α1–α2 interdomain tethers proposed to hamper antigen capture (17), if tethering interactions persist at both pHs. Remarkably, substoichiometric CD1e:CD1b ratios sufficed to mediate lipid transfer, contrasting with what has been reported for other LTPs. Thus, the transfer activity of saposins required equimolar saposin/CD1d ratios (8), whereas at least 10-fold molar excesses were needed in the case of NPC2 (9).
The 3D structure of CD1e strongly suggests an adaptation of groove design to an LTP function. The CD1e mitten-like groove architecture and volume (2,000 Å3, using a 1.7-Å radius probe) ensures good recognition of potential ligands of other CD1s. The A′ pocket of CD1e provides space to embed and tightly hold up to C30-long hydrophobic tails. In contrast, the wider entrance of the CD1e groove and lateral F′ pocket exposition might ensure that loose interactions are established with bound ligands. Such F′ pocket design could also endow CD1e with the capacity to interact with, and thus to compete with, other CD1s for structurally diverse ligands. In line with this possibility, CD1e was found to bind and to exchange di- and triacylated lipids. One should note that only five ionic residues are found in the α1 helix of CD1e, compared with 11–13 in other CD1 isoforms. Among them, only the polymorphic His71 and Arg75 lie sufficiently close to the CD1e groove portal to participate in ligand-holding interactions. Altogether, these structural features are predicted to enhance lipid association and dissociation rates in CD1e and permit an understanding of why lipids were not observed in the scCD1e structure despite being associated with the protein before crystallization. The weak electron density from ligands in the scCD1e structure is indeed a remarkable observation, which contrasts with the fact that ligand electron densities were invariantly observed in the grooves of all other reported CD1 molecules (28). This also raises the question of whether the wider cleft of CD1e is an intrinsic property of this isoform or, in contrast, could be consequence of such an empty groove state. Unfortunately, all our attempts to clarify this important point via the elucidation of crystal structures of purified scCD1e–lipid complexes (i.e., with SGL12 and hBMP) proved unsuccessful. Finally, an alternative explanation to the weak electron density might be proposed. Indeed, PC/SM/ULe ligands might be expected to adopt variable conformations and orientations within the groove if the greater interhelical separation results in weaker contacts between ligand acyl tails and hydrophobic groove residues. In our opinion, this hypothesis is unlikely because fatty acid tail portions should still be detected in the relatively narrow A′ pocket, similar to what was reported for endogenous PC and phosphatidylethanolamine (PE) ligands bound with variable orientations to bovine CD1b3 (29).
Lipid polar heads probably define the selectivity of CD1e lipid extraction from membranes and lipid exchange with CD1b, as has been proposed for other LTPs (8). However, the fact that SGL12 was more efficiently unloaded from CD1b than SGL1 indicates that fatty acids are also sensed by CD1e. Interestingly, we recently solved the crystal structure of the T-cell–stimulatory CD1b–SGL12 complex and found that the methyl-branched portion remains exposed above the A′ pocket. It can therefore be argued that such motifs might be recognized not only by T-cell receptor (TCR) but also by CD1e. Alternatively, the presence of methyl branches could induce conformational changes in exposed CD1b residues or differences of polar head positioning that would be sensed by CD1e. Further investigations will be necessary to determine the structural parameters recognized by this protein.
In summary, the groove design of CD1e, poorly intricate and presenting a large laterally exposed portal surrounded by relatively few residues capable of holding antigen polar heads, combined with the particular traffic of CD1e within DCs and the fact that soluble forms are generated in CD1b+ compartments, strongly suggests that CD1e has evolved to act as a privileged CD1-related LTP. The CD1e groove architecture ensures partial ligand selectivity overlap with other human CD1 molecules while permitting sufficiently fast rates of lipid association and dissociation. In this manner, human CD1e, in combination with other lysosomal LTPs, may be predicted to control the repertoire of lipids associating with CD1b and possibly those associating with other CD1 molecules, thereby influencing the presentation of lipid antigens to T cells.
Materials and Methods
Reagents, cell lines, and detailed experimental procedures are described in SI Materials and Methods.
Preparation of scCD1e.
A β2m–CD1e–mouse Fc IgG1 fusion molecule was expressed in pFuse-mIgG1 (InvivoGen). The human β2m sequence was followed by a DDDDKGSSSSDDDDK connecting peptide linked to amino acids 32–303 of prepro-CD1e, followed by a tobacco etch virus protease recognition site and the Fc fragment of a mouse IgG1. Stably transfected M10 cells expressing the sequence encoding the fusion protein were isolated and grown to confluence in 6,360-cm2 Cellstack tissue culture flasks (Corning, Sigma) containing 1 L RPMI 1640 and 5% FCS. The cells were maintained for 7–10 d, and the medium was collected every day. The filtered culture media were concentrated 50-fold by ultrafiltration on 10-kDa molecular weight cutoff (MWCO) membranes (Millipore). Fully deglycosylated scCD1e was prepared as follows.
Step 1.
Glycosylated scCD1e–Fc fusion protein was purified by affinity chromatography, using a column obtained by reaction between the anti-CD1e antibody 20.6 (27) and NHS-activated Sepharose 4 (GE Healthcare). After loading the cell supernatants and performing a PBS wash, elution was carried out with 100 mM glycine (pH 2.9). After immediate neutralization with Hepes (pH 7.5), protein fractions were pooled, concentrated, and buffer-exchanged against 10 mM Na acetate/20 mM NaCl/0.5 mM EDTA (pH 4.5), using 10-kDa MWCO centrifuge filters.
Step 2.
Purified scCD1e-Fc was treated overnight at 30 °C with an Endo F3–MBP fusion molecule (15). The pH was adjusted to 8.3 with 1 M Tris, DTT, and EDTA (1-mM final concentration of each) added and the mixture shaken in the presence of turboTEV (GenWay) for 24 h at 30 °C. Fully deglycosylated scCD1e, contaminated with ∼15% partially glycosylated species, was immunopurified as in step 1, concentrated, and buffer-exchanged against 20 mM Na citrate (pH 6.0).
Step 3.
The last sample was treated successively (4-h intervals) at 37 °C with neuraminidase, β(1,4)-galactosidase and β-N-acetyl-glucosaminidase (all from NEB). After overnight reaction, the pH was adjusted to 5.0 (1 M Na citrate), and Endo F1 (QA-Bio) was added and left to react for 24 h at 30 °C. The pH was neutralized (1 M Hepes), and fully deglycosylated scCD1e was immunopurified as in step 1.
Lipid Binding to CD1 Molecules.
Lipid loading onto CD1 molecules was monitored by IEF (SI Materials and Methods and ref. 15).
Purification of CD1–Lipid Complexes.
Aliquots containing a 10-fold molar excess of lipid over CD1 (100–500 μg) were shaken (90 min, 37 °C) as described (15). In the case of rsCD1b–SGL complexes, the mixture included 10 mM sodium taurocholate, and shaking was continued for 5 h. The samples were buffer-exchanged against buffer A: 25 mM Tris·HCl (pH 8.1), 25 mM bis·Tris·HCl (pH 6.5), or 25 mM piperazine/HCl (pH 5.7) for rsCD1e-2, scCD1e, or rsCD1b, respectively. Protein–lipid complexes were then injected into a chromatofocusing MonoP column (GE Healthcare) and eluted with buffer B: 10-fold diluted polybuffer 96/HCl (pH 6.5) for rsCD1e-2 or 10-fold diluted polybuffer 74/HCl (pH 4.3) for scCD1e and rsCD1b. Protein-containing fractions were pooled, concentrated, and buffer-exchanged in three cycles of 10-fold dilution-concentration against 10 mM Na acetate/50 mM NaCl/1 mM EDTA (pH 5.0), using 10-kDa MWCO centrifugation filters.
Stability of CD1–Lipid Complexes and LTP-Mediated Lipid Displacement.
Aliquots (6 μg) of purified CD1–lipid complexes (15–20 μM final concentration) were shaken (800 rpm, 37 °C) in glass tubes in the presence or absence of 0.5–2× concentrations of liposomes composed of PC/PE/SM/cholesterol (Chol) (1× corresponding to 500/200/200/250 μM final concentration) in 50 mM Na acetate/50 mM NaCl/1 mM DTT/1 mM EDTA (pH 5.0). In CD1e-mediated ligand displacement experiments with CD1b–lipid complexes, mixtures containing rsCD1b–lipid (7 μM), 0.5× liposomes, and an excess of each protein (indicated in the legends for Fig. 3 and Fig. S6) were shaken in the same buffer at 800 rpm and 20 °C. After 30 min (unless otherwise indicated), the samples were cooled on ice and analyzed by IEF as described in SI Materials and Methods.
Crystallization of Deglycosylated CD1e.
A sample of fully deglycosylated scCD1e was buffer-exchanged in five cycles of 10-fold dilution concentration against 10 mM Na acetate (pH 4.0) and concentrated to 5.8 mg/mL. Crystals grew after 3 d at 20 °C, using the hanging-drop vapor-diffusion method, from drops containing 1.0 μL scCD1e and 1.0 μL precipitant [10% PEG6000, 0.2 M Na malonate, 0.1 M Mg (valerate)2, 0.1 M Na borate (pH 3.60)].
Structure Determination, Analysis, and Presentation.
Diffracted intensities were collected from a single crystal at the European Synchrotron Radiation Facility. Other details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to the staff of the European Synchrotron Radiation Facility for excellent data collection facilities. This work benefited from the constant support of the Centre National de la Recherche Scientifique, the Agence Nationale de la Recherche Emergence (ANR-07-EMPB-029-01 and ANR-05-MIIM-006), the Etablissement Français du Sang-Alsace, the Swiss National Foundation (Grant 3100A0 122464/1), and the European Union (FP6 TBVAC program). G.G. was supported by Association de Recherche et de Développement en Médecine et en Santé Publique.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, http://www.pdb.org (PDB ID code 3S6C).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105627108/-/DCSupplemental.
References
- 1.De Libero G, Mori L. Recognition of lipid antigens by T cells. Nat Rev Immunol. 2005;5:485–496. doi: 10.1038/nri1631. [DOI] [PubMed] [Google Scholar]
- 2.Barral DC, Brenner MB. CD1 antigen presentation: How it works. Nat Rev Immunol. 2007;7:929–941. doi: 10.1038/nri2191. [DOI] [PubMed] [Google Scholar]
- 3.de la Salle H, et al. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310:1321–1324. doi: 10.1126/science.1115301. [DOI] [PubMed] [Google Scholar]
- 4.Moody DB, Porcelli SA. Intracellular pathways of CD1 antigen presentation. Nat Rev Immunol. 2003;3:11–22. doi: 10.1038/nri979. [DOI] [PubMed] [Google Scholar]
- 5.Maitre B, et al. The assembly of CD1e is controlled by an N-terminal propeptide which is processed in endosomal compartments. Biochem J. 2009;419:661–668. doi: 10.1042/BJ20082204. [DOI] [PubMed] [Google Scholar]
- 6.Maitre B, et al. Control of the intracellular pathway of CD1e. Traffic. 2008;9:431–445. doi: 10.1111/j.1600-0854.2008.00707.x. [DOI] [PubMed] [Google Scholar]
- 7.Angenieux C, et al. The cellular pathway of CD1e in immature and maturing dendritic cells. Traffic. 2005;6:286–302. doi: 10.1111/j.1600-0854.2005.00272.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhou D, et al. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 2004;303:523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schrantz N, et al. The Niemann-Pick type C2 protein loads isoglobotrihexosylceramide onto CD1d molecules and contributes to the thymic selection of NKT cells. J Exp Med. 2007;204:841–852. doi: 10.1084/jem.20061562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winau F, et al. Saposin C is required for lipid presentation by human CD1b. Nat Immunol. 2004;5:169–174. doi: 10.1038/ni1035. [DOI] [PubMed] [Google Scholar]
- 11.Kang SJ, Cresswell P. Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat Immunol. 2004;5:175–181. doi: 10.1038/ni1034. [DOI] [PubMed] [Google Scholar]
- 12.Zeng Z, et al. Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 13.Scharf L, et al. The 2.5 Å structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity. 2010;33:853–862. doi: 10.1016/j.immuni.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van den Heuvel RH, Heck AJ. Native protein mass spectrometry: From intact oligomers to functional machineries. Curr Opin Chem Biol. 2004;8:519–526. doi: 10.1016/j.cbpa.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Alles LF, et al. Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b. EMBO J. 2006;25:3684–3692. doi: 10.1038/sj.emboj.7601244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zajonc DM, et al. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat Immunol. 2005;6:810–818. doi: 10.1038/ni1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Relloso M, et al. pH-dependent interdomain tethers of CD1b regulate its antigen capture. Immunity. 2008;28:774–786. doi: 10.1016/j.immuni.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng TY, et al. Role of lipid trimming and CD1 groove size in cellular antigen presentation. EMBO J. 2006;25:2989–2999. doi: 10.1038/sj.emboj.7601185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernst WA, et al. Molecular interaction of CD1b with lipoglycan antigens. Immunity. 1998;8:331–340. doi: 10.1016/s1074-7613(00)80538-5. [DOI] [PubMed] [Google Scholar]
- 20.Bushmarina N, et al. Increased flexibility and liposome-binding capacity of CD1e at endosomal pH. FEBS J. 2011;278:21022–2033. doi: 10.1111/j.1742-4658.2011.08118.x. [DOI] [PubMed] [Google Scholar]
- 21.Guiard J, et al. Fatty acyl structures of mycobacterium tuberculosis sulfoglycolipid govern T cell response. J Immunol. 2009;182:7030–7037. doi: 10.4049/jimmunol.0804044. [DOI] [PubMed] [Google Scholar]
- 22.Joyce S, et al. Natural ligand of mouse CD1d1: Cellular glycosylphosphatidylinositol. Science. 1998;279:1541–1544. doi: 10.1126/science.279.5356.1541. [DOI] [PubMed] [Google Scholar]
- 23.Park JJ, et al. Lipid-protein interactions: Biosynthetic assembly of CD1 with lipids in the endoplasmic reticulum is evolutionarily conserved. Proc Natl Acad Sci USA. 2004;101:1022–1026. doi: 10.1073/pnas.0307847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giabbai B, et al. Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: A molecular basis for NKT cell activation. J Immunol. 2005;175:977–984. doi: 10.4049/jimmunol.175.2.977. [DOI] [PubMed] [Google Scholar]
- 25.Gadola SD, et al. Impaired selection of invariant natural killer T cells in diverse mouse models of glycosphingolipid lysosomal storage diseases. J Exp Med. 2006;203:2293–2303. doi: 10.1084/jem.20060921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schumann J, et al. Differential alteration of lipid antigen presentation to NKT cells due to imbalances in lipid metabolism. Eur J Immunol. 2007;37:1431–1441. doi: 10.1002/eji.200737160. [DOI] [PubMed] [Google Scholar]
- 27.Angenieux C, et al. Characterization of CD1e, a third type of CD1 molecule expressed in dendritic cells. J Biol Chem. 2000;275:37757–37764. doi: 10.1074/jbc.M007082200. [DOI] [PubMed] [Google Scholar]
- 28.Zajonc DM, Wilson IA. Architecture of CD1 proteins. Curr Top Microbiol Immunol. 2007;314:27–50. doi: 10.1007/978-3-540-69511-0_2. [DOI] [PubMed] [Google Scholar]
- 29.Girardi E, et al. Crystal structure of bovine CD1b3 with endogenously bound ligands. J Immunol. 2010;185:376–386. doi: 10.4049/jimmunol.1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.