Abstract
The homohexameric (L)Sm protein Hfq is a central mediator of small RNA-based gene regulation in bacteria. Hfq recognizes small regulatory RNAs (sRNAs) specifically, despite their structural diversity. This specificity could not be explained by previously described RNA-binding modes of Hfq. Here we present a distinct and preferred mode of Hfq–RNA interaction that involves the direct recognition of a uridine-rich RNA 3′ end. This feature is common in bacterial RNA transcripts as a consequence of Rho-independent transcription termination and hence likely contributes significantly to the general recognition of sRNAs by Hfq. Isothermal titration calorimetry shows nanomolar affinity between Salmonella typhimurium Hfq and a hexauridine substrate. We determined a crystal structure of the complex that reveals a constricted RNA backbone conformation in the proximal RNA-binding site of Hfq, allowing for a direct protein contact of the 3′ hydroxyl group. A free 3′ hydroxyl group is crucial for the high-affinity interaction with Hfq also in the context of a full-length sRNA substrate, RybB. The capacity of Hfq to occupy and sequester the RNA 3′ end has important implications for the mechanisms by which Hfq is thought to affect sRNA stability, turnover, and regulation.
Keywords: RNA chaperone, regulation of translation, RNA degradation, prokaryotes
Hfq is an abundant and widely conserved RNA-binding protein in bacteria and a major player in the RNA-based regulation of gene expression, such as in the adaptive response to cell stress or in the induction of virulence (1, 2).
Hfq was originally identified as a host factor in Escherichia coli for the replication of the Qβ phage (3), where it binds to the C-rich 3′ end of plus-strand viral RNA (4). Subsequently, physiological roles of Hfq were described in the regulation of mRNA translation and in mRNA degradation, where it modulates the processing of RNA 3′ ends (1, 5–8). The most prominent function of Hfq, however, is its interaction with small regulatory RNAs (sRNAs) that act in trans and that are differentially expressed under various metabolic and environmental conditions (9, 10). They globally regulate gene expression via base-pairing to frequently entire sets of partially complementary mRNAs (11, 12). Hfq stabilizes sRNAs in the absence of their targets (13). Hfq was also found to facilitate base-pairing to the mRNAs and help trigger subsequent steps, such as the repression of translation and/or the acceleration of decay, but also mRNA activation (11, 14). Despite their structural diversity, the recognition of many sRNAs by Hfq is highly specific and even works across species barriers (15). It is an intriguing question how this selectivity is achieved.
Crystal structures reveal that bacterial Hfq adopts an (L)Sm fold and forms homohexameric rings, whereas related (L)Sm proteins [Sm proteins and Sm-like (LSm) proteins] in archaea and eukaryotes are found to form homomeric or heteromeric heptamers, respectively (16). Two distinct RNA-binding sites have been described on opposite faces of the Hfq ring (17). The first, so-called distal binding site binds ARN repeats (A, adenine; R, purine; N, any nucleotide) and was cocrystallized with an oligo-(A)15 substrate. This RNA-binding mode is thought to be relevant in the processing of RNA 3′ ends (18). The second, so-called proximal binding site has been implicated in sRNA recognition. In analogy to the eukaryotic Sm heteroheptamer, where the RNA threads through the central pore of the Sm ring (19), the proximal site of Hfq has been assumed to bind internal A/U-rich sequences. Hence it was cocrystallized with a 5′AU5G3′ substrate, where the 3′ terminal nucleotide was found to be exposed (20).
Here we present a significantly different mode of Hfq–RNA interaction that involves the direct recognition of the 3′-terminal hydroxyl group, and we demonstrate that the proximal RNA-binding site of Hfq prefers to accommodate U-rich RNA 3′ ends over internal A/U-rich sequences. This result helps to explain the selectivity of Hfq for many sRNAs, because U-rich RNA 3′ ends are common to sRNAs as a consequence of Rho-independent transcription termination (21). Furthermore, the sequestration of the RNA 3′ hydroxyl group within Hfq suggests a protection of the respective 3′ end from enzymatic modification with important implications for Hfq function.
Results and Discussion
Hfq Prefers 3′-Terminal over Internal Uridines in Its Proximal RNA-Binding Site.
To characterize the RNA-binding properties of Salmonella typhimurium Hfq protein (St Hfq) we initially tested a series of RNA oligonucleotides containing an internal U-rich sequence found in RybB RNA, a well-characterized sRNA in Salmonella (22). RybB (Fig. 1A) is a small model sRNA that is induced in response to envelope stress and targets several outer membrane protein mRNAs, ultimately leading to their destabilization and degradation. As a typical sRNA, RybB is an independent, unprocessed bacterial transcription unit that consists of a 5′-terminal mRNA targeting region (23) followed by the RNA “body” that ends in a hairpin followed by a 3′-terminal oligo-U stretch as a consequence of intrinsic, Rho-independent transcription termination in bacteria (21).
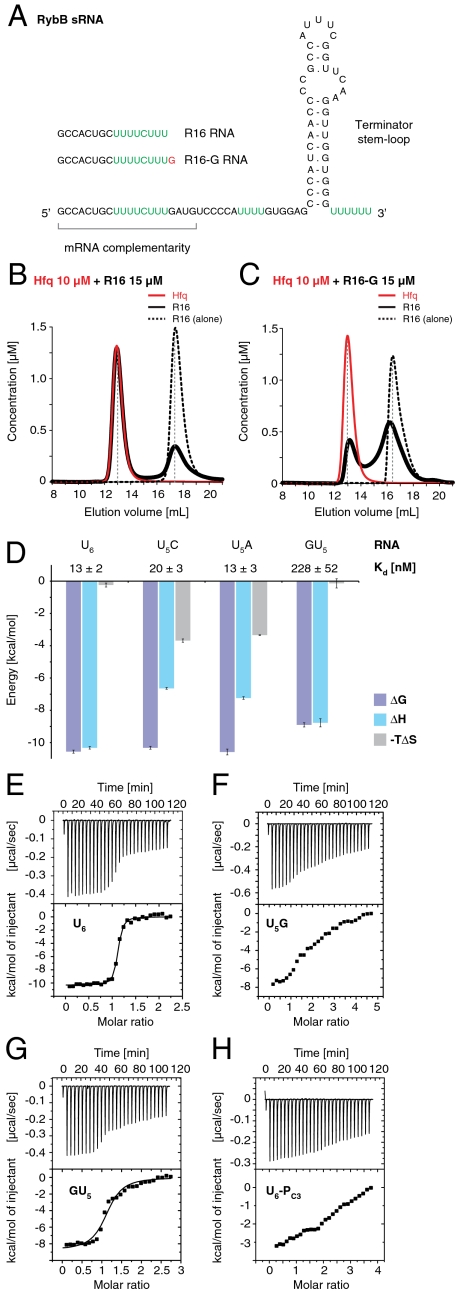
Fig. 1.
High-affinity binding of St Hfq to U-rich RNA 3′ ends. (A) Secondary structure of RybB sRNA. U-rich sequences prone to bind Hfq are highlighted in green. Oligonucleotides derived from the RybB 5′ end (R16 and R16-G) are indicated. (B and C) Analytical size exclusion chromatography of Hfq (red, solid lines, 10 μM at start) in the presence of RNA substrate (black, solid lines, 15 μM at start). Elution profiles show apparent concentrations for hexameric Hfq6 rings and RNA, calculated from the relative absorption properties of the components. Elution profiles for RNA substrate alone (black, dashed lines, 15 μM at start) are superimposed. R16 RNA (B) forms a stable, equimolar complex with Hfq6, whereas R16-G (C) with its additional 3′-terminal guanosine binds only weakly and dissociates during gel filtration. (D) ITC with hexanucleotide substrates. Favorable binding energies (ΔG, purple) are plotted as negative bars with the corresponding equilibrium dissociation constants (Kd) indicated on top, and together with the respective enthalpic (ΔH, blue) and entropic (-TΔS, gray) contributions. Error bars are standard fitting errors. (E–H) Individual titration experiments, including low-affinity substrates with unfavorable 3′ ends (F, U5G and H, U6-PC3) that do not fit a single-site binding model.
We identified a 16-mer RNA (R16, 5′ GCCACUGCUUUUCUUU 3′) that corresponds to the first 16 nucleotides of RybB and which formed a defined equimolar complex with St Hfq in analytical size exclusion chromatography experiments (Fig. 1B). Surprisingly, the inclusion of additional RybB nucleotides at the 3′ end of the oligonucleotide (R16-GATG; Fig. S1A) completely blocked the interaction and indeed already a single 3′ guanosine (R16-G; Fig. 1C) was sufficient to impair RNA binding. R16 binding required an intact proximal RNA-binding site on St Hfq because a previously characterized F42A mutation (17) abolished the interaction (Fig. S1B). The interaction was specific for RNA because a thymine-containing DNA oligonucleotide (D16) failed to bind, and it required the U-rich sequence at the 3′ end because a U4CU3 to A4GA3 mutation abolished binding as well (Fig. S1 C and D). Together, these results indicated a preferred RNA-binding mode for Hfq that is specific for the RNA 3′ end.
Hfq Strongly Discriminates Against Guanosines in the 3′-Terminal Position.
To quantify the significance of the 3′-terminal nucleotide for RNA binding, we used isothermal titration calorimetry (ITC) and compared the Hfq affinity of a homogeneous hexauridine (U6) substrate to substrates with single nucleotide substitutions (U5C, U5A, U5G, and GU5). RNA oligonucleotides terminating in C or A bound St Hfq in the low nanomolar range (13–20 nM), similarly to the U6 substrate (Fig. 1 D–G and Fig. S1 F and G). This finding is particularly surprising for U5C because, for Staphylococcus aureus Hfq (Sa Hfq), it was proposed that cytosine would be discriminated against (20). The present data demonstrate that this is not the case for St Hfq. Notably however, the detailed binding modes of the U5C and U5A substrates apparently differ from the U6 RNA, as indicated by the altered enthalpic and entropic contributions to complex formation (Fig. 1D).
The introduction of guanosine nucleotides had much more significant effects. A 3′-terminal guanosine in the U5G substrate strongly reduced St Hfq binding, as observed previously with R16-G (Fig. 1C). We estimate Hfq affinity for U5G to be lower than for U6 by at least two orders of magnitude, but nonspecific substrate binding became too prominent at the required ligand concentrations to accurately fit the data (Fig. 1F). In contrast, a 5′-terminal guanosine (GU5) reduced binding affinity only about 20-fold (230 nM; Fig. 1 D and G), showing that not only the identity of the base matters, but also its position in the RNA chain, such that manipulations at the 5′ end of the hexanucleotide substrate are tolerated more readily than at the 3′ end. Correspondingly, an extension of the RNA chain at the 5′-terminal nucleotide remains compatible with high-affinity binding, as demonstrated for the R16 substrate (Fig. 1B).
The strong interference of the 3′-terminal guanosine could be explained by an exclusion of the nucleotide together with the RNA 3′ end from the proximal binding site of Hfq, as observed in the crystal structure of Sa Hfq in complex with the AU5G substrate (20). Consistent with this hypothesis, and similar to the U5G substrate, the low-affinity binding of AU5G could not be fitted properly anymore in ITC (Fig. S1H) and it was estimated at only 2.5 μM by gel shift analysis (17). These observations suggest that AU5G is not the optimal substrate for the proximal site, and that the Sa Hfq/AU5G structure is not suited to explain the 3′-end-specific RNA binding. Therefore, we determined the crystal structure of St Hfq bound to U6 RNA.
High-Resolution Crystal Structure of St Hfq Bound to a Hexauridine Substrate.
Crystals of Hfq72 (C-terminally truncated after S72) in complex with U6 RNA (5′ OH-UpUpUpUpUpU-OH 3′) diffracted to 1.3-Å resolution with a crystallographic sixfold axis running through the center of the ring. Consequently, the electron density for the RNA backbone is averaged and forms a closed circle on the proximal side of the protein hexamer (Fig. 2A). The structure was thus refined as a single St Hfq72 monomer per asymmetric unit bound to a single uridine ligand with covalent bonds to its nucleotide neighbors and with a phosphate occupancy of 5/6. The absence of significant residual density indicates that the 5′- and 3′-terminal nucleotides are indeed oriented very similarly to their internal counterparts. To reveal structural changes upon RNA binding, we also crystallized St Hfq in the absence of RNA. These crystals contained one hexameric ring per asymmetric unit and diffracted X-rays to a resolution of 1.15 Å, the highest resolution for an (L)Sm ring protein up to date. Both structures are of excellent quality with Rfree values of 20.7% and 18.2%, respectively, for the free and ligand-bound forms (Table S1).
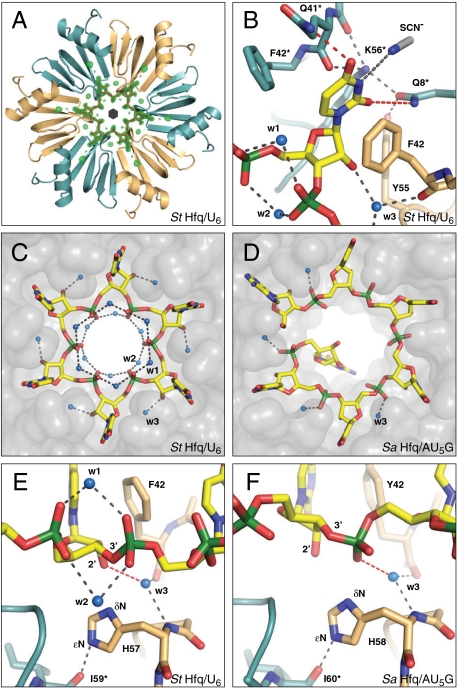
Fig. 2.
Crystal structure of the St Hfq/U6 RNA complex. (A) Electron difference density (green) for the RNA ligand (red), contoured at three sigmas over the mean. Shown is the Hfq hexamer as ribbons with the monomers colored alternately orange and cyan. The black hexagon symbolizes the sixfold axis relating the asymmetric units (Hfq monomers). (B) Recognition of the uracil base, with direct contacts to Hfq colored in red. (C and D) Recognition of the phosphoribose backbone, with Hfq represented as a transparent surface. (C) The constricted conformation of U6, stabilized by water molecules (w1 and w2) that bridge neighboring phosphates. (D) The dilated conformation of AU5G, with an exposed 3′-terminal guanosine (ref. 20; Protein Data Bank ID 1KQ2). (E and F) Differences in RNA backbone recognition, with crucial hydrogen bonds of water w3 in red. (E) Constricted conformation. Water w3 contacts the ribose, positioning it over H57. (F) Dilated conformation. Water w3 contacts the phosphate, preventing a direct recognition of the ribose. RNA and important amino acids are shown as sticks, water molecules as blue spheres, and hydrogen bonds as dashed gray lines. Nitrogen, blue; oxygen, red; sulfur, gray; phosphorus, green.
The accuracy of the two structures with their precise side-chain orientations and water positions allows a critical reassessment of nucleotide recognition by Hfq. The binding pocket for the uracil base between neighboring monomers is essentially preformed in the absence of a ligand and filled with water molecules (Fig. S2 A and B). Upon binding, the Q41 and F42 residues are slightly adjusted, such that the aromatic ring of F42 provides a stacking platform for the base (Fig. 2B and Fig. S2C). Additionally, Y55 and K56* (the asterisk indicates residues from the neighboring monomer) fix Q8* to contact the uracil O2 oxygen, while Q41* contacts the O4 oxygen. Importantly, both contacts are significantly different from ideal hydrogen bond geometry, with angles deviating substantially and in opposite directions from the plane of the uracil base (∼30° for O2 and ∼50° for O4). The N3 nitrogen binds to a thiocyanate ion from the crystallization condition, and hence is not contacted directly by the protein (Fig. 2B and Fig. S2B). As a consequence of this special binding geometry, the base-binding pocket could also accommodate a cytosine (if Q41* is flipped) or an adenine (if the thiocyanate ion is displaced; ref. 20). Only guanine would clash into Q8* with its exocyclic amino group as judged from a superposition with the corresponding base. Hence, the geometry of the base-binding pocket explains the binding data (Fig. 1) and supports the view that guanosines are indeed prevented from entering the proximal binding pocket of Hfq.
The Phosphoribose Backbone of the Hexauridine Substrate Is Bound in a Constricted Conformation.
A comparison of the St Hfq/U6 complex with the Sa Hfq/AU5G complex (20) demonstrates that the conformation of the phosphoribose backbone in the St Hfq/U6 complex is dramatically different (Fig. 2 C and D). This difference cannot be explained by differences between the proteins, because the residues fixing the phosphoribose backbone are structurally identical and because potential differences in base discrimination due to replacement of Q41 and F42 in St Hfq by K41 and Y42 in Sa Hfq are negligible.
Compared to the Sa Hfq/AU5G complex, the phosphates in the St Hfq/U6 complex are closer to the center of the ring and the free phosphate oxygens no longer contact the protein but are oriented toward the central pore. In this constricted conformation, the RNA is stabilized by additional water molecules (w1 and w2) that bridge both phosphate oxygens to their nucleotide neighbors, thus correlating the backbone conformation of adjacent residues (Fig. 2E). It is not possible to establish this hydrogen-bonding pattern in the dilated RNA conformation of the Sa Hfq/AU5G complex (Fig. 2F).
Furthermore, the constricted conformation allows a more direct fixation of the ribose in a shifted position that partially overlaps with the phosphate site from the dilated conformation (Fig. 2 E and F). The sugar pucker is clearly C3′ endo as in fact in the Sa Hfq/AU5G complex. Importantly, a very well-defined water molecule, w3, establishes a hydrogen bond to the 2′ oxygen of the ribose. This water molecule is fixed in its highly conserved position already in the absence of RNA (Fig. S2A) and is also present in the Sa Hfq/AU5G complex, where it mediates the interaction with the phosphate instead.
Finally, in the constricted conformation, the δN nitrogen of H57 is located in contact distance from both the 2′ and 3′ oxygens of the ribose (3.2 and 3.4 Å, respectively). This highly conserved residue is crucial for the stability of the Hfq ring (24) and donates a hydrogen bond from its ϵN nitrogen to the I59* carbonyl oxygen of the neighboring monomer (Fig. 2E). Consequently, the δN of H57 more likely acts as a hydrogen-bond acceptor, but the angle toward the ribose O2′ hydroxyl group is far from ideal and the O3′ oxygen in the RNA backbone is unprotonated. For the 3′-terminal ribose in the RNA chain, this situation is different, however, because in contrast to the internal riboses, its 3′ hydroxyl group can donate a hydrogen bond to the δN of H57 (Fig. 3A). Hence we propose that the RNA 3′ end is directly recognized by H57 from one of the six Hfq monomers.
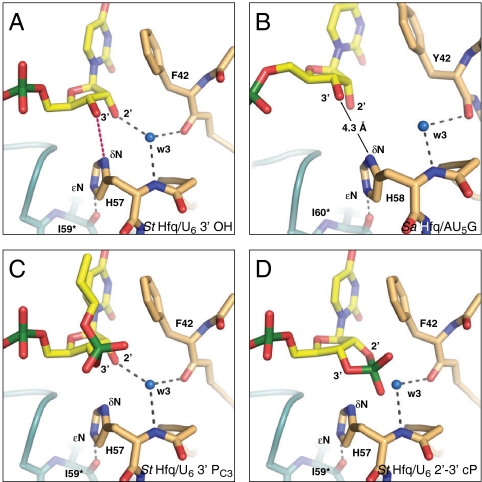
Fig. 3.
Model for the recognition of the RNA 3′-terminal hydroxyl group (A) Constricted conformation. The position of the ribose allows a specific hydrogen bond (red) from the 3′ hydroxyl group to the H57 δN. (B) Dilated conformation. The 3′ hydroxyl group would be too far away for a direct recognition by Hfq (model based on an internal nucleotide of Protein Data Bank ID 1KQ2; ref. 20). (C and D) Constricted conformation modeled with 3′-terminal phosphate groups. (C) Propyl-phosphate in the position of a downstream nucleotide. (D) A 2′–3′ cyclic phosphate according to Protein Data Bank ID 1HQ1. Although both substrates would be sterically compatible, they likely fail to trigger the constricted conformation due to the missing 3′-terminal hydrogen bond. See Fig. 2 for further details.
The Free 3′-Terminal Hydroxyl Group Is Crucial for the High-Affinity Interaction with Hfq.
Importantly, only the constricted conformation with its highly correlated nucleotide positions provides a rationale for the preferential and high-affinity recognition of U-rich RNA 3′ ends. In contrast to the dilated conformation observed in the Sa Hfq/AU5G complex, the constricted conformation demonstrates the specific recognition of the ribose via its 2′ hydroxyl group, the mutual stabilization of the phosphates and, crucially, allows for the formation of a 3′-terminal hydrogen bond to H57 (Fig. 3 A and B). Indeed, this terminal hydrogen bond may be required to trigger the constricted conformation, which otherwise may not be accessible. Furthermore, in the case of a 3′ continuing RNA chain, there may also be steric problems to adopt the constricted conformation.
To directly test for the relevance of a free 3′ hydroxyl end, we added a propyl-phosphate (a monophosphate-monopropylester) to the 3′ end of the hexauridine substrate (U6-PC3, Fig. 1H). This group was designed to fill the unoccupied slot of the missing hexauridine 5′ phosphate in the constricted conformation and hence to be sterically compatible with this conformation in the absence of a free 3′ hydroxyl group (Fig. 3C). Clearly, the addition of the propyl-phosphate reduced RNA affinity as dramatically as the inclusion of a 3′-terminal guanosine (U5G, AU5G; Fig. 1F and Fig. S1H). Consistently, we also lose high-affinity binding in size exclusion chromatography, when R16 terminates in a 2′–3′ cyclic phosphate (R16-cP; Fig. S1E). Similar to the PC3 modification, this terminal group is unable to donate hydrogen bonds but should fit in the proximal Hfq RNA-binding site without serious clashes (Fig. 3D). Together, these results demonstrate the need for a 3′ hydroxyl group and support the view that the constricted conformation indeed cannot be triggered in its absence.
The Uridine-Rich RNA 3′ End Contributes Significantly to the Dynamic Recognition of sRNAs by Hfq.
From a physiological point-of-view, the high affinity of Hfq for U-rich RNA 3′ ends is intriguing because it represents a molecular feature shared by the large majority of sRNAs as a consequence of Rho-independent transcription termination (21). To determine whether the RNA 3′ end of our initial model sRNA, RybB, is indeed recognized by the proximal RNA-binding site of Hfq we set up a direct competition experiment. We reconstituted St Hfq/RybB complexes in vitro and used 5′-Cy3-labeled R16 (Cy3-R16) as a specific competitor for the RNA 3′ end that could be traced selectively in analytical size exclusion chromatography.
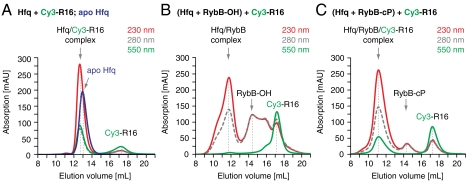
When RybB contained its native 3′ hydroxyl end, an excess (1.3-fold) of Cy3-R16 was not able to compete for Hfq binding and eluted separately from the Hfq/RybB complex (Fig. 4B), whereas, in the absence of RybB, Cy3-R16 readily bound Hfq (Fig. 4A). This result shows that RybB indeed blocks the proximal RNA-binding site. The blockage is not due to contaminating 5′ truncation products of RybB that were present in the chemically synthesized sample, as shown by independent experiments with unlabeled R16, where chromatography fractions were analyzed for protein and RNA on gels (Fig. S3A).
Fig. 4.
Small regulatory RNAs (RybB) occupy the Hfq proximal site with their U-rich 3′ end. Analytical size exclusion chromatography of Hfq in the presence of RybB RNA and of 5′ Cy3-labeled R16 RNA (Cy3-R16, monitored at 550 nm). Elution volumes are indicated by gray vertical lines, together with the molecular species. (A) Hfq6 rings (5 μM) in the presence of Cy3-R16 (10 μM). Cy3-R16 coelutes with the protein. The elution profile of Hfq6 in the absence of RNA is superimposed as a blue line (230 nm). (B) Hfq6 rings (5 μM) in the presence of RybB-OH (7 μM) and Cy3-R16 (10 μM). Chemically synthesized RybB-OH has a free 3′ hydroxyl group and elutes with trailing peaks (5′ truncation products resulting from chemical synthesis). RybB-OH coelutes with Hfq6 and prevents Cy3-R16 from binding Hfq6. Cy3-R16 does not coelute with RybB-OH. (C) Hfq6 rings (5 μM) in the presence of RybB-cP (7 μM) and Cy3-R16 (10 μM). RybB-cP terminates in a 2′-3′ cyclic phosphate, but still coelutes with Hfq6. However, it allows the coassociation of Cy3-R16 to form a ternary complex that elutes earlier than the binary Cy3-R16 Hfq6 complex alone (A).
To demonstrate that the Hfq proximal site is really occupied by the 3′-terminal end of RybB, we prepared RybB terminating in a 2′–3′ cyclic phosphate (RybB-cP) and repeated the competition experiment. This time R16 bound Hfq in addition to RybB and coeluted in a ternary complex (Fig. 4C and Fig. S3B). Consequently, the RybB-cP RNA 3′ end no longer occupies the proximal site with high affinity, whereas the natural 3′ end obviously did.
Very importantly, the experiment with RybB-cP also reveals that RybB, like probably many other sRNAs, uses at least two distinct contact sites on Hfq that are largely independent. These are the proximal binding site for the RNA 3′ end and at least one other site for the RybB body that further increases the affinity. The presence of separate, independent, and consecutively engageable binding sites means that the displacement of one sRNA molecule by another one can happen sequentially, with intermediate states where two sRNA molecules are bound on a single Hfq ring. It provides a molecular rationale for the rapid, concentration-dependent recycling of sRNAs on Hfq that is observed in a competitive situation (25), and it suggests that more than the proximal site alone should be considered in the respective model calculations. Clearly, these additional interactions require further investigation.
Implications of RNA 3′-End Binding by Hfq.
The high affinity of Hfq for RNA 3′ ends relies on a very unusual mode of terminal recognition, not in a specific pocket as one might expect (compare, e.g., Teplova et al. (2006), ref. 26, for the La protein), but in the context of a specific RNA backbone conformation that only forms at the RNA 3′ end. Indeed, this mode of RNA interaction has not previously been described for any member of the Sm or LSm ring families and it questions whether internal U-rich sequences bind to the proximal site of Hfq at all. The concept of internal sequence recognition was based on the interaction of small nuclear RNAs with eukaryotic Sm rings (19, 20), but may have to be revisited also for the eukaryotic LSm rings (LSm1–7 and LSm2–8). They have also been described to recognize RNA 3′ ends and they may be much closer to Hfq than to their Sm homologs with respect to proximal site specificity (27).
Given the apparent competition of RNA molecules for Hfq in the cell (25, 28), we speculate that the recognition of U-rich sRNA 3′ ends is the predominant function of the Hfq proximal site, whereas additional U-rich sequences in the sRNA body would be accommodated elsewhere on the Hfq ring. The specific interaction with the RNA 3′ end anchors the sRNA on the proximal face of Hfq and hence explains why this face is preferred. It also helps to explain the selectivity of Hfq for many sRNAs despite their structural diversity, because the high affinity of the interaction provides a competitive advantage of these sRNAs over other RNA species and RNA processing products that lack a U-rich 3′ end derived from a Rho-independent terminator. Clearly, however, the U-rich 3′ end cannot be the only Hfq-binding determinant in sRNAs because we show already with RybB RNA that it still interacts when the 3′ hydroxyl group is blocked by a 2′–3′ cyclic phosphate and even when the Hfq proximal site is occupied by a competing RNA 3′ end (Fig. 4C and Fig. S3B). Furthermore, the majority of mRNAs also have Rho-independent transcription terminators that result in U-rich RNA 3′ ends prone to bind Hfq via the proximal site.
Deep sequencing yielded a quantitative picture of Hfq-bound RNAs in the cell (9). The data show that the most highly enriched sRNAs on Hfq display long, single-stranded U-rich tails. In contrast, RNA molecules that act independently from Hfq either have processed 3′ ends that entirely lack a terminator structure or have their terminal uridines base-paired to upstream sequences and hence not easily available for Hfq binding (Table S2). These observations indicate that indeed rare sRNAs with an optimal 3′ end can outcompete more abundant sRNAs with a less favorable 3′ end.
Furthermore, the sequestration of the 3′-terminal hydroxyl group within the Hfq proximal site is expected to shield the group from enzymatic modifications that would reduce the affinity of the respective RNA substrate for Hfq. Consequently, this protection of the 3′ end will not only help stabilize the respective RNA (13), but also suggests a pathway for freshly transcribed sRNAs to actively switch off the effects of other, preexisting sRNAs in a rapid way, simply by competing them out from Hfq and exposing them to accelerated degradation. Indeed, Hfq has been found to protect the primary RNA 3′ end of both mRNAs from oligoadenylation by poly(A) polymerase I (5) and sRNAs from degradation mediated by polynucleotide phosphorylase (29).
In summary, RNA 3′-end binding emerges as an important function of Hfq and will have to be considered in future models of Hfq-mediated mechanisms. Clearly as well, Hfq can bind RNA in different modes with different mechanistic outcome. For example, Hfq has been described to promote the 3′ oligoadenylation of rpsO mRNA, leading to facilitated degradation (6, 7). This apparently conflicting function can easily be reconciled with the protective role of Hfq on primary terminator ends because oligoadenylation happens primarily on processed RNA 3′ ends where the terminator structure has been removed or modified (5, 6). Moreover, oligoadenylated 3′ ends are expected to interact with the distal face of Hfq, where the 3′ hydroxyl group is unprotected. Finally, our data indicate that there must yet be additional RNA-binding sites on Hfq that help to bind the sRNA body and that we presume to be responsible for the protection of the body from endonucleolytic cleavage by RNAse E (30, 31). It shall be interesting to learn where these additional sites are located and which conformational arrangements within the Hfq/sRNA complex accompany target mRNA binding.
Materials and Methods
Preparation of RNA and Protein Material.
Synthetic RNA oligonucleotides were purchased as desalted or HPLC-purified material. RybB-cP (nucleotides G1-U78; ref. 22) and R16-cP RNA were transcribed in vitro from a modified pSP64 plasmid containing a 3′-terminal hepatitis delta virus ribozyme that autocleaves cotranscriptionally, leaving a 2′–3′ cyclic phosphate on the target RNA. Target RNA was purified over denaturing polyacrylamide gels as described previously (32).
St Hfq constructs (St Hfq, GAM1-E102 and St Hfq72, GAM1-S72; UniProt ID P0A1R0) were expressed as NusA fusions from a pETM60 vector in E. coli BL21(DE3) GOLD cells. NusA-Hfq was purified from the cleared lysate by Ni2+ affinity chromatography. After proteolytic removal of the affinity tag, Hfq was further purified by heparin affinity and size exclusion chromatography.
Isothermal Titration Calorimetry .
RNA-binding affinities were determined by ITC using a VP-ITC microcalorimeter (MicroCal) essentially as described previously (32). Hfq (6 μM, 1.44 mL) were titrated with 60 or 120 μM RNA (28 × 10 μL injections) in chromatography buffer. The data were fitted to a single-site model (MicroCal).
Analytical Size Exclusion Chromatography.
RNA was first annealed (65 °C for 10 min, slow cooled to room temperature) in chromatography buffer [100 mM NaCl, 10 mM Tris•HCl (pH = 8.0), 10 mM MgCl2], then incubated with limiting amounts of St Hfq (10 min), and finally loaded onto the chromatography column (150-μL sample on a Superdex200 10/300 GL column mounted on an ÄKTA™ Purifier-10; GE Healthcare). Starting concentrations are given in the figure legends. Elution was monitored by UV absorption at 230, 260, and 280 nm (with Cy3-R16 RNA: 230, 280, 550 nm). Apparent concentrations were calculated from the relative absorption properties of the components as described before (32). For competition experiments with Cy3-R16, a limiting amount of Hfq was preincubated with RybB (10 min) and then challenged by an excess of Cy3-R16 (10 min) before chromatography.
Crystallization, Data Collection, and Refinement of St Hfq72 and St Hfq72/U6.
The best-diffracting crystal of St Hfq72 was obtained by sitting drop vapor diffusion, mixing 0.5 μL sample [200 μM, in 10 mM Tris•Cl (pH = 8.0), 100 mM NaCl] and 0.5 μL reservoir [100 mM Hepes (pH = 7.0), 0.5% Jeffamine, 1.1 M malonate] over 80 μL reservoir. For the St Hfq72/U6 complex, the sample additionally contained 220 μM U6 RNA over a reservoir of 0.2 M NaSCN and 20% PEG 3350. Cryoprotection was achieved by adding glycerol to a final concentration of 20%. Diffraction data were collected on beamline PXII of the Swiss Light Source. The structure of St Hfq72 was solved by molecular replacement with Escherichia coli Hfq (Protein Data Bank ID 1HK9), whereas St Hfq72 was used as a model for St Hfq72/U6. Auto-built models were finished manually and refined with anisotropic B factors. Processing of the St Hfq72/U6 data in a lower symmetry space group (P1) did not resolve the RNA 5′ and 3′ ends in the respective difference density. Consequently, the uridine ligand was built into the P6 difference density after completion of the model for St Hfq72 and refined with covalent bonds to its nucleotide neighbors. Further experimental procedures are given in the SI Material and Methods.
Supplementary Material
Acknowledgments.
We thank Regina Büttner for technical assistance and the staff at the Swiss Light Source (Villigen, Switzerland) for assistance during data collection. We are grateful to Pavel Afonine for tips on refinement and to Steffen Schmidt for computing infrastructure and software support. We thank Jörg Vogel for Hfq and RybB plasmids and Elisa Izaurralde and members of the department for helpful comments and discussions. This work was funded by the Deutsche Forschungsgemeinschaft Priority Program SPP1258 (Sensory and Regulatory RNAs in Prokaryotes).
Note.
A related paper by Otaka et al. in this issue of PNAS entitled “PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action” complements the current findings by very important in vivo evidence for a functional role of the sRNA terminator oligoU tail in sRNA-mediated riboregulation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2YLB and 2YLC for the St Hfq72 apo structure and the St Hfq72/U6 complex, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103420108/-/DCSupplemental.
References
- 1.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr Opin Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- 4.Barrera I, Schüppli D, Sogo JM, Weber H. Different mechanisms of recognition of bacteriophage Q beta plus and minus strand RNAs by Q beta replicase. J Mol Biol. 1993;232:512–521. doi: 10.1006/jmbi.1993.1407. [DOI] [PubMed] [Google Scholar]
- 5.Le Derout J, et al. Hfq affects the length and the frequency of short oligo(A) tails at the 3′ end of Escherichia coli rpsO mRNAs. Nucleic Acids Res. 2003;31:4017–4023. doi: 10.1093/nar/gkg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 7.Folichon M, Allemand F, Régnier P, Hajnsdorf E. Stimulation of poly(A) synthesis by Escherichia coli poly(A)polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J. 2005;272:454–463. doi: 10.1111/j.1742-4658.2004.04485.x. [DOI] [PubMed] [Google Scholar]
- 8.Worrall JA, et al. Reconstitution and analysis of the multienzyme Escherichia coli RNA degradosome. J Mol Biol. 2008;382:870–883. doi: 10.1016/j.jmb.2008.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sittka A, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang A, et al. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 11.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res Microbiol. 2009;160:278–287. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fröhlich KS, Vogel J. Activation of gene expression by small RNA. Curr Opin Microbiol. 2009;12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 15.Sittka A, Sharma CM, Rolle K, Vogel J. Deep sequencing of Salmonella RNA associated with heterologous Hfq proteins in vivo reveals small RNAs as a major target class and identifies RNA processing phenotypes. RNA Biol. 2009;6:266–275. doi: 10.4161/rna.6.3.8332. [DOI] [PubMed] [Google Scholar]
- 16.Wilusz CJ, Wilusz J. Eukaryotic Lsm proteins: Lessons from bacteria. Nat Struct Mol Biol. 2005;12:1031–1036. doi: 10.1038/nsmb1037. [DOI] [PubMed] [Google Scholar]
- 17.Mikulecky PJ, et al. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq–RNA complex: A bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson KS, von Hippel PH. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc Natl Acad Sci USA. 1995;92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc Natl Acad Sci USA. 2010;107:20435–20440. doi: 10.1073/pnas.1009784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol Cell. 2008;32:827–837. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 24.Moskaleva O, et al. The structures of mutant forms of Hfq from Pseudomonas aeruginosa reveal the importance of the conserved His57 for the protein hexamer organization. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:760–764. doi: 10.1107/S1744309110017331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–2626. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teplova M, et al. Structural basis for recognition and sequestration of UUU(OH) 3′ termini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol Cell. 2006;21:75–85. doi: 10.1016/j.molcel.2005.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilusz CJ, Wilusz J. New ways to meet your (3′) end oligouridylation as a step on the path to destruction. Genes Dev. 2008;22:1–7. doi: 10.1101/gad.1634508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papenfort K, et al. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol. 2009;74:139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- 29.Andrade JM, Arraiano CM. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA. 2008;14:543–551. doi: 10.1261/rna.683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Folichon M, et al. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Bläsi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller M, Weigand JE, Weichenrieder O, Suess B. Thermodynamic characterization of an engineered tetracycline-binding riboswitch. Nucleic Acids Res. 2006;34:2607–2617. doi: 10.1093/nar/gkl347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.