Abstract
Anopheles gambiae, the primary African vector of malaria parasites, exhibits numerous rhythmic behaviors including flight activity, swarming, mating, host seeking, egg laying, and sugar feeding. However, little work has been performed to elucidate the molecular basis for these daily rhythms. To study how gene expression is regulated globally by diel and circadian mechanisms, we have undertaken a DNA microarray analysis of An. gambiae under light/dark cycle (LD) and constant dark (DD) conditions. Adult mated, non–blood-fed female mosquitoes were collected every 4 h for 48 h, and samples were processed with DNA microarrays. Using a cosine wave-fitting algorithm, we identified 1,293 and 600 rhythmic genes with a period length of 20–28 h in the head and body, respectively, under LD conditions, representing 9.7 and 4.5% of the An. gambiae gene set. A majority of these genes was specific to heads or bodies. Examination of mosquitoes under DD conditions revealed that rhythmic programming of the transcriptome is dependent on an interaction between the endogenous clock and extrinsic regulation by the LD cycle. A subset of genes, including the canonical clock components, was expressed rhythmically under both environmental conditions. A majority of genes had peak expression clustered around the day/night transitions, anticipating dawn and dusk. Genes cover diverse biological processes such as transcription/translation, metabolism, detoxification, olfaction, vision, cuticle regulation, and immunity, and include rate-limiting steps in the pathways. This study highlights the fundamental roles that both the circadian clock and light play in the physiology of this important insect vector and suggests targets for intervention.
Keywords: biological rhythm, clock gene, cytochrome P450, melanization, mosquito
The Anopheles gambiae mosquito is the major African vector of malaria parasites, which are responsible for almost 1 million deaths annually, mostly of small children. Malaria has been controlled primarily through insect-control strategies, in particular with the application of insecticide through indoor residual spraying and insecticide-treated bed nets (ITNs), which protect sleeping human hosts from infection at night when mosquitoes are active (1). Mosquito physiology and behavior are under rhythmic control, organized in a time-of-day–specific manner. In An. gambiae these behaviors include dusk mating swarms and nocturnal flight activity, feeding on sugar and on blood-meal hosts, and oviposition. Larval–pupal ecdysis and subsequent eclosion occur during the late day and late day/early night, respectively (2–4). Eukaryotic organisms possess a circadian (“about a day”) clock regulating daily rhythms in biochemistry, physiology, and behavior. The clock is cell autonomous and at the molecular level comprises a series of transcriptional–translational feedback loops whose completion takes ∼24 h (5).
DNA microarray time courses in numerous organisms (6–14) reveal that 2–12% of the transcriptome is under circadian clock or diel control in individual tissues, and this regulation is highly tissue specific (15, 16). The full complement of rhythmic genes exhibiting a 24-h period length is generated by a combination of two processes. The first is an endogenous circadian clock that persists under constant environmental temperature and light conditions. Genes regulated by the clock are known as “clock-controlled genes” (CCGs), and rhythms persisting in constant dark (DD) may be referred to as “circadian.” The second is a direct action of the environmental light/dark (LD) cycle on the organism that generates additional rhythms in gene expression and suppresses a proportion of rhythms generated by the endogenous circadian clock mechanism (Fig. S1). This direct LD cycle mechanism has been described in Drosophila only recently and is poorly understood at a mechanistic level but obviously includes photoreception, including a contribution from the compound eyes (8, 11, 14). We may describe such rhythms driven by the LD cycle as “diel.”
Although a wealth of knowledge exists regarding the behavioral rhythms in An. gambiae, work on the molecular basis of the clock in any mosquito species remains sparse. Studies have begun to describe important aspects of the canonical clock components, specifically the cloning of the Aedes aegypti timeless (tim) gene, and the expression profiling of the clock genes in Ae. aegypti and Culex quinquefasciatus (17, 18), and the functional analysis of the cryptochrome proteins CRY1 and CRY2 was performed in An. gambiae among several other insect groups (19, 20). These studies reveal a molecular clock that is similar to that of other insect species but with distinct differences as compared with Drosophila. Genetic analysis of Anopheles cruzii tim reveals geographic differences in its sequence (21), and in Wyeomyia smithii the profile of tim expression varies according to latitude and developmental state (22). The role of specific An. gambiae clock genes in the light-inhibition of blood feeding behavior was revealed by DNA microarray analysis and RNAi-mediated gene silencing (4).
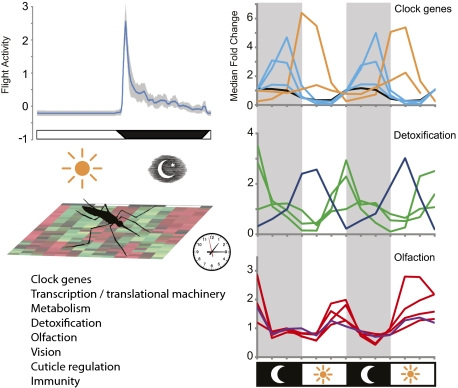
Here we describe a circadian transcriptome analysis performed on heads and bodies of mated female An. gambiae. This species and developmental stage was chosen because it displays pronounced nocturnal locomotor behavior and is of high epidemiological relevance (2). Our analysis, which was conducted in both an environmental LD cycle and DD conditions, allowed gene set-wide transcriptional analysis of An. gambiae rhythms, letting us explore the extent of its biochemistry, physiology, and behavior that is under circadian and LD cycle control (Fig. 1), and greatly expands the existing gene-expression work performed in the species (e.g., refs. 4, 23–25). Our results indicate that at least 15.8% of the An. gambiae gene set, across a wide range of biological functions and processes, is under diel and/or circadian control.
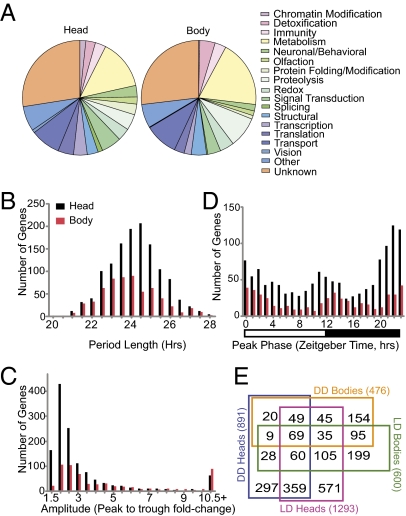
Fig. 1.
An. gambiae transcription analysis reveals a great number and diversity of rhythmic genes. (A) Manual annotation of rhythmic genes in both heads and bodies reveals a wide range of biological functions among rhythmic genes under LD conditions. (B) Under LD conditions, period length of rhythmic genes (mean ± SEM) was 24.0 ± 0.04 h in the head and 23.6 ± 0.06 h in the body. (C) Rhythmic gene amplitude determined as peak-to-trough fold change. In LD heads, rhythms had a medial 2.09-fold change; however, a significant number of rhythmic genes had >10-fold amplitude in expression. (D) Peaks of transcriptional expression occur at the dawn and dusk transitions. Day and night are indicated by the horizontal white/black bar below the histogram. Data in B–D are binned according to their value up to and including the values provided on the x axes. (E) Venn diagram showing the number of genes identified as rhythmically expressed under four experimental conditions and their respective overlaps with the other experimental conditions.
Results and Discussion
Global Transcription Analysis.
To perform a comprehensive determination of An. gambiae diel and circadian gene expression, we profiled genome-wide expression patterns in the heads and bodies of adult mated but non–blood-fed female mosquitoes every 4 h over 2 d while the mosquitoes were maintained under a 12-h/12-h LD cycle or DD conditions. RNA samples were examined using Affymetrix Plasmodium/Anopheles high-density oligonucleotide probe arrays, and data were analyzed statistically using the COSOPT cosine wave-fitting algorithm (6, 12, 14, 26). We identified 1,293 rhythmic transcripts in the head and 600 in the body under LD conditions with period lengths between 20–28 h, which represent 9.7% and 4.5%, respectively, of the total An. gambiae gene set. Under DD conditions, we identified 891 rhythmic transcripts in the head and 476 in the body with an 18.5–26.5 h period length (median and mean period length in DD was 21.8 and 21.9 ± 0.04 h SEM, respectively) (Fig. 1, Fig. S2, and Dataset S1). These results are consistent with a short 23.27 ± 0.03 h SEM period length observed in flight-activity rhythms that reflects the endogenous period length of the An. gambiae circadian clock (Fig. S3). Most of rhythmically expressed genes we identified had amplitude changes less than threefold, although some genes had amplitude changes >10-fold (Fig. 1C and Fig. S2C). Both dawn and dusk are enriched in the number of genes peaking in expression at these times (Fig. 1D and Fig. S2D). A subset of 537 head and 208 body genes were rhythmically expressed under both LD and DD conditions, and 69 genes rhythmic in both head and body tissues under both LD and DD conditions (Fig. 1E). Rhythmic genes represent diverse biological functions, including transcriptional/translational machinery, metabolism, detoxification, olfaction, vision, cuticle/peritrophic membrane regulation, and immunity (Fig. 1A and Fig. S2A).
Similar to transcriptome analysis in Drosophila, we have found genes rhythmic in both head and body as well as genes uniquely rhythmic in individual tissues (Fig. 1E) (8, 11, 14). There is evidence in Diptera (Drosophila) and other taxonomic groups for a separate mechanism by which genes and downstream physiology are expressed rhythmically only under LD conditions versus constant conditions, suggesting a direct light-induced or -suppressed pathway separate from a clock-controlled pathway (8, 13, 14, 27–30). This theory has been tested empirically in Drosophila (11), and our Anopheles data, with 59% of rhythmic head genes rhythmic only under LD conditions, are consistent with this model of 24-h rhythmic control (Fig. S1).
Temporal expression patterns of several genes, including clock genes and CCGs, were assayed by quantitative RT-PCR (qRT-PCR) and further validated the microarray measurements. The expression profiles of the genes were similar in both phase and amplitude between the two techniques (Fig. S4). Finally, we have presented our data in a publicly accessible database (http://www.nd.edu/∼bioclock) that can be searched via a text-based query for temporal expression patterns of a given gene in both head and body under LD and DD conditions. The output of the search is a graphical representation of the expression data along with associated COSOPT algorithm-derived statistics and gene annotations. Peak time-of-day expression is reported in LD conditions as Zeitgeber time (ZT), with ZT0 defined as the end of the dawn transition and ZT12 defined as the time of lights off at the end of the dusk transition. Under constant conditions, we report circadian time (CT) with CT0 defined as the end of subjective dawn inferred from the previous LD cycle. In this paper, we have followed the gene-naming guidelines suggested in both VectorBase and Christophides et al. (31). For genes that already have been given a symbolic name that has been submitted to VectorBase by either the community or by UniProt, we use that name. For genes that have been named in publications but not submitted to VectorBase, we have retained those names and used the “ag” prefix. For those An. gambiae genes without formal names either in VectorBase or in publications, we have identified them by using the name of the most similar ortholog recorded in VectorBase with a gene symbol name or determined from searches using DAVID and InterProScan.
Clock Genes.
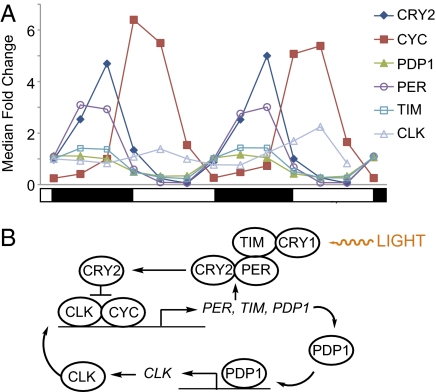
Many of the genes and the organization of the circadian clock are similar across taxonomic groups (5). However, there appear to be differences among insect orders (19, 32–34), and our data presented here, among other data, suggest that the An. gambiae circadian system, as well as that of other mosquito species (18), more closely resembles other insects (e.g., butterfly) than it does its fellow Dipteran, Drosophila (Fig. 2B). Of the canonical clock genes, period (agPER, AGAP001856), timeless (agTIM, AGAP008288), the transcriptional repressor cryptochrome 2 (agCRY2, AGAP004261), PAR-domain protein 1 (agPDP1, AGAP006376), and cycle (agCYC, AGAP005655) were found to be rhythmically expressed under LD and DD in both heads and bodies, consistent with findings in Ae. aegypti and Cx. quinquefasciatus (Fig. 2A, Fig. S5A, and Dataset S1) (18, 19). Moreover, these genes were rhythmic with high amplitudes ranging from a 1.9- to a 89-fold change and with predicted phase relationships (i.e., agCRY2, agTIM, and agPER sharing a similar peak phase and in antiphase with agCYC). Unlike Drosophila, but similar to Ae. Aegypti, Cx. quinquefasciatus, and Sarcophaga crassipalpis, expression of CRY1 (AGAP001958) was not found using our criteria to be rhythmic in either heads or bodies under LD or DD conditions (Fig. S5B) (7, 14, 18, 34). Perhaps because of deficiencies in the microarray design, we were unable to detect the expression of clock (agCLK, AGAP005711), which we expected also to be rhythmic from work in other mosquito species (18). Therefore, using qRT-PCR, we confirmed that agCLK is in fact rhythmic, peaking in the early morning in the LD head (Fig. 2A and Fig. S4). The profiles of agCLK and agCYC are approximately phase coincident and therefore fit the predicted model for the molecular clock (Fig. 2B). [Information on identification of An. gambiae clock genes can be found in refs. 4 and 19 and in Fig. S6A. Fig. S6B presents our identification of agTIM and agPDP1.] In line with predictions from other insects (18, 19, 32–34), these data suggest a transcriptional–translation feedback mechanism comprising the rhythmic positive elements CLK and CYC, the negative elements CRY2, TIM, and PER, and an interlocking loop containing PDP1.
Fig. 2.
The molecular clock of An. gambiae. (A) Transcriptional profiles of the canonical clock genes in heads under LD conditions. Under all conditions and tissues, clock genes exhibited robust high-amplitude rhythms with patterns and phases predicted from other insect models (see Fig. S5A for clock gene profiles in bodies in LD). The clock (agCLK) profile is from qRT-PCR analysis. Note: CRY1 was nonrhythmic and thus was not included (Fig. S5B). Data have been normalized to median fold change, and SD error bars have been omitted for viewing purposes. Day and night are indicated by the horizontal white/black bar below the chart. (B) Proposed model of the An. gambiae molecular clock comprising a transcriptional/translational feedback mechanism and derived from work presented here and by others (4, 5, 18, 19, 32, 33).
Transcriptional/Translational Machinery.
Circadian regulation exists at multiple levels from transcription through posttranslational modifications of gene expression (15, 35). Examples include cyclic mRNA levels of Drosophila period in the absence of transcriptional rhythms that are caused by posttranscriptional regulation of mRNA stability (36). The Arabidopsis gene glycine-rich RNA-binding protein 7 (GRP7) is circadianly autoregulated through generation of GRP7 splice variants of varying stability (37). Further, chromatin remodeling may serve to propagate circadian time cues. The Drosophila Clock gene itself is a rhythmically transcribed gene with histone acetyl-transferase activity. Transcriptional profiling of Drosophila revealed chromosomal spacing of rhythmic genes consistent with circadian chromatin remodeling driving genome-wide circadian expression patterns (7, 9, 38).
In our study, we found examples of genes at all these levels that may contribute to circadian/diel regulation. We identified the nearest An. gambiae orthologs to Drosophila chromatin-related genes that exhibit a rhythmic pattern peaking late in the night, including histone deacetylase 3 (AGAP001143; circadian); a histone H3-Lys9 methyltransferase (AGAP003597; diel); a trithorax gene required for trimethylation of histone H3-Lys4, ash2 (AGAP003705; circadian); and a member of the Brahma (SWI2/SNF2) chromatin remodeling factor complex, Bap60 (AGAP005336; diel) (Fig. 3A) (39–41). Following transcription, we note diel-expressed head genes related to mRNA splicing, including putative orthologs to Drosophila genes B52 (AGAP004592), Srp54 (AGAP002265), and U2 small nuclear riboprotein auxiliary factor 38 (AGAP002956) also peaking ∼4 h before dawn. These factors are known through RNAi screens of Drosophila to be required for the generation of particular splice variants (42). Finally, we found that the entire translational machinery appears to peak in expression at the end of night. Numerous translation-initiation factors are expressed in a rhythmic manner, including the eukaryotic initiation factors (EIFs) EIF3C (AGAP004725) and the putative orthologs to Drosophila eIF6 (AGAP001380), eIF5B (AGAP004824), and EfTuM (AGAP006996) and with genes including EIF3L (AGAP006130) and EIF3H (AGAP009204) rhythmic under diel conditions in the head.
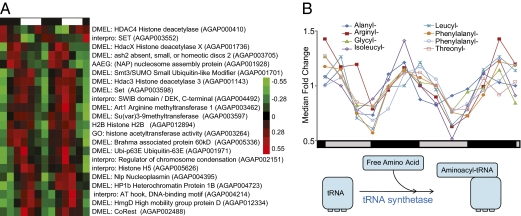
Fig. 3.
Components of transcription and translation are rhythmically expressed. (A) Hierarchical clustering of rhythmic chromatin-related genes in LD heads. Red indicates higher expression, and green indicates lower expression versus the mean value for each gene. Genes with no prefix are An. gambiae gene names. Genes with species prefixes are the closest predicted named orthologs. Day and night are indicated by the horizontal white/black bar above the heatmap. (B) Aminoacyl-tRNA synthetases are circadianly expressed in the head. Free amino acid is “primed” to tRNA by tRNA synthetases specific for each amino acid. Eight of the 12 synthetase genes rhythmic in the head and/or body under diel conditions are also expressed in a circadian manner in the head. Data have been normalized to median fold change. Circadian day and night are indicated by the horizontal gray/black (subjective day/subjective night) bar below the chart. Genes are indentified based on DAVID predictions.
Remarkably, using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics tool, we identified 12 putative tRNA synthetases in the head and/or body that “prime” amino acids to tRNA to be rhythmic and also that peak primarily at the end of the night under diel conditions. Eight of these synthetases also were expressed in a circadian pattern in the head under DD conditions (Fig. 3B). Finally, we found genes involved in posttranslational modification to be under rhythmic control including the nearest An. gambiae orthologs to Drosophila UDP-glucose-glycoprotein glucosyltransferase (AGAP003560, ZT0.9/CT1.4) and oligosaccharyl transferase 3 (AGAP002396, ZT3.1).
Observed rhythms in expression levels of translational machinery at the end of night are congruent with a mosquito that is preparing for amplified levels of protein synthesis during the daytime. These rhythms could be beneficial in the rebuilding of cellular products while the mosquito rests and in the large amounts of protein synthesis involved in egg development, especially vitellogenesis, that follow a blood meal. Our results demonstrate there may be multiple layers of circadian/diel control at the transcriptional, translational, and posttranslational level including alternate splicing and ribosomal machinery which produce, enhance, or modify 24-h rhythms in gene expression. These processes in turn drive rhythms of cellular effectors that may not be predictable from mRNA levels alone.
Metabolism.
Metabolic demands vary drastically between rest and nocturnal mosquito flight activity (43, 44). Moreover, An. gambiae sugar and blood feeding is restricted to the night phase (2, 3). With these predictable temporal changes in nutrient availability and respiratory demands, it is no surprise that many key rate-limiting steps in anabolic/catabolic metabolic pathways are under circadian or diel control. Indeed, in numerous organisms, pathways such as oxidative phosphorylation, gluconeogenesis, and lipogenesis have been found to be rhythmically expressed (11, 12, 45, 46). In mosquitoes, flight activity is metabolically maintained more by sugar than by lipid reserves, which serve primarily to maintain more basal metabolic processes; excess carbohydrates are converted to triglycerides in anophelines (47, 48). Using DAVID to predict gene function, we found genes involved in glycolysis, the citric acid cycle, oxidative phosphorylation, and fatty acid oxidation, among other processes, to be rhythmically expressed (Fig. 4A and Figs. S7 and S8). Additionally, genes associated with control of intermediary metabolism and feeding, nutritional homeostasis, energy sensing, and nutrient mobilization were rhythmic in head and/or body. These genes include neuropeptide F (NPF, AGAP004642) and target of rapamycin (agTOR, AGAP007873) (49), as well as takeout 1 (agTO1, AGAP004263), agTO2, and/or agTO3 (AGAP012703/AGAP004262) (4), and orthologs to Drosophila AMP-activated protein kinase (AGAP002686) and lipid storage droplet-1 (AGAP002890) (Dataset S1) (4, 50, 51). The rhythm in agTOR expression is of particular interest because the nutrient-sensing TOR-signaling pathway in Drosophila is capable of modulating canonical clock components and influencing circadian behavior (46). In the mosquito body, we also note rhythmic expression, peaking in midnight, of the adipokinetic hormone receptor gene (agAKHR, synonymous with gonadotropin-releasing hormone receptor, GPRGNR1, AGAP002156) (52). Adipokinetic hormone 1 (AKH1) has been shown in An. gambiae to promote levels of the flight fuel trehalose in the hemolymph and to deplete glycogen reserves (53). This observation suggests that agAKHR is up-regulated at night, coinciding with peak flight activity (Fig. S3). Unfortunately, a probe was not available for AKH1 (AGAP008834) itself. Consistent with our findings, Anopheles freeborni synthesizes glycogen during the day when agAKHR levels would be low (47). Rhythmic expression of the takeout genes is also of interest, because their expression in An. gambiae is up-regulated by sugar starvation, down-regulated in the head by light, and up-regulated in the body by blood feeding. Furthermore, RNAi gene silencing suggests that all three agTO genes modulate the drive to blood-feed (4). Perhaps it is not surprising that these genes are rhythmically expressed in An. gambiae, that exhibits distinct cycles in sugar and blood feeding. It is plausible that the difference in the peak phases for the agTO genes in the head (∼ZT12) versus the body (∼ZT21) may underlie the apparent differential tissue-specific roles in activation and suppression of feeding.
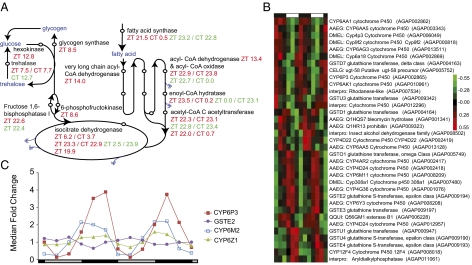
Fig. 4.
Key enzymes in core metabolic pathways and in metabolic detoxification are under circadian/diel conditions. (A) Red type indicates head peak phases; green type indicates body peak phase; blue type indicates metabolic molecules; double blue arrows indicate products that supply oxidative phosphorylation. See Figs. S7 and S8 for the full complement of rhythmic genes in these metabolic pathways. Additional peak phases are listed for enzymes with more than one associated rhythmic gene. (B) Hierarchical clustering of rhythmic detoxification genes in LD heads. Red indicates higher and green indicates lower expression versus the mean value for each gene. Day and night are indicated by the horizontal white/black bar above the heatmap. Genes with no prefix are An. gambiae gene names. Genes with species prefixes are the closest predicted named orthologs. (C) DD body expression profiles of GSTE2, a GST whose up-regulation can confer resistance to the insecticide DDT (59), and three P450 genes, CYP6Z1, agCYP6P3, and agCYP6M2, whose up-regulation is implicated in pyrethroid insecticide resistance (60, 62). Data have been normalized to median fold change. Circadian day and night are indicated by the horizontal gray/black (subjective day/subjective night) bar below the chart.
Expression of other rhythmic genes is consistent with rhythms in hemolymph trehalose including the genes encoding the proenzyme trehalase (AGAP012053, Drosophila ortholog), which converts trehalose into glucose, and hexokinase (AGAP011208, Drosophila ortholog), which converts glucose into glucose-6-phosphate, facilitating a steep concentration gradient across the cell membrane (Fig. 4A and Figs. S7A and S8). Trehalase, which is vital for flight activity in many insects (54), peaks at ZT7.5, prior to the initiation of the flight-activity phase. Such a rhythm in hemolymph trehalose has been noted in the cricket Gryllus domesticus (55). Furthermore, hexokinase peaks at ZT12.8, timing that is consistent both with the mosquito imbibing a sugar meal in the early night and with massive energy expenditures in flight activity at dusk. Additionally, both fructose-1,6-bisphosphatase (AGAP009173, ZT22.3, Drosophila ortholog) and 6-phosphofructokinase (AGAP007642, ZT8.6, Drosophila ortholog) are rhythmic and in opposite phases. These genes encode enzymes catalyzing opposing unidirectional reactions, with fructose-1,6-bisphosphate promoting gluconeogenesis and possible storage of sugar as trehalose and glycogen, and 6-phosphofructokinase promoting the utilization of sugar by glycolysis and the citric acid cycle (56).
In the head, we found free fatty acid oxidation has numerous rhythmically expressed enzymes, many of which are expressed in a circadian manner and are rhythmic in the body as well (Fig. 4A and Figs. S7A and S8). In the head, these include three genes encoding the oxidation step, the long (AGAP008769, ZT14.0; Drosophila ortholog) and medium (AGAP005662; ZT13.4; Drosophila ortholog) forms of acyl-CoA dehydrogenase, and acyl-CoA oxidase (AGAP011798, ZT22.9; Drosophila ortholog). Additionally, enoyl-CoA hydratase (AGAP011833; ZT23.5; Caenorhabditis elegans ortholog), whose product catalyzes the hydration step, and two paralogs encoding the thiolytic acetyl-CoA C-acetyltransferase (AGAP011329; ZT22.0/AGAP001318; ZT22.3; DAVID predictions), are also rhythmic. Various components of the citric acid cycle are also rhythmic in the An. gambiae head, including the gene encoding isocitrate dehydrogenase, a major control point in the citric acid cycle, with three rhythmic paralogs in the head (AGAP006660; ZT23.3/AGAP003168; ZT6.2/AGAP007786; ZT19.9; Drosophila and C. elegans orthologs) (43, 57). In the body, one isocitrate dehydrogenase (AGAP006660) paralog is also rhythmic, peaking at ZT2.5. Finally, energy-storing processes are rhythmic, with head glycogen synthase (AGAP002586; ZT8.5; Drosophila ortholog) rhythmic and fatty acid synthase (AGAP009176; C. elegans ortholog) circadianly expressed in both the head (ZT21.5/CT0.5) and body (ZT23.2/CT22.8). Up-regulation of both free fatty acid oxidation and synthesis pathways at the end of night is congruent with a female anticipating either receiving excess nutrients from a blood/sugar meal to store as fat reserves or using reserves during the following day if unsuccessful at feeding. Similarly, glycogen synthase, peaking before dusk, encodes a proenzyme, so its availability could allow the mosquito to process quickly excess sugar from a sugar meal consumed at the start of night. Finally, orthologs of the components of oxidative phosphorylation, four genes encoding subunits of the NADH dehydrogenase complex, three subunits of cytochrome C oxidase, and four F-type ATPase subunits, are rhythmic under diel conditions in the head at the end of night/early morning (Fig. S7B). These data collectively suggest that metabolic activity in An. gambiae is temporally coordinated, allowing the mosquito to anticipate the differing energy demands of activity and rest and the variations in anabolic and catabolic states that occur across the 24-h period (46).
Metabolic Detoxification.
Our analysis revealed 60 genes to be rhythmic that are known or predicted to be associated with metabolic detoxification. Metabolic resistance to insecticides is caused predominantly by elevated activity of cytochrome P450 mono-oxygenases (P450s), GSTs, and carboxylesterases (COEs) (58–60), many of which we found to display highly rhythmic expression patterns (Fig. 4B). Specifically, 35 P450s, 16 GSTs, and two COEs across both tissues and environmental conditions were identified, accounting for approximately one third of the total number of known or putative An. gambiae P450 and GST genes (61). These genes include GSTE2 (AGAP009194), a known GST whose up-regulation can confer resistance to the insecticide dichlorodiphenyltrichloroethane (DDT) (59), and three P450 genes [CYP6Z1 (AGAP008219), agCYP6P3 (AGAP002865), and agCYP6M2 (AGAP008212)], whose up-regulation is implicated in pyrethroid insecticide resistance (Fig. 4C) (60, 62). These enzymes were found to be expressed under circadian conditions in bodies as well as in heads and/or in bodies under diel conditions (Dataset S1). Rhythmic expression of detoxifying genes in An. gambiae likely translates into rhythmic patterns of time-of-day susceptibility to insecticide, as found in numerous insects including the mosquito Ae. aegypti (63, 64).
Interestingly, the metabolic detoxifying genes we identified do not cluster uniformly in time of peak expression. However, the three P450s implicated in pyrethroid resistance (CYP6Z1, agCYP6P3, and agCYP6M2) all share a peak in expression during the early night phase, perhaps in anticipation of a blood meal. Such a pattern has been reported in the house fly Musca domestica, with the daily peak in eating behavior corresponding with the time of greatest resistance to insecticide (65).
Vesicular-Type ATPase.
The multisubunit vesicular-type ATPase (V-ATPase) uses ATP to transport H+ actively and is found in Ae. aegypti in the osmoregulatory tissues including stomach, Malpighian tubules, anterior hindgut, and rectum (66). In the moth Spodoptera littoralis, regulation of the V-ATPase B-subunit and associated pH within the vas deferens are under circadian control, contributing to the regulation of spermatozoa maturation (67). In the body under LD conditions, using DAVID to identify orthologs, we found that 9 of the 12 subunits are rhythmic and in phase, peaking at ∼ZT11.5 (Fig. S9). Up-regulation of the V-ATPase complex could occur in anticipation of significant osmotic changes induced by a blood or sugar meal. Important roles for the V-ATPase in mosquito infections have been reported. Pharmacological or RNAi inhibition of the V-ATPase complex or subunits, respectively, results in diminished replication of both dengue and Japanese encephalitis viruses in Ae. aegypti cells (68, 69). Moreover, in An. gambiae and Ae. aegypti the region of the mosquito midgut epithelium that is the primary site of invasion for Plasmodium ookinetes is enriched for cells that overexpress V-ATPase (70). This observation leads to the intriguing possibility that mosquito pathogens may exploit these potential rhythms in V-ATPase synthesis during their replication and invasion processes.
Olfaction.
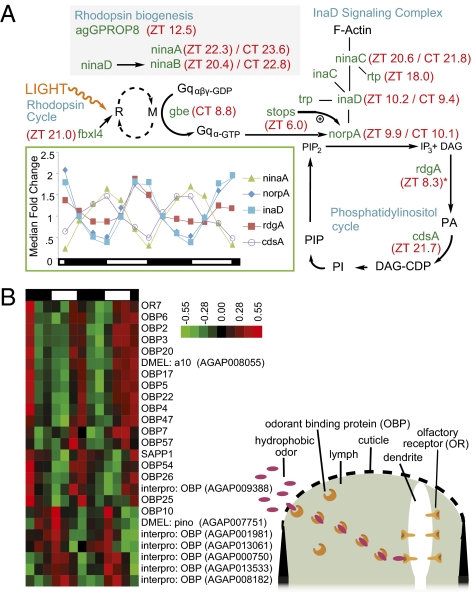
Olfaction is of importance to the mosquito in detecting blood-feeding hosts and sources of sugar feeding as well as in oviposition site selection (2, 71, 72). Additionally, daily rhythms of biting behavior have long been known (2, 4). Rhythmic antennal electrophysical responses to CO2 have been reported in tsetse flies and to the fruit odor ethyl acetate in Drosophila (73, 74). We identified 25 olfaction-related genes that were rhythmic under LD conditions in the head, including 15 known odorant-binding proteins (OBPs), a sensory appendage protein (SAP), and an odorant receptor, OR7 (AGAP002560) (Fig. 5B). OBPs and SAPs are soluble proteins that facilitate the activation of olfactory receptors by transporting odor molecules through the mucous layer to the receptors in the olfactory membrane (24, 75, 76). These data suggest that changes in OBP levels may modify the sensitivity of the olfactory sensilla, located primarily in the antennae, maxillary palps, and proboscis.
Fig. 5.
Components of the visual and olfactory systems are highly rhythmic. (A) Twelve genes in the Drosophila visual signaling transduction pathway were identified as rhythmic in LD and or/DD heads. Positions of rhythmic genes within the visual signaling transduction pathway are shown. Peak phase under LD and DD conditions is indicated in red next to gene names as Zeitgeber time (ZT) and circadian time (CT). Blue type indicates subcomponents of the visual transduction pathway. The Drosophila ortholog to the eye-specific G protein β-subunit gene gbe (AGAP001506) was found to be rhythmic in DD conditions only. *Note that Drosophila ortholog to the eye-specific diacylglycerol kinase gene retinal degeneration A (rdgA) (AGAP000519) is included in the pathway despite having a mean normalized fluorescence expression level <20. The signaling pathway was adapted from the Drosophila pathway described by Zuker (84) and Katz and Minke (85) and is assumed to be similar in Anopheles (123). Many of these genes continue to be rhythmic in DD conditions (Dataset S1), revealing that visual transduction is under circadian clock control. Gene identification was predicted from the highest percent similarity An. gambiae ortholog. M, active metarhodopsin; R, rhodopsin; PI, phosphatidylinositol; DAG-CDP, cytidine diphosphate diacylglycerol; PIP, phosphatidylinositol phosphate; PA, phosphatidic acid. (Inset) Transcription profiles of selected rhythmic vision genes under LD conditions. Expression values are normalized to the median value across the time course of each gene. Day and night are indicated by the horizontal white/black bar below the chart. (B) Hierarchical clustering of rhythmic olfaction genes under LD conditions in the head. Red indicates higher expression, and green indicates lower expression versus the mean value for each gene. Day and night are indicated by the horizontal white/black bar above the heatmap. A majority of the OBPs are expressed in synchrony, with a peak phase at ∼ZT12.
Interestingly, the known OBPs and SAP oscillate in two phases, one peaking 3 h before dusk and the other at the end of dusk/early night (Fig. 5B), perhaps preparing the system for an increase in arrival of odor molecules during the nocturnal host- and nectar-seeking behavior. The rhythmically expressed OBP genes OBP3, OBP17, OBP20, and OBP47 are particularly interesting, because they are enriched in the female head and antennae as compared with male tissues and female bodies (24, 75). This difference suggests a more prominent role for these specific OBPs in the mechanism of host detection, because host preference and blood feeding are restricted to female mosquitoes. Almost all genes related to olfaction in the head were no longer scored as rhythmically expressed in DD, suggesting that they are driven primarily by the LD cycle. This proposition is supported further by the finding that the expression of several of the rhythmic OBPs (specifically OBP22, OBP25, OBP26, and OBP47) are down-regulated in the head following acute light treatment presented during early night (4).
Therefore we would expect a diel but not a circadian rhythm in sensitivity to host and other cues. In contrast, OBPs, presumably located in the olfactory structures on the legs (75), are rhythmically expressed in the body samples under both LD and DD conditions or only in DD (Dataset S1), suggesting a different mechanism of temporal regulation involving the circadian clock. The master An. gambiae gustatory and odorant receptor, OR7 (olfactory coreceptor Agam/Orco, dsOr83b homolog), which is the required heterodimer for all odorant receptor transduction (77), was also rhythmic in the head under diel conditions, peaking similarly to the OBPs at ZT10.9 (Fig. 5B) near the end of the light phase. Rhythmicity of the OR7 could prove to be another potential gate for diel control of olfactory sensitivity (77) and is particularly interesting, because it contributes to the detection of the N,N-diethyl-m-toluamide (DEET) insect repellent (78, 79). Finally, the An. gambiae ortholog to Drosophila norpA (AGAP001936), which encodes a phospholipase C-β and which in Drosophila is involved in both visual and odorant responses (specifically maxillary palps) (72), was identified as rhythmic, peaking at ZT9.9 (Fig. 5A). This result highlights the possibility of a rhythm in inositol-1,4,5-trisphosphate activity and therefore gating of olfactory sensitivity occurring additionally downstream of the OBPs and olfactory receptors.
Vision.
Numerous examples illustrate the importance of vision to mosquito behavior. Mosquitoes respond to changes in landscape detail and light levels. Species, including Anopheles crucians, are attracted to visually conspicuous objects at a long distance (80). Ae. aegypti orientate themselves in flight using visual cues, presumably to counteract displacement by the wind. Shifts in the “horizon” modulate flight activity under laboratory conditions (81). Anopheles stephensi and Anopheles funestus exhibit greater nocturnal flight activity in dim light than in darkness, and a full moon induced a dramatic (400%) increase in house-entering behavior compared with moonless nights (82, 83). We used the Drosophila visual signaling pathway (84–86) as a model to identify An. gambiae orthologs. Our analysis identified 12 putative An. gambiae visual system genes that are rhythmically expressed under LD and/or DD conditions (Fig. 5A). These include genes involved in rhodopsin biogenesis, ninaA (AGAP009991) encoding an eye-specific cyclophilin, and ninaB (AGAP008143) encoding a β,β-carotene-15,15′-oxygenase. They include the An. gambiae rhodopsin GPROP8 (AGAP006126, dsRh3 homolog) and fbxl4 (AGAP000471), a transcription factor involved in deactivation of rhodopsin. There is also rhythmicity of genes involved in the phototransduction cascade, including cdsA (AGAP007175), a photoreceptor-specific cytidine diphosphate diacylglycerol synthase in the phosphatidylinositol cycle, and inaD, which encodes a scaffold protein. INAD is of particular interest as a potential major regulating step in circadian control of the visual system, because it organizes components of the phototransduction cascade into a signaling complex containing the major light-gated ion channel, TRP, the protein kinase C (INAC), the phospholipase C (NORPA), diacylglycerol (DAG) kinase, and the founding member of a family of kinase/myosin hybrid proteins, NINAC (AGAP009730) (85). Not only is inaD (AGAP002145) rhythmic, peaking 1 h before start of dusk (ZT10.2); so are many of the genes encoding various proteins with which cdsA forms a complex [e.g., norpA (AGAP001936), peaking similarly at ZT9.9 (Fig. 5A)].
Recent work in Drosophila has identified that the slow termination of phototransduction (stops) gene product is critical for maintaining protein, but not mRNA, levels of NORPA. It is intriguing that stops (AGAP000213) is rhythmically expressed and peaks at midday (ZT6.0), suggesting that NORPA levels could be tightly controlled for circadian expression at transcriptional, posttranscriptional (through interaction with STOPS), and localization levels (through INAD). An additional binding partner to INAD is NINAC, which associates INAD to F-actin (85). We found that ninaC peaks later in the night than inaD, stops, and norpA, but similar to the time of peaking of another gene encoding a retinophilin (RTP) (AGAP003547). In Drosophila, NINAC and RTP are binding partners, and their protein levels are codependent (87). We find in An. gambiae that the gene expressions of these binding partners peak at a similar time (ZT20.6 and ZT18.0, respectively). These phase-concordant relationships in gene expression between specific partner proteins might contribute to a time-of-day–specific gating mechanism for tuning the sensitivity of the An. gambiae visual system to photic activation.
Moreover, the rhythm in norpA is intriguing, because in Drosophila NORPA has been shown to regulate splicing of the clock gene period, to be important for seasonal photoperiodic control of locomotor behavior, and to be required for the rhythmic expression of multiple clock-independent light-driven transcripts (11). Finally, early work in Ae. aegypti noted apparent dramatic diel changes in rhabdom volume (88). These changes might be driven by the circadian system through both NINAC and RTP, which are both required for normal rhabdomere volume in Drosophila (87, 89). Many of these visual system genes continue to be rhythmic in DD conditions, suggesting that components of visual transduction are under circadian clock control (Fig. 5A). By extension, we hypothesize that aspects of visual function such as sensitivity to light are similarly under circadian control, consistent with electrophysiological studies in numerous other insect species (90).
Cuticle/Peritrophic Membrane Regulation.
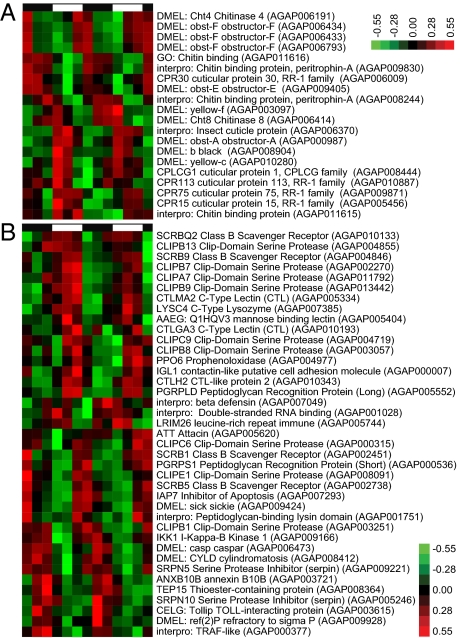
Structural cuticular proteins comprise nearly 2% of the An. gambiae gene set, with chitin serving as the major component of the mosquito cuticle, tracheal system, the peritrophic matrix that protects the midgut epithelium from damage upon blood feeding, and in egg chorion hardening (91, 92). Daily cuticle deposition in the inner apodemes has been noted in An. gambiae, forming discrete layers that are reliable indicators of mosquito age in days (93), and in many insect species this deposition is under circadian regulation (94, 95). In fact, in Drosophila this rhythm is regulated locally by a peripheral oscillator in the epidermis (95). In our analysis of An. gambiae body and head, we found numerous genes encoding proteins known to associate with chitin, such as cuticular proteins of the RR-1 and RR-2 families, cuticular proteins of low complexity, and orthologs to Drosophila Gasp and members of the obstructor and peritrophin A families of chitin-binding proteins (Fig. 6A and Dataset 1). A majority of cuticular protein genes exhibited rhythms under both LD and DD or only under diel conditions. For example, CPR75 and two orthologs to Drosophila obstructor-F (AGAP006433 and AGAP006434) in the body, and CPR79 (AGAP009877) and an ortholog to Drosophila Peritrophin A (AGAP000986) in the head, exhibited pronounced high-amplitude rhythms only in LD conditions (Fig. 6A and Dataset 1). A Drosophila ortholog of black (AGAP008904), also involved in the regulation of cuticular protein cross-linking and necessary for melanization, is rhythmically expressed in both tissues with a peak in the morning (Fig. 6A) (96, 97). Several genes associated with the production of melanin, important in cuticle tanning, chorion hardening, wound healing, and defense reactions (see “Immunity” below), were found to be rhythmically expressed in both head and body, as were Drosophila orthologs to several members of the yellow gene family associated with pigmentation (Fig. 6A and Dataset S1) (98, 99). Chitin synthesis itself may be under rhythmic control, because glucosamine 6-phosphate N-acetyltransferase (agGNA, AGAP010769), encoding a key enzyme of the hexosamine biosynthetic pathway, exhibits a diel rhythm in the head (Dataset S1) (100). Moreover, remodeling of chitin may be under rhythmic regulation, because the ortholog to Drosophila serpentine (AGAP011936), encoding a chitin-binding deacetylase protein involved in tracheal and cuticle growth, is under circadian control in the head (Dataset S1), and orthologs to Drosophila Chitinase 4 and Chitinase 8 are under circadian control in the body (Fig. 6A) (101). These daily oscillations in genes regulating both the deposition and remodeling of the chitin matrix and the synthesis of chitin-binding proteins are consistent with the daily cycles in cuticular growth layers observed in the thoracic apodemes of adult anopheline mosquitoes (93). The function of these cycles appears to be associated with age-related growth of the flight muscles, because the apodemes are the attachment site of the thoracic muscles, and apodeme length is correlated with flight muscle size (93). Furthermore, these data highlight the possibility of rhythmic tracheal growth, also incorporating chitin and binding proteins, as tracheal growth in the adult insect is correlated with age-associated increases in muscle density (102). Finally, these data may be important in cuticular resistance to insecticides, because the uptake of these lipophilic compounds used in malaria control is primarily through the appendages, and increased thickness or reduced permeability of the tarsal cuticle may contribute to the development of pyrethroid resistance in anophelines (60).
Fig. 6.
Components of the cuticle/peritrophic membrane and immune systems are highly rhythmic. (A) Hierarchical clustering of rhythmic chitin- and cuticular/peritrophic membrane-related genes in LD bodies. These genes primarily encode chitin-binding proteins and genes involved in the regulation of chitin remodeling and cuticule tanning. Red indicates higher expression, and green indicates lower expression versus the mean value for each gene. Day and night are indicated by the horizontal white/black bar above the heatmap. (B) Hierarchical clustering of rhythmic immunity genes in LD heads. These genes are primarily components of the immune deficiency (Imd) and melanization pathways and include class B scavenger receptors, several clip-domain serine proteases, a C-type lysozyme, peptidoglycan-recognition proteins (bacterial recognition), and C-type lectins. Genes with no prefix are An. gambiae gene names. Genes with species prefixes are the closest predicted named orthologs.
Immunity.
A circadian rhythm of susceptibility to bacterial infection in Drosophila has been demonstrated previously (103), and our microarray analysis revealed that such a pattern in susceptibility may exist in An. gambiae, with 39 different genes of known or putative immune function in the head and 20 such genes in the body showing diel expression patterns (Fig. 6B and Dataset S1). These genes are primarily in the immune deficiency (Imd) and melanization pathways and include class B scavenger receptors, several clip-domain serine proteases, a C-type lysozyme, peptidoglycan-recognition proteins (bacterial recognition), and several C-type lectins (23, 31). Of these genes, those involved in the melanization immune response, which encapsulates bacteria and Plasmodium parasites as well as producing toxic antimicrobial by-products, are prominent (96). Both in the head and body, the class B scavenger receptor SCRBQ2 (∼ZT23.5) and the clip-domain serine protease CLIPB8 (∼ZT7.5), which have gene products that promote the melanization response, show a diel rhythm (Fig. 6B) (104, 105). In addition melanization biosynthesis genes exhibit rhythmic expression patterns, including the prophenoloxidase (PPO)-encoding genes PPO6 (ZT9.3 in the head) (Fig. 6B), PPO5 (AGAP012616), and PPO9 (AGAP004978) (∼CT23 in the body), and an ortholog to Drosophila dopachrome-conversion enzyme yellow-f (AGAP003097, ZT2.0 in the body) (Fig. 6A) (96, 98, 106). The C-type lectin, CTLMA2 (ZT1.8 in the head and ZT5.0 in the body) and the clip-domain serine protease, CLIPA7 (ZT23.0 in the head), two immune genes whose gene products are capable of modulating the melanization response, were also found to have a diel expression pattern (107, 108). Our results lead to the interesting conclusion that time-of-day–specific effects should be considered when developing new biopesticide-based interventions. Additionally, extensive work in various Plasmodium species and hosts has demonstrated rhythmic variations in the ability of the parasite to infect mosquitoes, known as the “Hawking phenomenon.” It has been shown that the number of infective parasites tends to increase in the peripheral blood at times corresponding, presumably, to optimal vector-biting times (109, 110). Our results highlight the additional possibility that the parasite may respond not only to vector availability but also to daily rhythms in the innate immunity of the mosquito and its ability to be infected. Note, however, that parasite adaptation to a temporal pattern of mosquito immunity would be possible only if mosquitoes are biting over a long period of the 24-h cycle.
Conclusions
We have presented here the genome-wide diel and circadian transcriptional profiling of a disease vector that directly affects human health. Our analysis reveals that the mosquito expresses a great number and diversity of genes in a highly rhythmic manner. We have constructed a publicly accessible database (http://www.nd.edu/∼bioclock) that users can use to query for rhythmically regulated genes and to evaluate graphically the temporal expression patterns of genes of interest. Better understanding of the rhythmic nature of mosquito behavior and physiology, including sensory perception and susceptibilities to insecticide or immune challenge, may provide opportunities for novel malarial control strategies. Our data highlight a potential in An. gambiae for rhythmic susceptibility, prompting investigations into practical applications, including reexamination of timing for optimal application of insecticide-based interventions. Moreover, manipulation of genes relevant to key biological processes now is possible in light of developments in the generation of transgenic mosquitoes (1, 111). A consideration of circadian timing of mating may be important in the application of control strategies such as the sterile insect technique (SIT) and the introduction of pathogen-resistant strains (1, 111). Timing of nocturnal activities such as flight and biting vary among members of the An. gambiae complex and also among other Anopheles species and can impact the effectiveness of ITNs (112, 113). For example, a difference between reared and wild populations in the timing of mating activity has been proposed to contribute to the efficacy of the SIT technique (114). Moreover, there is evidence suggesting that use of ITNs may act as a selective pressure, potentially modifying the age and genetic composition of the population and selecting for individuals that initiate host-seeking behavior earlier in the night (112, 115, 116). Improved understanding of biological timing at the molecular level that underlies key physiological aspects of the An. gambiae malaria vector may prove to be important for successful implementation of control methods and future experimental design.
Materials and Methods
Biological Material.
Female mated but not blood-fed An. gambiae (Pimperena S form) mosquitoes [MRA-861, Malaria Research and Reference Reagent Resource Center (MR4), ATCC] were maintained at 85% relative humidity and 27 ± 1 °C in either DD conditions or a 12-h/12-h LD cycle (11 h full light (∼250 lx), 11 h darkness, and 1-h dawn and 1-h dusk transitions). Access to fructose was provided ad libitum. The colony was established in the laboratory from collections originally harvested from Pimperena, Mali, in 2005 (25, 117). Mosquitoes were maintained with females blood fed on a human donor (investigator's arm) for purposes of egg laying. Blood feeding occurred generally, although not exclusively, during the late day/early night phases of the LD cycle. As mosquitoes were maintained in the laboratory for many generations, there is some possibility that diel/circadian rhythms were influenced by the process of selection in colony adaptation and maintenance. Experiments began with females 7–9 d old, and 20–30 mosquitoes were collected on dry ice every 4 h for 48 h. Mosquitoes collected from each time point were pooled, with heads (including eyes, mouth parts, and antennae) separated from bodies (including appendages), and total RNA was purified by two rounds of TRIzol reagent (Invitrogen) extraction and sodium acetate precipitation. The quality and quantity of resulting RNA was assessed using a Nano Drop (Thermo Scientific) and RNA Nano LabChip for the Bioanalyzer (Agilent Technologies) and by qRT-PCR analysis of canonical clock genes (Fig. S4). ZT12 was defined as the time light was turned off in the LD cycle, and ZT0 was defined as end of the dawn transition. In CT, CT0 was defined as the end of subjective dawn, inferred from ZT0 of the previous LD cycle.
Locomotor Activity.
Individual mosquito locomotor/flight activity was measured with a Locomotor Activity Monitor 25 system (TriKinetics) and analyzed using the ClockLab analysis program (Actimetrics).
DNA Microarray Analysis.
DNA microarray analysis was performed in the Genomics Core Facility at the University of Notre Dame. Total RNA was reversed transcribed in two separate replicates using an 3′ IVT Express Kit (Affymetrix) and hybridized to a Plasmodium/Anopheles Affymetrix GeneChip Genome expression microarray (900511; Affymetrix) per the manufacturer's protocol. Expression values were normalized using the GC-RMA algorithm in GeneSpring GX11 (Agilent Technologies) against all other head or body samples. The Affymetrix GeneChip included 16,884 probe sets estimated to represent ∼14,900 predicted An. gambiae transcripts, with sequence information having been generated primarily from Ensembl (Build 2, 2003) and GenBank and dbEST. The current predicted entire An. gambiae gene set is 13,254 (determined as the number of Ensembl Gene identities from VectorBase at time of analysis), and 10,980 of the genes were represented on the microarray (10,172 unambiguously and 808 that could not be differentiated from at least one other gene).
Data Deposition.
The DNA microarray data reported in this paper have been deposited in GEO Express (accession no. GSE22585) and VectorBase Expression Data BioMart. Data are graphically available at http://www.nd.edu/∼bioclock.
Microarray Data Analysis.
Probes were scored as being rhythmic (i) if they were found using the COSOPT algorithm (6, 11, 13, 22) to have an average probability, multiple means corrected β (pMMCβ) value of < 0.1; (ii) had a mean fluorescence intensity >20 across all 13 time points in both replicates (a criterion that would exclude >99% of Plasmodium probe sets on the microarray); and (iii) had a period length of 18.5–26.5 h for constant-condition experiments or 20–28 h for LD experiments in both replicates. Probes were mapped to genes based on the Pest 3.5 assembly at VectorBase (118), and ambiguous probes mapping to multiple genes were excluded manually (agTO2 and agTO3 being exceptions). Presented pMMCβ values, phases, and period length estimates are the means of replicates. Genes were annotated primarily from information stored at VectorBase, often using the closest homolog from Ae. aegypti, Cx. quinquefasciatus, Drosophila, or C. elegans (in that order) but also using published literature and DAVID to match putative An. gambiae genes to enzymatic pathways (specifically tRNA, metabolism, oxidative phosphorylation, and V-ATPase) (118, 119). Where no An. gambiae or orthologous gene name was available, InterProScan (120) was used to annotate genes, and we provide a representative InterPro or the associated Gene Ontology (GO) term. Dataset S1 gives the full complement of data for each gene scored as rhythmic. Hierarchical cluster analysis was performed using Cluster 3.0 and visualized using Java TreeView (121). Data were log2 transformed, mean centered, and normalized across the time course for each gene. Rhythms of selected genes were confirmed using qRT-PCR (Fig. S4). While this manuscript was in preparation, a new algorithm, JTK_CYCLE, was developed for the automated discovery of rhythmic transcripts (122). We include JTK_CYCLE-derived values at http://www.nd.edu/∼bioclock.
qRT-PCR Analysis.
Total RNA was treated with DNaseI (Invitrogen) and used for cDNA synthesis using a TaqMan or high-capacity cDNA reverse transcriptase kit (Applied Biosystems), primed with random hexamers. PCR thermocycling and qRT-PCR were performed as previously described (6) using SYBR green reagents, an ABI PRISM 7700 or 7500 sequence-detection system, and quantification based on the generation of standard curves. Dissociation curves to test for primer dimers were generated using dissociation curve software (Applied Biosystems). Normalization of genes was calculated relative to ribosomal protein S7 (RPS7, AGAP010592). Primer sequences are provided in Fig. S4.
Supplementary Material
Acknowledgments
We thank N. J. Besansky, J. O'Tousa, B. Harker, J. Niedbalski, M. Kern, and B. Cassone for technical assistance; J. Hogenesch and M. Hughes for providing and assisting with the COSOPT algorithm; J. Hogenesch, A. Pizarro, S. Emrich, and R. Carmichael for providing and assisting with the implementation of the Circa database system; and C. Dong, M. Champion, K. Mecklenburg, and J. O'Tousa for comments on the manuscript. This work was supported by a fellowship (to S.S.C.R.) and a grant (to G.E.D.) from the Genomics, Disease Ecology and Global Health Strategic Research Initiative and the Eck Institute for Global Health, University of Notre Dame, and by Contract HHSN266200400039C from the National Institutes of Health/National Institute of Allergy and Infectious Disease to VectorBase.
Footnotes
The authors declare no conflict of interest.
Data deposition: The DNA microarray data reported in this paper have been deposited in GEO Express (accession no. GSE22585) and VectorBase Expression BioMart. Data are available graphically at http://www.nd.edu/∼bioclock, which allows graphical examination of the temporal expression data and access to statistical data for all Anopheles probe sets/transcripts represented on the microarray, not only those deemed rhythmic according to our criteria.
This article is a PNAS Direct Submission.
See Author Summary on page 12979.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100584108/-/DCSupplemental.
References
- 1.Enayati A, Hemingway J. Malaria management: Past, present, and future. Annu Rev Entomol. 2010;55:569–591. doi: 10.1146/annurev-ento-112408-085423. [DOI] [PubMed] [Google Scholar]
- 2.Clements AN. The Biology of Mosquitoes. Wallingford, UK: CABI; 1999. [Google Scholar]
- 3.Gary RE, Jr, Foster WA. Diel timing and frequency of sugar feeding in the mosquito Anopheles gambiae, depending on sex, gonotrophic state and resource availability. Med Vet Entomol. 2006;20:308–316. doi: 10.1111/j.1365-2915.2006.00638.x. [DOI] [PubMed] [Google Scholar]
- 4.Das S, Dimopoulos G. Molecular analysis of photic inhibition of blood-feeding in Anopheles gambiae. BMC Physiol. 2008;8:23. doi: 10.1186/1472-6793-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunlap JC, Loros JJ, Decoursey PJ. Chronobiology: Biological Timekeeping. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- 6.Duffield GE, et al. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 7.McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 8.Lin Y, et al. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:9562–9567. doi: 10.1073/pnas.132269699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueda HR, et al. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- 10.Claridge-Chang A, et al. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 11.Wijnen H, Naef F, Boothroyd C, Claridge-Chang A, Young MW. Control of daily transcript oscillations in Drosophila by light and the circadian clock. PLoS Genet. 2006;2:e39. doi: 10.1371/journal.pgen.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 13.Michael TP, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008;4:e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceriani MF, et al. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci. 2002;22:9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duffield GE. DNA microarray analyses of circadian timing: The genomic basis of biological time. J Neuroendocrinol. 2003;15:991–1002. doi: 10.1046/j.1365-2826.2003.01082.x. [DOI] [PubMed] [Google Scholar]
- 16.Doherty CJ, Kay SA. Circadian control of global gene expression patterns. Annu Rev Genet. 2010;44:419–444. doi: 10.1146/annurev-genet-102209-163432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentile C, et al. Cloning and daily expression of the timeless gene in Aedes aegypti (Diptera:Culicidae) Insect Biochem Mol Biol. 2006;36:878–884. doi: 10.1016/j.ibmb.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Gentile C, Rivas GBS, Meireles-Filho ACA, Lima JBP, Peixoto AA. Circadian expression of clock genes in two mosquito disease vectors: Cry2 is different. J Biol Rhythms. 2009;24:444–451. doi: 10.1177/0748730409349169. [DOI] [PubMed] [Google Scholar]
- 19.Yuan Q, Metterville D, Briscoe AD, Reppert SM. Insect cryptochromes: Gene duplication and loss define diverse ways to construct insect circadian clocks. Mol Biol Evol. 2007;24:948–955. doi: 10.1093/molbev/msm011. [DOI] [PubMed] [Google Scholar]
- 20.Oztürk N, Song SH, Selby CP, Sancar A. Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J Biol Chem. 2008;283:3256–3263. doi: 10.1074/jbc.M708612200. [DOI] [PubMed] [Google Scholar]
- 21.Rona LDP, Carvalho-Pinto CJ, Gentile C, Grisard EC, Peixoto AA. Assessing the molecular divergence between Anopheles (Kerteszia) cruzii populations from Brazil using the timeless gene: Further evidence of a species complex. Malar J. 2009;8:60. doi: 10.1186/1475-2875-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathias D, Jacky L, Bradshaw WE, Holzapfel CM. Geographic and developmental variation in expression of the circadian rhythm gene, timeless, in the pitcher-plant mosquito, Wyeomyia smithii. J Insect Physiol. 2005;51:661–667. doi: 10.1016/j.jinsphys.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Dimopoulos G, et al. Genome expression analysis of Anopheles gambiae: Responses to injury, bacterial challenge, and malaria infection. Proc Natl Acad Sci USA. 2002;99:8814–8819. doi: 10.1073/pnas.092274999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biessmann H, Nguyen QK, Le D, Walter MF. Microarray-based survey of a subset of putative olfactory genes in the mosquito Anopheles gambiae. Insect Mol Biol. 2005;14:575–589. doi: 10.1111/j.1365-2583.2005.00590.x. [DOI] [PubMed] [Google Scholar]
- 25.Cassone BJ, et al. Differential gene expression in incipient species of Anopheles gambiae. Mol Ecol. 2008;17:2491–2504. doi: 10.1111/j.1365-294X.2008.03774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Straume M. DNA microarray time series analysis: Automated statistical assessment of circadian rhythms in gene expression patterning. Methods Enzymol. 2004;383:149–166. doi: 10.1016/S0076-6879(04)83007-6. [DOI] [PubMed] [Google Scholar]
- 27.Albers HE, Liou SY, Ferris CF, Stopa EG, Zoeller RT. Neurochemistry of circadian timing. In: Klein DC, Moore RY, Reppert SM, editors. Suprachiasmatic Nucleus: The Mind's Clock. New York: Oxford Univ Press; 1991. [Google Scholar]
- 28.Inouye ST. Circadian rhythms of neuropeptides in the suprachiasmatic nucleus. Prog Brain Res. 1996;111:75–90. doi: 10.1016/s0079-6123(08)60401-x. [DOI] [PubMed] [Google Scholar]
- 29.Chen CH, Ringelberg CS, Gross RH, Dunlap JC, Loros JJ. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J. 2009;28:1029–1042. doi: 10.1038/emboj.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hastings JW. Biochemical aspects of rhythms: Phase shifting by chemicals. Cold Spring Harb Symp Quant Biol. 1960;25:131–143. doi: 10.1101/sqb.1960.025.01.012. [DOI] [PubMed] [Google Scholar]
- 31.Christophides GK, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 32.Meireles-Filho ACA, et al. The biological clock of an hematophagous insect: Locomotor activity rhythms, circadian expression and downregulation after a blood meal. FEBS Lett. 2006;580:2–8. doi: 10.1016/j.febslet.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 33.Sandrelli F, Costa R, Kyriacou CP, Rosato E. Comparative analysis of circadian clock genes in insects. Insect Mol Biol. 2008;17:447–463. doi: 10.1111/j.1365-2583.2008.00832.x. [DOI] [PubMed] [Google Scholar]
- 34.Goto SG, Denlinger DL. Short-day and long-day expression patterns of genes involved in the flesh fly clock mechanism: period, timeless, cycle and cryptochrome. J Insect Physiol. 2002;48:803–816. doi: 10.1016/s0022-1910(02)00108-7. [DOI] [PubMed] [Google Scholar]
- 35.Garbarino-Pico E, Green CB. Posttranscriptional regulation of mammalian circadian clock output. Cold Spring Harb Symp Quant Biol. 2007;72:145–156. doi: 10.1101/sqb.2007.72.022. [DOI] [PubMed] [Google Scholar]
- 36.So WV, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schöning JC, et al. Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 2007;52:1119–1130. doi: 10.1111/j.1365-313X.2007.03302.x. [DOI] [PubMed] [Google Scholar]
- 38.Nakahata Y, Grimaldi B, Sahar S, Hirayama J, Sassone-Corsi P. Signaling to the circadian clock: Plasticity by chromatin remodeling. Curr Opin Cell Biol. 2007;19:230–237. doi: 10.1016/j.ceb.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Möller A, Avila FW, Erickson JW, Jäckle H. Drosophila BAP60 is an essential component of the Brahma complex, required for gene activation and repression. J Mol Biol. 2005;352:329–337. doi: 10.1016/j.jmb.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Schotta G, et al. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beltran S, Angulo M, Pignatelli M, Serras F, Corominas M. Functional dissection of the ash2 and ash1 transcriptomes provides insights into the transcriptional basis of wing phenotypes and reveals conserved protein interactions. Genome Biol. 2007;8:R67. doi: 10.1186/gb-2007-8-4-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci USA. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sacktor B. Biochemical adaptations for flight in the insect. Biochem Soc Symp. 1976;41:111–131. [PubMed] [Google Scholar]
- 44.Gray EM, Bradley TJ. Metabolic rate in female Culex tarsalis (Diptera: Culicidae): Age, size, activity, and feeding effects. J Med Entomol. 2003;40:903–911. doi: 10.1603/0022-2585-40.6.903. [DOI] [PubMed] [Google Scholar]
- 45.Kohsaka A, Bass J. A sense of time: How molecular clocks organize metabolism. Trends Endocrinol Metab. 2007;18:4–11. doi: 10.1016/j.tem.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 46.Zheng X, Sehgal A. AKT and TOR signaling set the pace of the circadian pacemaker. Curr Biol. 2010;20:1203–1208. doi: 10.1016/j.cub.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holliday-Hanson ML, Yuval B, Washino RK. Energetics and sugar-feeding of field-collected anopheline females. J Vector Ecol. 1997;22:83–89. [PubMed] [Google Scholar]
- 48.Kaufmann C, Briegel H. Flight performance of the malaria vectors Anopheles gambiae and Anopheles atroparvus. J Vector Ecol. 2004;29:140–153. [PubMed] [Google Scholar]
- 49.Arsic D, Guerin PM. Nutrient content of diet affects the signaling activity of the insulin/target of rapamycin/p70 S6 kinase pathway in the African malaria mosquito Anopheles gambiae. J Insect Physiol. 2008;54:1226–1235. doi: 10.1016/j.jinsphys.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 50.Hahn DA, Denlinger DL. Energetics of insect diapause. Annu Rev Entomol. 2011;56:103–121. doi: 10.1146/annurev-ento-112408-085436. [DOI] [PubMed] [Google Scholar]
- 51.Sarov-Blat L, So WV, Liu L, Rosbash M. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell. 2000;101:647–656. doi: 10.1016/s0092-8674(00)80876-4. [DOI] [PubMed] [Google Scholar]
- 52.Kaufmann C, Brown MR. Adipokinetic hormones in the African malaria mosquito, Anopheles gambiae: Identification and expression of genes for two peptides and a putative receptor. Insect Biochem Mol Biol. 2006;36:466–481. doi: 10.1016/j.ibmb.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 53.Kaufmann C, Brown MR. Regulation of carbohydrate metabolism and flight performance by a hypertrehalosaemic hormone in the mosquito Anopheles gambiae. J Insect Physiol. 2008;54:367–377. doi: 10.1016/j.jinsphys.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Becker A, Schlöder P, Steele JE, Wegener G. The regulation of trehalose metabolism in insects. Experientia. 1996;52:433–439. doi: 10.1007/BF01919312. [DOI] [PubMed] [Google Scholar]
- 55.Nowosielski JW, Patton RL. Daily fluctuation in the blood sugar concentration of the house cricket, Gryllus domesticus L. Science. 1964;144:180–181. doi: 10.1126/science.144.3615.180. [DOI] [PubMed] [Google Scholar]
- 56.Berg JM, Tymoczko JL, Stryer L. Biochemistry. New York: W.H. Freeman; 2010. [Google Scholar]
- 57.Nation JL. Insect Physiology and Biochemistry. Boca Raton, FL: CRC; 2002. [Google Scholar]
- 58.Corbel V, et al. Multiple insecticide resistance mechanisms in Anopheles gambiae and Culex quinquefasciatus from Benin, West Africa. Acta Trop. 2007;101:207–216. doi: 10.1016/j.actatropica.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 59.Ranson H, et al. Identification of a novel class of insect glutathione S-transferases involved in resistance to DDT in the malaria vector Anopheles gambiae. Biochem J. 2001;359:295–304. doi: 10.1042/0264-6021:3590295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ranson H, et al. Pyrethroid resistance in African anopheline mosquitoes: What are the implications for malaria control? Trends Parasitol. 2011;27:91–98. doi: 10.1016/j.pt.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 61.Ranson H, et al. Evolution of supergene families associated with insecticide resistance. Science. 2002;298:179–181. doi: 10.1126/science.1076781. [DOI] [PubMed] [Google Scholar]
- 62.Nikou D, Ranson H, Hemingway J. An adult-specific CYP6 P450 gene is overexpressed in a pyrethroid-resistant strain of the malaria vector, Anopheles gambiae. Gene. 2003;318:91–102. doi: 10.1016/s0378-1119(03)00763-7. [DOI] [PubMed] [Google Scholar]
- 63.Yang YY, et al. Circadian control of permethrin-resistance in the mosquito Aedes aegypti. J Insect Physiol. 2010;56:1219–1223. doi: 10.1016/j.jinsphys.2010.03.028. [DOI] [PubMed] [Google Scholar]
- 64.Hooven LA, Sherman KA, Butcher S, Giebultowicz JM. Does the clock make the poison? Circadian variation in response to pesticides. PLoS ONE. 2009;4:e6469. doi: 10.1371/journal.pone.0006469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Batth SS. Pesticide effectiveness and biorhythm in the house fly. J Econ Entomol. 1972;65:1191–1193. doi: 10.1093/jee/65.4.1191. [DOI] [PubMed] [Google Scholar]
- 66.Patrick ML, Aimanova K, Sanders HR, Gill SS. P-type Na+/K+-ATPase and V-type H+-ATPase expression patterns in the osmoregulatory organs of larval and adult mosquito Aedes aegypti. J Exp Biol. 2006;209:4638–4651. doi: 10.1242/jeb.02551. [DOI] [PubMed] [Google Scholar]
- 67.Bebas P, Cymborowski B, Giebultowicz JM. Circadian rhythm of acidification in insect vas deferens regulated by rhythmic expression of vacuolar H(+)-ATPase. J Exp Biol. 2002;205:37–44. doi: 10.1242/jeb.205.1.37. [DOI] [PubMed] [Google Scholar]
- 68.Sessions OM, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nawa M. Effects of bafilomycin A1 on Japanese encephalitis virus in C6/36 mosquito cells. Arch Virol. 1998;143:1555–1568. doi: 10.1007/s007050050398. [DOI] [PubMed] [Google Scholar]
- 70.Cociancich SO, Park SS, Fidock DA, Shahabuddin M. Vesicular ATPase-overexpressing cells determine the distribution of malaria parasite oocysts on the midguts of mosquitoes. J Biol Chem. 1999;274:12650–12655. doi: 10.1074/jbc.274.18.12650. [DOI] [PubMed] [Google Scholar]
- 71.Takken W, Knols BG. Odor-mediated behavior of Afrotropical malaria mosquitoes. Annu Rev Entomol. 1999;44:131–157. doi: 10.1146/annurev.ento.44.1.131. [DOI] [PubMed] [Google Scholar]
- 72.Bock GR, Cardew G, Ciba Foundation Symposium . Olfactory Regulation of Mosquito–Host Interactions. New York: Wiley; 1996. [Google Scholar]
- 73.Barrozo RB, Schilman PE, Minoli SA, Lazzari CR. Daily rhythms in disease-vector insects. Biol Rhythm Res. 2004;35:79–92. [Google Scholar]
- 74.Van der Goes van Naters WM, Den Otter CJ, Maes FW. Olfactory sensitivity in tsetse flies: A daily rhythm. Chem Senses. 1998;23:351–357. doi: 10.1093/chemse/23.3.351. [DOI] [PubMed] [Google Scholar]
- 75.Li ZX, Pickett JA, Field LM, Zhou JJ. Identification and expression of odorant-binding proteins of the malaria-carrying mosquitoes Anopheles gambiae and Anopheles arabiensis. Arch Insect Biochem Physiol. 2005;58:175–189. doi: 10.1002/arch.20047. [DOI] [PubMed] [Google Scholar]
- 76.Pelosi P, Calvello M, Ban L. Diversity of odorant-binding proteins and chemosensory proteins in insects. Chem Senses. 2005;30(Suppl 1):i291–i292. doi: 10.1093/chemse/bjh229. [DOI] [PubMed] [Google Scholar]
- 77.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ditzen M, Pellegrino M, Vosshall LB. Insect odorant receptors are molecular targets of the insect repellent DEET. Science. 2008;319:1838–1842. doi: 10.1126/science.1153121. [DOI] [PubMed] [Google Scholar]
- 79.Liu C, et al. Distinct olfactory signaling mechanisms in the malaria vector mosquito Anopheles gambiae. PLoS Biol. 2010;8:e1000467. doi: 10.1371/journal.pbio.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bidlingmayer WL, Hem DG. The range of visual attraction and the effect of competitive visual attractants upon mosquito (Diptera, Culicidae) flight. Bull Entomol Res. 1980;70:321–342. [Google Scholar]
- 81.Kennedy JS. The visual responses of flying mosquitoes. Proc Zool Soc Lond A-GE. 1940;109:221–242. [Google Scholar]
- 82.Ribbands CR. Moonlight and house-haunting habits of female anophelines in West Africa. Bull Entomol Res. 1946;36:395–417. doi: 10.1017/s0007485300024056. [DOI] [PubMed] [Google Scholar]
- 83.Rowland M. Changes in the circadian flight activity of the mosquito Anopheles stephensi associated with insemination, blood-feeding, oviposition and nocturnal light-intensity. Physiol Entomol. 1989;14:77–84. [Google Scholar]
- 84.Zuker CS. The biology of vision of Drosophila. Proc Natl Acad Sci USA. 1996;93:571–576. doi: 10.1073/pnas.93.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Katz B, Minke B. Drosophila photoreceptors and signaling mechanisms. Front Cell Neurosci. 2009;3:2. doi: 10.3389/neuro.03.002.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han J, et al. The fly CAMTA transcription factor potentiates deactivation of rhodopsin, a G protein-coupled light receptor. Cell. 2006;127:847–858. doi: 10.1016/j.cell.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 87.Venkatachalam K, et al. Dependence on a retinophilin/myosin complex for stability of PKC and INAD and termination of phototransduction. J Neurosci. 2010;30:11337–11345. doi: 10.1523/JNEUROSCI.2709-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brammer JD, Stein PJ, Anderson RA. Effect of light and dark adaptation upon the rhabdom in the compound eye of the mosquito. J Exp Zool. 1978;206:151–156. [Google Scholar]
- 89.Mecklenburg KL, et al. Retinophilin is a light-regulated phosphoprotein required to suppress photoreceptor dark noise in Drosophila. J Neurosci. 2010;30:1238–1249. doi: 10.1523/JNEUROSCI.4464-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fleissner G, Fleissner G. Efferent Control of Visual Sensitivity in Arthropod Eyes: With Emphasis on Circadian Rhythms. Stuttgart: G. Fischer; 1987. [Google Scholar]
- 91.Merzendorfer H, Zimoch L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J Exp Biol. 2003;206:4393–4412. doi: 10.1242/jeb.00709. [DOI] [PubMed] [Google Scholar]
- 92.Willis JH. Structural cuticular proteins from arthropods: Annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem Mol Biol. 2010;40:189–204. doi: 10.1016/j.ibmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schlein Y, Gratz NG. Determination of the age of some anopheline mosquitoes by daily growth layers of skeletal apodemes. Bull World Health Organ. 1973;49:371–375. [PMC free article] [PubMed] [Google Scholar]
- 94.Neville AC. Circadian organization of chitin in some insect skeletons. Q J Microsc Sci. 1965;106:315–325. [Google Scholar]
- 95.Ito C, Goto SG, Shiga S, Tomioka K, Numata H. Peripheral circadian clock for the cuticle deposition rhythm in Drosophila melanogaster. Proc Natl Acad Sci USA. 2008;105:8446–8451. doi: 10.1073/pnas.0800145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Christensen BM, Li J, Chen CC, Nappi AJ. Melanization immune responses in mosquito vectors. Trends Parasitol. 2005;21:192–199. doi: 10.1016/j.pt.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 97.Phillips AM, Smart R, Strauss R, Brembs B, Kelly LE. The Drosophila black enigma: The molecular and behavioural characterization of the black1 mutant allele. Gene. 2005;351:131–142. doi: 10.1016/j.gene.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 98.Han Q, et al. Identification of Drosophila melanogaster yellow-f and yellow-f2 proteins as dopachrome-conversion enzymes. Biochem J. 2002;368:333–340. doi: 10.1042/BJ20020272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Drapeau MD. The family of yellow-related Drosophila melanogaster proteins. Biochem Biophys Res Commun. 2001;281:611–613. doi: 10.1006/bbrc.2001.4391. [DOI] [PubMed] [Google Scholar]
- 100.Kato N, Mueller CR, Wessely V, Lan Q, Christensen BM. Mosquito glucosamine-6-phosphate N-acetyltransferase: cDNA, gene structure and enzyme kinetics. Insect Biochem Mol Biol. 2005;35:637–646. doi: 10.1016/j.ibmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 101.Luschnig S, Bätz T, Armbruster K, Krasnow MA. serpentine and vermiform encode matrix proteins with chitin binding and deacetylation domains that limit tracheal tube length in Drosophila. Curr Biol. 2006;16:186–194. doi: 10.1016/j.cub.2005.11.072. [DOI] [PubMed] [Google Scholar]
- 102.Kirkton SD. Effects of insect body size on tracheal structure and function. In: Roach RC, Wagner PD, Hackett PH, editors. Hypoxia and the Circulation. New York: Springer Science and Business Media; 2007. [Google Scholar]
- 103.Lee JE, Edery I. Circadian regulation in the ability of Drosophila to combat pathogenic infections. Curr Biol. 2008;18:195–199. doi: 10.1016/j.cub.2007.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.González-Lázaro M, et al. Anopheles gambiae Croquemort SCRBQ2, expression profile in the mosquito and its potential interaction with the malaria parasite Plasmodium berghei. Insect Biochem Mol Biol. 2009;39:395–402. doi: 10.1016/j.ibmb.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Paskewitz SM, Andreev O, Shi L. Gene silencing of serine proteases affects melanization of Sephadex beads in Anopheles gambiae. Insect Biochem Mol Biol. 2006;36:701–711. doi: 10.1016/j.ibmb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 106.Müller HM, Dimopoulos G, Blass C, Kafatos FC. A hemocyte-like cell line established from the malaria vector Anopheles gambiae expresses six prophenoloxidase genes. J Biol Chem. 1999;274:11727–11735. doi: 10.1074/jbc.274.17.11727. [DOI] [PubMed] [Google Scholar]
- 107.Volz J, Müller HM, Zdanowicz A, Kafatos FC, Osta MA. A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell Microbiol. 2006;8:1392–1405. doi: 10.1111/j.1462-5822.2006.00718.x. [DOI] [PubMed] [Google Scholar]
- 108.Osta MA, Christophides GK, Kafatos FC. Effects of mosquito genes on Plasmodium development. Science. 2004;303:2030–2032. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- 109.Gautret P, Motard A. Periodic infectivity of Plasmodium gametocytes to the vector. A review. Parasite. 1999;6:103–111. doi: 10.1051/parasite/1999062103. [DOI] [PubMed] [Google Scholar]
- 110.Hawking F. The clock of the malaria parasite. Sci Am. 1970;222:123–131. doi: 10.1038/scientificamerican0670-123. [DOI] [PubMed] [Google Scholar]
- 111.Nolan T, et al. Developing transgenic Anopheles mosquitoes for the sterile insect technique. Genetica. 2011;139:33–39. doi: 10.1007/s10709-010-9482-8. [DOI] [PubMed] [Google Scholar]
- 112.Jones MDR, Gubbins SJ, Cubbin CM. Circadian flight activity in 4 sibling species of Anopheles gambiae complex (Diptera, Culicidae) Bull Entomol Res. 1974;64:241–246. [Google Scholar]
- 113.Harris AF, Matias-Arnéz A, Hill N. Biting time of Anopheles darlingi in the Bolivian Amazon and implications for control of malaria. Trans R Soc Trop Med Hyg. 2006;100:45–47. doi: 10.1016/j.trstmh.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 114.Matsumoto A, et al. Period gene of Bactrocera cucurbitae (Diptera: Tephritidae) among strains with different mating times and sterile insect technique. Ann Entomol Soc Am. 2008;101:1121–1130. [Google Scholar]
- 115.Mbogo CNM, Baya NM, Ofulla AVO, Githure JI, Snow RW. The impact of permethrin-impregnated bednets on malaria vectors of the Kenyan coast. Med Vet Entomol. 1996;10:251–259. doi: 10.1111/j.1365-2915.1996.tb00739.x. [DOI] [PubMed] [Google Scholar]
- 116.Mathenge EM, et al. Effect of permethrin-impregnated nets on exiting behavior, blood feeding success, and time of feeding of malaria mosquitoes (Diptera: Culicidae) in western Kenya. J Med Entomol. 2001;38:531–536. doi: 10.1603/0022-2585-38.4.531. [DOI] [PubMed] [Google Scholar]
- 117.Lawniczak MK, et al. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science. 2010;330:512–514. doi: 10.1126/science.1195755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lawson D, et al. VectorBase: A data resource for invertebrate vector genomics. Nucleic Acids Res. 2009;37(Database issue):D583–D587. doi: 10.1093/nar/gkn857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 120.Hunter S, et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009;37(Database issue):D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yau KW, Hardie RC. Phototransduction motifs and variations. Cell. 2009;139:246–264. doi: 10.1016/j.cell.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]