Abstract
Major bacterial small RNAs (sRNAs) regulate the translation and stability of target mRNAs through base pairing with the help of the RNA chaperone Hfq. The Hfq-dependent sRNAs consist of three basic elements, mRNA base-pairing region, Hfq-binding site, and rho-independent terminator. Although the base-pairing region and the terminator are well documented in many sRNAs, the Hfq-binding site is less well-defined except that Hfq binds RNA with a preference for AU-rich sequences. Here, we performed mutational and biochemical studies to define the sRNA site required for Hfq action using SgrS as a model sRNA. We found that shortening terminator polyU tail eliminates the ability of SgrS to bind to Hfq and to silence ptsG mRNA. We also demonstrate that the SgrS terminator can be replaced with any foreign rho-independent terminators possessing a polyU tail longer than 8 without losing the ability to silence ptsG mRNA in an Hfq-dependent manner. Moreover, we found that shortening the terminator polyU tail of several other sRNAs also eliminates the ability to bind to Hfq and to regulate target mRNAs. We conclude that the polyU tail of sRNAs is essential for Hfq action in general. The data also indicate that the terminator polyU tail plays a role in Hfq-dependent stabilization of sRNAs.
Keywords: riboregulation, RNA 3′ end, RNase E, RyhB
A major class of bacterial small RNAs (sRNAs) binds to the RNA chaperone Hfq and acts as an RNA regulator by affecting the translation and stability of target mRNAs through imperfect base pairing with the help of Hfq (1–3). Hfq-dependent sRNAs consist of three basic elements, mRNA base-pairing region, Hfq-binding site, and rho-independent transcription terminator. The base-pairing region is a region complementary to the target mRNA and is well-defined in many sRNAs. Its role is to form an RNA–RNA hybrid with the target mRNA. The rho-independent terminator, characterized as a GC-rich palindrome sequence followed by a run of U residues, is also clearly defined. The obvious role of the rho-independent terminator is to terminate transcription resulting in distinct sRNA molecules. Another important role of the terminator is to stabilize the transcribed sRNAs (4, 5). Among the three elements of Hfq-dependent sRNAs, the Hfq-binding site is the least well-defined. Hfq has been shown to bind to sRNAs with a preference for AU-rich sequences, and Hfq is essential in vivo for pairing of sRNAs with target mRNAs (6, 7). Identification and characterization of Hfq-binding site within sRNAs are certainly important for better understanding of the mechanism by which Hfq facilitates the base pairing between sRNAs and cognate mRNAs.
Escherichia coli SgrS is one of the well-characterized Hfq-binding sRNAs (8, 9). It is induced in response to glucose–phosphate stress such as accumulation of glucose-6-phosphate and down-regulates the expression of ptsG encoding the membrane component of the major glucose transporter (10). The 31-nt-long stretch (nucleotides 157–187) in the 3′ region of SgrS is partially complementary to a 32-nt-long region of ptsG mRNA including the SD sequence and AUG start codon (10). We demonstrated recently that the 14-nt stretch between nucleotides 168 and 181 is the minimal base-pairing region for SgrS function (11). SgrS possesses a rho-independent terminator consisting of a GC-rich stem–loop followed by maximum eight consecutive U residues (10). In contrast, the Hfq-binding site on SgrS is totally unknown. The aim of the present study is to define the site required for Hfq action on sRNAs by using SgrS as a model sRNA. We introduced various mutations in the sgrS gene, and the effects of these mutations on the abilities of SgrS to down-regulate the target ptsG mRNA and to bind Hfq were analyzed. The results indicate that the polyU tail of the rho-independent terminator of SgrS is essential for Hfq binding and therefore for riboregulation. We also demonstrate that the polyU tail is required for the function of other Hfq-binding sRNAs.
Results
The Functional Hfq-Binding Site Is Located in the 3′ Portion of SgrS.
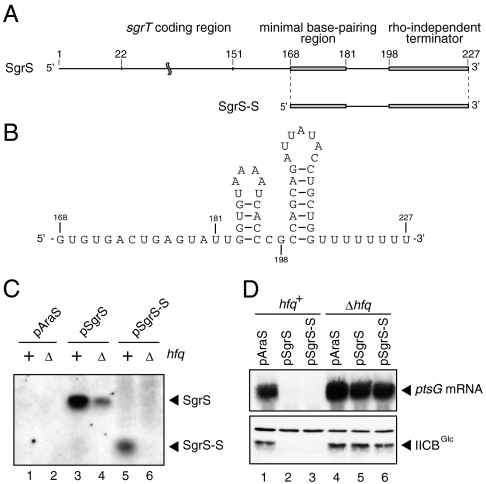
The nucleotide sequences around the rho-independent terminator region of sRNA genes used in this study are shown in Table S1. For simplicity, we refer to the transcript from wild-type sgrS as SgrS and those from sgrS mutants, for example, as SgrS-S and SgrS-7U, although the actual transcripts are expected to be heterogeneous. SgrS is 227 nt in length assuming that transcription termination occurs at the last U within the polyU tail (Fig. 1A). The minimal base-pairing region of SgrS is located at nucleotides 168–181 in the 3′ portion (11), whereas the 5′ portion of SgrS encodes a polypeptide of 43 amino acids designated SgrT (12). As the first step to define the functional Hfq-binding site on SgrS, we deleted nucleotides 1–167 of the sgrS gene under the arabinose-inducible promoter on the low copy number plasmid (Fig. 1A). The resulting sgrS-S is expected to generate SgrS-S of 60 nt (Fig. 1B). Plasmid pSgrS-S carrying the sgrS-S was introduced into both hfq+ and hfq- cells. As controls, cells were also transformed with plasmid pSgrS or the empty vector pAraS. Cells were grown in the presence of arabinose and expression of sRNAs was analyzed by Northern blotting. The sgrS and sgrS-S genes produce SgrS and SgrS-S in hfq+ cells, respectively, as expected (Fig. 1C, lanes 3 and 5). The expression level of SgrS-S was significantly lower compared to the full-length SgrS (Fig. 1C, lanes 3 and 5). It is possible that removal of residues 1–167 of SgrS may affect transcript stability resulting in the reduced expression of SgrS-S. It should be noted that the expression of SgrS-S was markedly reduced in hfq- cells (Fig. 1C, lane 6), suggesting that SgrS-S is also capable of binding to Hfq. We then tested the effect of SgrS-S expression on ptsG mRNA and its protein product IICBGlc. SgrS-S caused an efficient translational inhibition and rapid degradation of ptsG mRNA in an Hfq-dependent manner as the full-length SgrS did (Fig. 1D). Thus, SgrS-S retains the full activity to down-regulate ptsG mRNA. This implies that the long 5′ portion of SgrS is dispensable for Hfq action. In other words, the functional Hfq-binding site of SgrS should be located somewhere between nucleotides 168 and 227.
Fig. 1.
Properties of the 5′ truncated SgrS (SgrS-S). (A) Organization of SgrS and SgrS-S. SgrS is 227 nt in length when its transcription terminates at the last U within the polyU tail (10). The 5′ portion (22–151) of SgrS encodes a polypeptide of 43 amino acids designated SgrT (12). The 3′ portion of SgrS consists of the minimal base-pairing region (168–181) (11), a terminator hairpin including polyU stretch (198–227), and a spacer (182–197). The truncated form of SgrS (SgrS-S) lacks nucleotides 1–167. (B) Nucleotide sequence and secondary structure of SgrS-S. The sequence is based on the assumption that the transcription terminates at the last U within the polyU tail. (C) Expression of SgrS and SgrS-S. IT1568 (hfq+) and TM589 (Δhfq) cells harboring indicated plasmids were grown in LB medium. Total RNAs were prepared to A600 = 0.6 and subjected to Northern blot analysis using the SgrS probe 2. The following amounts of RNAs were loaded to the gel: lanes 1–4, 0.25 μg; lanes 5 and 6, 1.0 μg. (D) Effects of SgrS and SgrS-S on ptsG expression. IT1568 and TM589 cells harboring indicated plasmids were grown in LB medium. Total RNAs were prepared to A600 = 0.6 and 7 μg of each RNA sample was subjected to Northern blot analysis using the ptsG probe. Total proteins were also prepared and samples equivalent to 0.025 A600 units were subjected to Western blot analysis using anti-IIB antibodies.
An AU-Rich Sequence in the Hairpin Loop Is Not Involved in Hfq Action.
The 3′ portion of SgrS consists of the base-pairing region (168–181), the terminator hairpin including polyU stretch (198–227), and the spacer (182–197) as shown in Fig. 1A. We assumed that the rho-independent terminator is involved in Hfq action because it contains two potential Hfq-binding sites, an AU-rich sequence of 6-nt length in the hairpin loop and the polyU tail (see Fig. 1B). To test whether these potential Hfq-binding sites are required for Hfq action, we have introduced mutations in these sites and examined their effects on SgrS function. First, the AU-rich sequence in the loop was converted to a GC-rich sequence (SgrS-LM; Table S1 and Fig. S1). The resulting SgrS-LM retained the full Hfq-dependent silencing ability (Fig. S1). We also constructed plasmids carrying each of two additional mutants (sgrS-SM1 and sgrS-SM2) in which the sequence in the stem was changed without losing five GC and two AU base pairs (Table S1 and Fig. S1). Again these two SgrS mutants exhibited the full Hfq-dependent regulatory function (Fig. S1). Thus, no specific sequence within the terminator hairpin of SgrS, including the AU-rich sequence in the loop, is required for Hfq action.
Shortening PolyU Tail of SgrS Impairs Hfq-Dependent Regulatory Function.
Next, we examined whether the terminator polyU tail of SgrS is involved in Hfq action. We constructed a series of sgrS mutants in which the terminator T stretch is sequentially deleted (Table S1 and Fig. 2A). Each of these plasmids was introduced in both hfq+ and hfq- cells. The expression and the ability of these SgrS variants to silence ptsG mRNA were tested by Northern blotting and by Western blotting (Fig. 2B). A complexity associated with the shortening of the polyT tail is that it affects transcription termination and the abundance of transcripts. The shortening of the polyT sequence to 7 had little effect on the silencing ability, although it moderately reduced the abundance of terminated transcript presumably due to a moderate transcriptional read-through (Fig. 2B, lane 3). When the polyU sequence was shortened to 5 or 4, a significant transcriptional read-through occurred and the silencing ability of SgrS was essentially eliminated (Fig. 2B, lanes 5 and 6). The shortening of the polyU tail to 6 caused an intermediate effect regarding transcription termination and silencing ability (Fig. 2B, lane 4). There are two possible explanations regarding why the polyU shortening leads to the reduction in ptsG mRNA silencing. One possibility is that SgrS variants are still active, but their levels are not sufficient for the regulation of ptsG mRNA. Alternatively, the polyU shortening itself impairs the Hfq-binding ability of SgrS or otherwise impairs SgrS activity. In other words, the long polyU tail is required for the Hfq action. The two scenarios are not mutually exclusive, and both could contribute to the loss of silencing ability. It should be noted that no significant reduction of both terminated and elongated RNAs generated from sgrS-5T and sgrS-4T mutants was observed in hfq- cells (for example, compare lanes 6 and 12), suggesting that SgrS variants with shortened polyU tails lose the ability to bind to Hfq.
Fig. 2.
Properties of SgrS variants possessing short polyU tails. (A) Nucleotide sequences of the rho-independent terminators of SgrS variants. The inverted repeat sequences are underlined. The sequences are based on the assumption that the transcription of the corresponding genes terminates at the last U within the polyU tail. (B) Effects of shortening of polyU tail on the expression and function of SgrS. IT1568 and TM589 cells harboring indicated plasmids were grown in LB medium. At A600 = 0.6, total RNAs were prepared and each RNA sample was subjected to Northern blot analysis using the SgrS probe 1 and the ptsG probe. tmRNA was used as a loading control. The following amounts of RNAs were loaded: SgrS variants, 0.25 μg; the ptsG mRNA, 7 μg; tmRNA, 0.25 μg. Total proteins were also prepared and samples equivalent to 0.025 A600 units were subjected to Western blot analysis using anti-IIB antibodies.
SgrS Variant with a Short PolyU Tail Loses Hfq-Dependent Silencing Function.
To examine whether SgrS variants with shortened polyU tails are still active or not, we tried to increase the expression level of SgrS variants. For this, we constructed sgrS-LS4T in which the inverted repeat sequence in the sgrS-4T terminator is extended by adding 4G and 4C residues (Table S1 and Fig. 3A). We expected that a longer GC-rich stem of terminator hairpin could help transcription termination even when the polyU stretch is shortened. As a control, sgrS-LS8T possessing eight consecutive T residues was also constructed (Table S1 and Fig. 3A). The expression and the silencing ability of transcripts were tested by Northern blotting and by Western blotting. As expected, transcriptional read-through of SgrS-LS4U was dramatically reduced and the terminated form of SgrS-LS4U was stably expressed in hfq+ cells (Fig. 3B, Upper, lane 3). Interestingly, the hfq mutation did not affect the expression levels of SgrS-LS4U and SgrS-LS8U (Fig. 3B, Upper, lanes 5 and 6), suggesting that extension of the terminator stem of SgrS not only overcomes the transcriptional read-through but also stabilizes the transcript even in the absence of Hfq. Then, we tested the ability of SgrS-LS4U and SgrS-LS8U to silence the ptsG mRNA. SgrS-LS8U exhibits the full activity to down-regulate the ptsG mRNA in hfq+ cells but not in hfq- cells (Fig. 3B, Middle and Lower, lanes 2 and 5), indicating that the extension of the terminator stem itself does not affect the Hfq-dependent silencing ability of SgrS. Importantly, SgrS-LS4U failed to down-regulate the ptsG mRNA even in hfq+ cells although it is fairly well expressed (Fig. 3B, Middle and Lower, lane 3). Taken together, we conclude that SgrS variants are no longer active regarding the Hfq-dependent silencing function when the polyU tail is shortened to 4.
Fig. 3.
Effects of the extension of the terminator stem on the properties of SgrS. (A) Nucleotide sequences around the rho-independent terminators of SgrS-LS4U and SgrS-LS8U. The bold letters represent the bases corresponding to the extended stem. The inverted repeat sequences are underlined. (B) Expression and function of SgrS-LS4U and SgrS-LS8U. T1568 and TM589 cells harboring indicated plasmids were grown in LB medium. At A600 = 0.6, total RNAs were prepared, and 0.25 or 7 μg of each RNA sample was subjected to Northern blot analysis using the SgrS probe 1 or the ptsG probe. Total proteins were also prepared and samples equivalent to 0.025 A600 units were subjected to Western blot analysis using anti-IIB antibodies.
SgrS Variants with a Short PolyU Tail Lose the Hfq-Binding Ability.
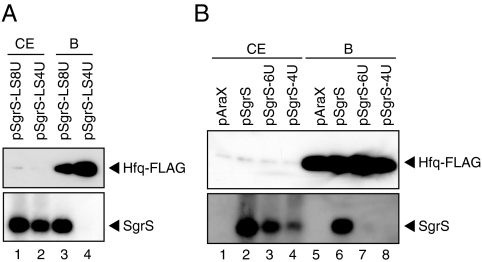
It is highly possible that SgrS variants with a short polyU tail are inactive because they lose Hfq-binding ability. To test directly the Hfq-binding ability of SgrS variants in vivo, we performed a pull-down assay using cells expressing Hfq-FLAG. Cell extracts were incubated with anti-FLAG M2-agarose beads. Proteins bound to the agarose beads were analyzed by Western blotting using anti-FLAG antibodies. The affinity-purified Hfq-FLAG was treated with phenol and subjected to Northern blotting. A significant amount of SgrS-LS8U but not SgrS-LS4U copurified Hfq-FLAG (Fig. 4A, lanes 3 and 4). We also performed the pull-down assay in cells expressing SgrS, SgrS-6U, or SgrS-4U. A significant amount of SgrS but not SgrS-4U copurified with Hfq-FLAG (Fig. 4B, lanes 6 and 8). A small amount of SgrS-6U copurified with Hfq-FLAG (Fig. 4B, lane 7). Thus, shortening the polyU tail of SgrS eliminates the Hfq-dependent silencing ability by impairing the Hfq-binding activity in vivo. We conclude that the long terminator polyU tail of SgrS is essential for Hfq binding.
Fig. 4.
Analysis of in vivo binding of SgrS variants to Hfq. TM771 (ΔsgrS hfq-FLAG-cat) cells harboring indicated plasmids were grown in LB medium to A600 = 0.6. Cell extracts were prepared and subjected to the pull-down assay using anti-FLAG agarose as described in Materials and Methods. Crude extracts (CE) (5 μL) and bound fractions (B) (5 μL) were analyzed by Western blotting using anti-Hfq antibodies. For analysis of RNAs, crude extracts (5 μL) and bound fractions (5 μL) were treated with phenol and subjected to Northern blotting using the SgrS probe 1.
SgrS Variants in Which the Terminator Is Replaced by Foreign Terminators Are Functional.
To test further the role of terminator polyU tail in Hfq action, we constructed additional sgrS mutants in which the terminator of sgrS is replaced by several rho-independent terminators derived from other genes. All of chimeric SgrS RNAs are well expressed in hfq+ cells, whereas the expression levels are markedly reduced in hfq- cells, suggesting that they are able to bind to Hfq (Fig. S2). Then, we tested the effect of SgrS variants on the expression of ptsG mRNA and IICBGlc. All of these SgrS variants led to an efficient translational inhibition and rapid degradation of ptsG mRNA in hfq+ but not in hfq- cells (Fig. S2). Thus, the chimeric SgrS RNAs retain the full activity to down-regulate the ptsG mRNA in an Hfq-dependent manner, indicating that any rho-independent terminators could support the Hfq action at least when the polyU tail is long enough.
Shortening of PolyU Tail of RyhB Impairs Hfq-Dependent Regulatory Function.
To test whether the polyU tail is required for Hfq binding and for riboregulation by other sRNAs, we have cloned the ryhB gene on pAraX. RyhB down-regulates the sodB mRNA encoding superoxide dismutase (13). The maximum length of the polyU tail of RyhB is 9 (Table S1 and Fig. 5A). Then, ryhB-4T in which the polyT tail was shortened to 4 was constructed (Table S1 and Fig. 5A). The plasmids carrying the ryhB or ryhB-4T gene were introduced in both hfq+ and hfq- cells. The ryhB gene produced significant levels of RyhB in hfq+ cells, whereas the expression level of RyhB-4U was significantly reduced (Fig. 5B, Upper, lanes 2 and 3). In addition, the expression levels of RyhB but not RyhB-4U are markedly reduced in hfq- cells (Fig. 5B, Upper, lanes 5 and 6), suggesting that RyhB-4U loses the Hfq-binding ability. Then, we tested the effect of expression of RyhB-4U on the target sodB mRNA. As expected, the full-length RyhB efficiently reduced the expression of sodB mRNA in hfq+ but not in hfq- cells (Fig. 5B, Lower, lanes 2 and 5). On the other hand, RyhB-4U was not able to silence the target mRNA even in an hfq+ background (Fig. 5B, Lower, lane 3). These data suggest that RyhB-4U loses the silencing ability by losing Hfq-binding ability. However, the expression level of RyhB-4U was markedly reduced compared to RyhB, raising again the possibility that RyhB-4U is active but its abundance is not sufficient to silence the sod mRNA.
Fig. 5.
Effects of polyU tail shortening of on the expression and function of RyhB. (A) Nucleotide sequences around the rho-independent terminators of RyhB and RyhB-4U. (B) Expression and function of RyhB and RyhB-4U. IT1568 and TM589 cells harboring indicated plasmids were grown in LB medium. At A600 = 0.6, total RNAs were prepared, and 1 or 5 μg of each RNA sample was subjected to Northern blot analysis using the RyhB and sodB probes. (C) Effect of overexpression of RyhB and RyhB-4U on the sodB mRNA. IT1568 and TM589 cells harboring indicated plasmids were grown in LB medium. At A600 = 0.6, total RNAs were prepared, and 1 or 5 μg of each RNA sample was subjected to Northern blot analysis using the RyhB and sodB probes. (D) Size analysis of RyhB and RyhB-4U. Total RNAs (5 μg) prepared from IT1568 cells harboring pRyhB (lane 2) or pT-RyhB-4U (lane 1) were fractionated on a 10% polyacrylamide/8M urea gel and was subjected to Northern blot analysis using the RyhB probe. RNA size markers are indicated on the left. (E) Binding of RyhB and RyhB-4U to Hfq. TM615 (hfq-FLAG-cat) cells harboring indicated plasmids were grown in LB medium to A600 = 0.6. Cell extracts were prepared and subjected to the pull-down assay using anti-FLAG agarose as described in Materials and Methods. Crude extracts (5 μL) and bound fractions (5 μL) were analyzed by Western blotting using anti-Hfq antibodies. For analysis of RNAs, crude extracts (5 μL) and bound fractions (5 μL) were treated with phenol and subjected to Northern blotting using the RyhB probe.
PolyU Tail of RyhB Is Essential for Hfq Binding and for Riboregulation.
To examine whether RyhB-4U is active or not, we tried to increase the expression level by constructing the multicopy plasmid pT-RyhB-4U carrying the ryhB-4T gene. As a control, plasmid pT-RyhB carrying the wild-type ryhB gene was also constructed. Each plasmid was introduced in both hfq+ and hfq- cells. The expression and the ability to silence sodB mRNA of RyhB and RyhB-4U were tested by Northern blotting. The abundance of RyhB was markedly elevated in cells harboring pT-RyhB resulting in an increased silencing of sodB mRNA (Fig. 3C, lane 2). On the other hand, RyhB-4U failed to down-regulate the sodB mRNA even in cells harboring pT-RyhB-4U in which the abundance of RyhB-4U was comparable to that of the wild-type RyhB expressed from pRyhB (Fig. 3C, compare lanes 3 and 4). This suggests that RyhB-4U is no longer active by losing its Hfq-binding ability. In fact, the pull-down assay demonstrated that RyhB but not RyhB-4U copurified with Hfq (Fig. 5E). We conclude that the terminator polyU tail of RyhB is essential for Hfq-binding and therefore for riboregulation. Furthermore, we tried to see a size difference between RyhB and RyhB-4U on a polyacrylamide gel because they are relatively short (RyhB is 95 nt in length when the transcription termination occurs at the last U within the polyU tail). Two RNAs were fractionated on a polyacrylamide/urea gel and the gel was subjected to Northern blotting (Fig. 5D). The data clearly indicate that RyhB-4U is shorter than RyhB on average, validating that the 4U construct has a shortened polyU tail.
PolyU Tail Is Required for Riboregulation by MicA and MicF.
We also constructed MicA-4U and MicF-4U and examined their properties. The full-length MicA and MicF, which have 5UC4U and 8U tail, respectively, silenced significantly the expression of target ompA and ompF mRNAs, whereas MicF-4U and MicA-4U were not able to silence the target mRNAs (Fig. S3). Thus, it is highly possible that the terminator polyU tail is essential for the regulatory function of Hfq-binding sRNAs in general.
Discussion
The RNA chaperon Hfq is essential for trans-acting sRNAs to regulate the translation and stability of target mRNAs in gram-negative bacteria such as E. coli and Salmonella. The primary role of Hfq is to facilitate base pairing between sRNAs and target mRNAs to regulate their translation, mostly negatively and also positively in some cases (2, 3). Another role of Hfq is to recruit RNase E near target mRNAs to destabilize sRNA-mRNA hybrids (14). In addition, Hfq is known to stabilize sRNAs by protecting from the attack of ribonucleases (1, 15). An important unsolved question concerning the mechanism of sRNA action is how Hfq promotes the base pairing. The identification of Hfq-binding sites on sRNAs is certainly crucial for understanding of the mechanism of Hfq action. Biochemical studies including RNase footprinting have demonstrated that Hfq binds preferentially to AU-rich sequences in several sRNAs such as OxyS (16), Spot42 (17), DsrA (18), RyhB (19), and RybB (20). The identified Hfq-binding sites are located in the internal portion of sRNA molecules near stem–loop structures. However, essentially no mutational study regarding the Hfq-binding sites has been reported even in these cases. Thus, it remains obscure whether or not the physically identified Hfq-binding sites really act as the functional Hfq-binding sites. In addition, little is known about the Hfq-binding site on many other sRNA including SgrS.
In this work, we tried to identify the functional Hfq-binding site primarily by mutational approach using SgrS as a model sRNA. A key finding in the present study is that the shortening terminator polyU tail of SgrS dramatically reduces the ability to down-regulate ptsG mRNA (Fig. 2). A complication in interpreting these data was that the shortening of the polyU tail severely reduced the abundance of SgrS variants. By extending the terminator stem, we were able to show that the shortening of the polyU tail up to 4 completely eliminates the ability of SgrS to silence ptsG mRNA (Fig. 3). Then, we showed by pull-down assay that the shortening of polyU tail up to 4 completely eliminates the Hfq-binding ability of SgrS (Fig. 4). Thus, we conclude that the terminator polyU tail of SgrS is essential for Hfq action. Moreover, we found that shortening of the terminator polyU in several other sRNAs also eliminated their abilities to bind to Hfq and to regulate the target mRNAs (Fig. 5 and Fig. S3). Taken together, we conclude that the polyU tail of the rho-independent terminator of sRNAs is essential for Hfq action in general. In fact, the rho-independent terminators of genes encoding Hfq-binding sRNAs possess more than seven consecutive T residues in many cases (Table S2).
We showed that the polyU tail is indeed shortened in the RyhB-4U construct compared to RyhB (Fig. 5D), although we have not determined yet the actual 3′ ends of sRNAs. The 3′ end of any RNAs including sRNA is expected to be heterogeneous due to several reasons. First, transcription termination occurs within the T stretches of the rho-independent terminator resulting in transcripts with different 3′ ends (21). Transcriptional read-through, and exonucleolytic processing (22) and/or polyadenylation of transcripts (23) would also contribute to the 3′ end heterogeneity. In this regard, it is interesting to note that polynucleotide phosphorylase has been shown to be involved in sRNA function (24). The determination of the 3′ ends of sRNA and its variants in different genetic backgrounds should be useful to clarify the actual 3′ end heterogeneity and to understand how the 3′ end of sRNA is generated.
The finding that the terminator polyU tail of sRNAs is essential for Hfq binding and for riboregulation strongly suggests that the polyU tail acts as a critical Hfq-binding site. This view is consistent with previous reports that Hfq binds preferentially to AU-rich sequences, in particular polyU, near hairpin structures (16–19). However, it remains to be seen whether the polyU tail acts as a direct target of Hfq binding or it acts indirectly to support the sRNA-Hfq interaction by some other ways. Another important question is whether the terminator polyU tail alone is sufficient for the functional Hfq-sRNA interaction. Considering that the known Hfq-binding sites are located in the internal of sRNA molecules in several sRNAs, it is more likely that both the terminator polyU tail and an internal AU-rich sequence are necessary for Hfq action. In fact, two additional AU-rich sequences (UAUU from nucleotide 179 to 182 and UAAAAU from nucleotide 187 to 192 exist in the 3′ internal the spacer region of SgrS). It is certainly interesting to examine whether these internal AU-rich sequences are required for Hfq to stably bind to SgrS. Alternatively, the polyU tail may act as an entry site for Hfq, and then Hfq can move to an internal site. In this regard, it is interesting to note that there exist mutations in an internal AU-rich sequence near the stem–loop structure of sRNA ChiX (25). These mutations destabilize ChiX and abolish its regulatory function, although this AU-rich sequence has not been proved to be an Hfq-binding site. In any case, further biochemical and mutational studies are needed to clarify whether Hfq binds directly to the terminator polyU tail and how the terminator polyU tail contributes to the functional Hfq-sRNA interaction. Likewise, it is interesting to examine whether the known internal Hfq-binding sites on several sRNAs are required for the functional Hfq-sRNA interaction. Experiments to address these important questions are in progress.
Finally, the present study suggests a unique view regarding how Hfq binding leads to the stabilization of sRNAs. It is well known that the expression level of Hfq-binding sRNAs is markedly reduced in an hfq- background because sRNAs are stabilized by Hfq (1, 15). It is believed that the Hfq binding stabilizes sRNAs by blocking the attack of RNase E at least in some sRNAs (15). Interestingly, SgrS no longer requires Hfq for stabilization when the terminator hairpin is intrinsically stabilized by extending the stem (Fig. 4). Although it is possible that the extension of the stem somehow blocks SgrS from the attack of RNase E, it is reasonable to assume that the Hfq binding to the polyU tail contributes to the stabilization of sRNAs by impeding the 3′-exonuclease attack at least in SgrS. In any case, how the Hfq binding stabilizes sRNAs is another intriguing issue to be studied.
The rho-independent terminator is widely associated with many mRNAs and stable RNAs (21, 26). The major role of the terminator polyU tail is surely to act as a punctuation signal of transcription termination. The present study not only has unveiled a hidden important regulatory role of the polyU tail of rho-independent terminator of sRNAs, but it also raises the possibility that the terminator polyU tail along with Hfq plays additional regulatory roles in RNA metabolism beyond transcription termination in general. In this regard, it is interesting to note that Hfq was shown to stimulate polyadenylation of the lpp mRNA at its 3′ end by polyA polymerase through preferential binding to the polyU tail of rho-independent terminator (23).
Materials and Methods
Bacterial Strains and Plasmids.
The E. coli K12 strains and plasmids used in this study are listed in Table S3. IT1568 (W3110 mlc) was used as a parent wild-type strain. To construct TM771, the hfq-FLAG-cat allele of TM615 (14) was moved to TM542 (27) by P1 transduction. Details of construction of plasmids are presented in SI Materials and Methods.
Northern Blotting.
Cells carrying the sRNA expressing plasmids were grown at 37 °C in LB medium supplemented with 0.4% arabinose and antibiotics to midlog phase. Total RNAs were isolated as described (28). RNA samples were resolved mostly by 1.2% agarose gel electrophoresis in the presence of formaldehyde and blotted on to Hybond-N+ membrane (Amersham Biosciences). The RNAs were visualized using digoxigenin reagents and kits for nonradioactive nucleic acid labeling and detection system (Roche Molecular Biochemicals) according to the procedure specified by the manufacturer. The DNA probes used are described in SI Materials and Methods.
Western Blotting.
Cells carrying the sRNA expressing plasmids were grown at 37 °C in LB medium supplemented with 1.0% arabinose and antibiotics to midlog phase. The cultures (0.5 mL) were centrifuged and the cell pellets were suspended in 100 μL of SDS-PAGE loading buffer (62.5 mM Tris-HCl at pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, and 0.1% bromophenol blue). The sample was heated at 100 °C for 5 min and subjected to a polyacrylamide-0.1% SDS gel electrophoresis and transferred to Immobilon membrane (Millipore). The 12% and 15% polyacrylamide gels were used to fractionate IICBGlc and Hfq, respectively. The membranes were treated either with anti-FLAG monoclonal antibody (Sigma), or anti-IIBGlc polyclonal antibodies (29). Signals were visualized by the Lumi-Light Western Blotting Substrate (Roche).
Pull-Down Assay.
Cells were grown in 200 mL of LB medium to A600 of 0.6, harvested, and washed with 10 mL of STE buffer (100 mM NaCl, 10 mM Tris-HCl pH 8.0, and 1 mM EDTA). The cell pellets were suspended in ice cold 10 mL of IP buffer (20 mM Tris-HCl pH 8.0, 0.1 M KCl, 5 mM MgCl2, 10% glycerol, and 0.1% Tween20). The cell suspension was sonicated and centrifuged at 10,000 × g for 30 min at 4 °C. The supernatant (crude extract) was incubated with 50 μL of anti-FLAG M2-agarose suspension (Sigma) for 30 min at 4 °C. The mixture was filtered by using a mini chromatography column (Bio-Rad). The agarose beads were washed by 10 mL of IP buffer 2 times. The proteins bound to the beads were eluted with 50 μL of IP buffer containing 0.4 mg/mL FLAG peptide (Sigma) and used as bound fraction (B). The samples were analyzed by Western blotting. To analyze RNAs, the crude extract (10 μL) and the bound fraction (10 μL) were treated with phenol, precipitated, and washed with ethanol. Each precipitate was dissolved in 10 μL of RNA buffer (0.02 M sodium acetate, pH 5.5, 0.5% SDS, and 1 mM EDTA). Five microliters of RNA sample were subjected to Northern blotting.
Supplementary Material
Acknowledgments.
We thank Susan Gottesman for comments on the manuscript. This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to H.A. and T.M) and Uehara Memorial Foundation (to T.M.).
Note.
A related paper by Sauer and Weichenrieder in this issue of PNAS entitled “Structural basis for RNA 3′-end recognition by Hfq” complements the current findings by showing that Hfq binds strongly U-rich RNA 3′ ends in vitro.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107050108/-/DCSupplemental.
References
- 1.Gottesman S. The small RNA regulators of Escherichia coli: Roles and mechanisms. Annu Rev Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 2.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gottesman S, Storz G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2010;1(a003798):1–16. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abe H, Aiba H. Differential contributions of two elements of rho-independent terminator to transcription termination and mRNA stabilization. Biochimie. 1996;78:1035–1042. doi: 10.1016/s0300-9084(97)86727-2. [DOI] [PubMed] [Google Scholar]
- 5.Aiba H, Hanamura A, Yamano H. Transcriptional terminator is a positive regulatory element in the expression of the Escherichia coli crp gene. J Biol Chem. 1991;266:1721–1727. [PubMed] [Google Scholar]
- 6.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr Opin Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: A key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 8.Vanderpool CK. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr Opin Microbiol. 2007;10:146–151. doi: 10.1016/j.mib.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 9.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 11.Maki K, Morita T, Otaka H, Aiba H. A minimal base-pairing region of a bacterial small RNA SgrS required for translational repression of ptsG mRNA. Mol Microbiol. 2010;76:782–792. doi: 10.1111/j.1365-2958.2010.07141.x. [DOI] [PubMed] [Google Scholar]
- 12.Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci USA. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 17.Moller T, et al. Hfq: A bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 18.Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geissmann TA, Touati D. Hfq, a new chaperoning role: Binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balbonti R, et al. Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol Microbiol. 2010;78:380–394. doi: 10.1111/j.1365-2958.2010.07342.x. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds R, Bermudez-Cruz RM, Chamberlin MJ. Parameters affecting transcription termination by Escherichia coli RNA polymerase. I. Analysis of 13 rho-independent terminators. J Mol Biol. 1992;224:31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- 22.Abe H, Abo T, Aiba H. Regulation of intrinsic terminator by translation in Escherichia coli: Transcription termination at a distance downstream. Genes Cells. 1999;4:87–97. doi: 10.1046/j.1365-2443.1999.00246.x. [DOI] [PubMed] [Google Scholar]
- 23.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 24.De Lay N, Gottesman S. Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA. 2011;17:1172–1189. doi: 10.1261/rna.2531211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figueroa-Bossi N, et al. Caught at its own game: Regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009;23:2004–2015. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.d’Aubenton Carafa Y, Brody E, Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 27.Kawamoto H, et al. Implication of membrane localization of target mRNA in the action of a small RNA: Mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev. 2005;19:328–338. doi: 10.1101/gad.1270605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 29.Tanaka Y, Kimata K, Aiba H. A novel regulatory role of glucose transporter of Escherichia coli: Membrane sequestration of a global repressor Mlc. EMBO J. 2000;19:5344–5352. doi: 10.1093/emboj/19.20.5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.