Abstract
Adaptive variation in craniofacial structure contributes to resource specialization and speciation, but the genetic loci that underlie craniofacial adaptation remain unknown. Here we show that alleles of the hedgehog pathway receptor Patched1 (Ptch1) gene are responsible for adaptive variation in the shape of the lower jaw both within and among genera of Lake Malawi cichlid fish. The evolutionarily derived allele of Ptch1 reduces the length of the retroarticular (RA) process of the lower jaw, a change predicted to increase speed of jaw rotation for improved suction-feeding. The alternate allele is associated with a longer RA and a more robustly mineralized jaw, typical of species that use a biting mode of feeding. Genera with the most divergent feeding morphologies are nearly fixed for different Ptch1 alleles, whereas species with intermediate morphologies still segregate variation at Ptch1. Thus, the same alleles that help to define macroevolutionary divergence among genera also contribute to microevolutionary fine-tuning of adaptive traits within some species. Variability of craniofacial morphology mediated by Ptch1 polymorphism has likely contributed to niche partitioning and ecological speciation of these fishes.
Keywords: adaptive radiation, development, genetics, quantitative trait loci, trophic morphology
The astounding diversity of vertebrate craniofacial morphology reflects adaptation to a wide variety of resources and environments. Evolution of craniofacial structure has been key to niche specialization and speciation in vertebrates (1), most famously exemplified by the varied beak morphology of Darwin's finches (2). The radiation of finches demonstrates both species-defining craniofacial divergence (2) and rapid adaptation of craniofacial morphology within species over the course of a few years in response to changes in resource availability (3). Although shifts in the expression of key craniofacial genes have been correlated with differences in trophic morphology among species of birds and cichlid fishes (4–8), the specific genetic loci producing these differences remain uncharacterized. As a result, it is not clear whether interspecific divergence and intraspecific adaptation share the same molecular genetic basis.
Diversification of trophic morphology in the adaptive radiation of cichlids in Lake Malawi (East Africa) is also correlated with specialized modes of feeding and resource partitioning and has likely contributed to their rapid speciation (9, 10). Because craniofacial structure arises from a complex and dynamic developmental program with both pleiotropic and modular effects (11), the expectation is that it should evolve via continuous fine-tuning steps. In accordance with this prediction, we previously demonstrated that morphological differences between closely related cichlid genera are the result of directional selection on numerous genetic loci of small to moderate effect (8, 12). These studies identified numerous quantitative trait loci (QTL) for craniofacial morphology segregrating in the F2 hybrid progeny of the genera Labeotropheus and Metriaclima (8, 12). Although Labeotropheus and Metriaclima are closely related rock-dwelling cichlid genera, they are divergent in craniofacial morphology and microhabitat utilization (10, 13). Labeotropheus is a specialist algal scraper with robust jaws adapted for biting, whereas Metriaclima is a generalist with gracile jaws better adapted for suction feeding (Fig. 1 A and B). These two genera represent opposite ends of the benthic/limnetic ecomorphological continuum that characterizes East African cichlid radiations (10) as well as many other notable divergences among teleosts at both the population and species level (14–19). Identifying the molecular genetic basis for these ecomorphologic shifts therefore has the potential to inform a more comprehensive understanding of teleost diversity in general.
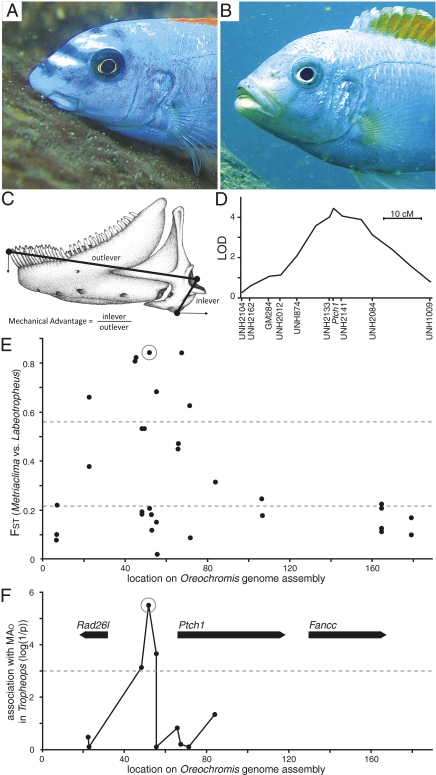
Fig. 1.
Ptch1 locus characterization in families, genera, and populations. Craniofacial appearance of (A) Labeotropheus trewavasae (image courtesy of Justin Marshall) and (B) Metriaclima mbenjii. (C) Lower jaw phenotype measures; RA and inlever length is equivalent. (D) QTL mapping interval for MAO on cichlid LG12. (E) Population differentiation (FST) between Labeotropheus and Metriaclima (n = 24 each) for SNPs at the Ptch1 locus. Dashed lines indicate two experimental measures of mean FST; the lower line from global comparison of Labeotropheus vs. Metriaclima across many populations (20), and the upper line from comparisons of randomized Labeotropheus and Metriaclima population pairs from distinct sites (48). (F) Significance of association between SNPs at the Ptch1 locus and MAO in the genus Tropheops (Wald test, n = 59); dashed line indicates a P value of 0.001. Genes in the region annotated with bold arrows. The SNP used to indicate long and short alleles of Ptch1 is circled in E and F.
A recent genome scan study identified single nucleotide polymorphisms (SNPs) exhibiting unusually high differentiation (FST) between Labeotropheus and Metriaclima (20). The high FST of these SNPs suggests they may be linked to genetic loci associated with evolutionary divergence between the two genera. Here we compared the genomic locations of the high FST SNPs and previously identified QTL (8, 12) as a starting point to identify the specific genes contributing to morphological divergence between the genera. We pinpoint alleles of the Ptch1 gene as contributing to morphological divergence between the two genera and further show that the same alleles of Ptch1 segregate within other species, producing continuing adaptive effects on craniofacial structure.
Results and Discussion
In a genome scan comparison of Labeotropheus and Metriaclima (20), SNP Aln100281_1741 had an FST of 0.86 between the two genera, and a comparative BLAST search placed the SNP in the first intron of the Ptch1 homolog. We identified a sequence scaffold containing Aln100281 and Ptch1 from the draft genome assembly of another cichlid fish, the Nile tilapia (Oreochromis niloticus). With additional comparative genomics we placed this scaffold on linkage group (LG) 12 of the cichlid genetic map, within a previously identified QTL interval associated with 8.6% of variance in length of the retroarticular process (RA) (8). This QTL was one of five identified for RA length (8). Functionally the inlever for the action of lower jaw depression (Fig. 1C), RA length modulates the functional tradeoff between force and velocity during jaw rotation (21, 22). No association was found between the LG12 QTL and outlever length of the lower jaw (8). Shortening of the RA while outlever length remains uniform is predicted to increase the speed of jaw opening, and the LG12 QTL also explains 11.3% of the functional phenotype, the mechanical advantage of jaw opening (MAO; Fig. 1C). We refer to the Labeotropheus genotype at the LG12 locus as the “long”-RA allele, and that of Metriaclima as the “short”-RA allele, to reflect the morphological phenotypes associated with each. The long allele acts dominantly to increase RA length and MAO (8).
Population Genetic Mapping of the Causative Interval.
We developed additional markers across the Ptch1 genome scaffold by resequencing a panel of Malawi cichlid DNA to identify polymorphisms (Table S1). Genotyping the F2 mapping family for a marker adjacent to Ptch1 confirmed its location on LG12 and centers the QTL interval squarely over this locus (Fig. 1D). We then used a DNA panel consisting of four Labeotropheus and four Metriaclima wild-caught populations to characterize patterns of sequence divergence in the region (Table S2). The SNP Aln100281_1741 continued to show high FST between genera in the populations included in the panel. Several markers located as far as 20 kb upstream of Ptch1 also exhibited extremely high FST, approaching alternate fixation (Fig. 1E). The consistency of genetic divergence between Labeotropheus and Metriaclima across species and populations suggests that variation in this region underlies a fundamental phenotypic difference between the two genera.
Although genetic differentiation at Ptch1 is coincident with the QTL for RA length, the Labeotropheus and Metriaclima panel does not allow correlation of genotype to a specific phenotype within populations because alternate alleles are essentially fixed within each genus. However, these alleles are segregating within populations of a third genus, Tropheops, which exhibits a range of trophic morphology roughly intermediate between Metriaclima and Labeotropheus (23, 24). Thus, we were able to directly measure association between individual MAO phenotypes and markers across the high FST region, using a panel of four natural populations of Tropheops (Table S2). The association peaks at an SNP roughly 15 kb upstream of Ptch1 and falls below significance before the Ptch1 start codon (Fig. 1F). The genotype–phenotype association within and among Tropheops species provides strong evidence that the Ptch1 locus is modulating RA length and MAO and narrows the interval of interest to the region upstream of the gene (Figs. 1F and 2). The upstream SNPs serve to label long and short alleles at the locus; as in the F2 mapping family, the long allele seems to be dominant over the short allele in natural Tropheops populations (Fig. 2).
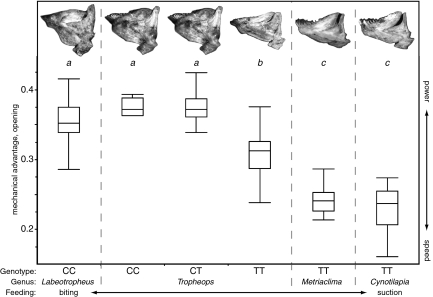
Fig. 2.
Morphology and function of the lower jaw by genus and Ptch1 genotype. MAO values of wild individuals (Table S2) indicated by box-plot, with letters above designating statistical groups by one-way ANOVA and Tukey's honest significance test (P < 0.05). Lower jaw images from representative individuals at top; each jaw has an MAO matching the mean of the corresponding genus and genotype. Genotype refers to high-association SNP (circled in Fig. 1 E and F). Box boundaries indicate quartiles, and whiskers indicate range of MAO data.
Ptch1 Expression Differences Correlate with Developmental Divergence.
We did not find any nonsynonymous polymorphisms in Ptch1 coding sequence. The lack of structural differences in the protein is not surprising given the fundamental importance of Ptch1 to a variety of developmental processes (25). Because signatures of genetic divergence immediately upstream of Ptch1 suggest adaptation via cis-regulatory elements, we examined Ptch1 expression by in situ hybridization. Dimensionality of the lower jaw is determined early, and in comparisons of Labeotropheus and Metriaclima differences in the shape of the lower jaw precursor (Meckel's cartilage) are evident as early as 5 d postfertilization (dpf), stage 10 (8, 26). By the following day (stage 12) many of the bones that constitute the feeding apparatus have begun to condense, and at this early stage we observed discrete nodes of Ptch1 expression in areas where bone development has or will be initiated (Fig. 3 B–D and F–H). Notably, species exhibit differences in Ptch1 expression that correlate with biting and suction feeding jaw morphologies. In the long RA species Labeotropheus fuelleborni (LF) there is marked expression of Ptch1 surrounding the lower jaw at this stage, with nodes of robust expression in or adjacent to areas of skeletal differentiation, including the regions where the dentary and RA will form (Fig. 3 B and D). In contrast, levels of Ptch1 are much lower in the short RA species Metriaclima zebra (MZ) in the same context (Fig. 3 F and H). Unlike expression at the developing lower jaw, levels of Ptch1 expression are qualitatively similar between LF and MZ in the developing fin-ray elements of the tail (Fig. 3 C and G).
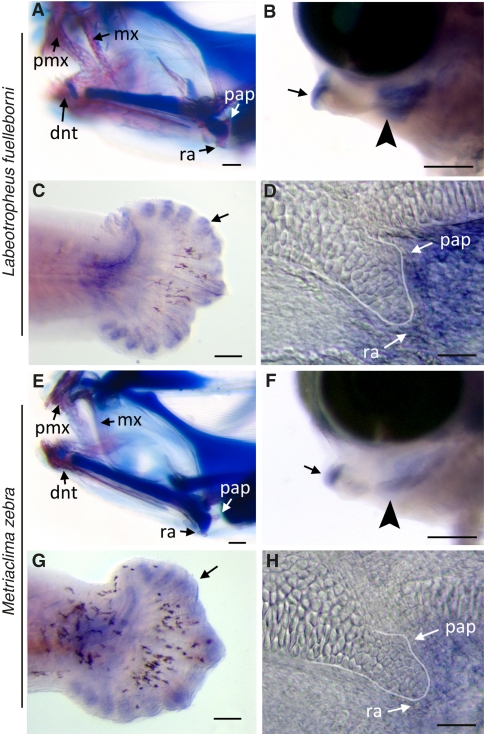
Fig. 3.
Interspecific differences in Ptch1 expression during cichlid craniofacial development. Craniofacial outcomes in (A) LF and (E) MZ larvae at 13 dpf; LF larvae exhibit accelerated bone (pink) development compared with MZ; cartilage stains blue. Ptch1 in situ labeling in representative 6-dpf (stage 12) (B–D) LF and (F–H) MZ larvae: (B and F) Lateral whole-mount view of lower jaw with nodes of Ptch1 expression, particularly in mesenchymal cells at the distal (arrow) and proximal (arrowhead) ends of the lower jaw precursor where the dentary and RA process will form, respectively. (C and G) Ptch1 labeling is qualitatively similar in developing fin-ray elements of tail (arrows). (D and H) Flat-mount preparation of the jaw joint in lateral view, showing the cartilaginous precursor of the RA process (outlined) relative to node of Ptch1 expression. dnt, dentary; mx, maxilla; pap, posterior articulation process; pmx, premaxilla. (Scale bars, 200 μm in A, B, E, and F; 100 μm in C and G; and 10 μm in D and H.)
To more accurately describe regional differences in levels of expression of Ptch1, as well as its downstream target Gli1, we performed a pixel density analysis on stage-matched LF and MZ larvae as previously described (27, 28). We find that a significantly greater proportion of pixels were labeled in the jaws of LF compared with MZ for both Ptch1 and Gli1 (Fig. S1 and Table S3). No difference was observed for either factor in the caudal fin (Fig. S1 and Table S3). Additionally, a quantitative PCR assay for Ptch1 transcript in microdissected tissues from 6.5-dpf larvae demonstrated higher levels of Ptch1 transcript in the lower jaw of LF relative to MZ, but there was no difference in Ptch1 transcript levels in the trunk and tail by species (Table S4). Together these data strongly support the assertion that PTCH1-mediated hedgehog signaling is up-regulated in the jaws of LF compared with MZ, specifically at the time and within the area where the RA develops.
Manipulation of Hedgehog Pathway Signaling Affects Bone Development and Recapitulates Natural Interspecific Variation in Bone Shape.
Our expression results are consistent with recent work demonstrating a role for hedgehog signaling in dermal bone development (29, 30) and predict that modulation of this pathway can lead to variation in jaw shape. To test this prediction we treated LF larvae with cyclopamine, an antagonist of hedgehog pathway signaling (31), at a critical stage of craniofacial bone development and Ptch1 expression (stage 12). During normal development, both Ptch1 and its downstream target Gli1 are expressed in areas adjacent to or coincident with the bone differentiation marker Col1a1 (Fig. S1 A–L). In treated larvae, we find that expression of both Ptch1 and Gli1 is dramatically and globally reduced (Fig. S1 P–W and Table S3). We also find that bone development is delayed in areas where Ptch1 is normally expressed (Fig. S1 X–AA and Table S3). Specifically, less Col1a1 expression was observed around the developing RA, whereas more cells were expressing Col1a1 around the branchiostegal rays and within the caudal fin (Fig. S1 Y–AA and Table S3), suggesting that these structures were in a more undifferentiated state after cyclopamine treatment relative to control animals. Expression of Col1a1 around the dentary was relatively unaffected by cyclopamine treatment (Fig. S1X); however, this is likely because development of this structure was well underway at the stage when animals were treated. Overall, these patterns of expression are consistent with the phenotypic outcome of cyclopamine treatment. Specifically, we find that cyclopamine-treated larvae exhibit significantly reduced RA length (and MAO), whereas the dentary and overall length of the lower jaw, which is determined primarily by outgrowth of Meckel's cartilage that occurs earlier than the stages examined here, remains roughly the same (Fig. 4 A–C and Table S5). In terms of gene expression patterns (Fig. S1 and Table S3), relative bone development (Fig. S1 and Table S3), and MAO (Fig. 4 A–C and Table S5), cyclopamine-treated LF recapitulate a suction-feeding, MZ-like lower jaw phenotype. Although rates of overall development do not differ between LF and MZ during early larval stages, we suggest that key differences in lower jaw shape are due at least in part to PTCH1-mediated differences in the rate of bone development within the RA. The exact cellular mechanism(s) through which hedgehog signaling influences differences in bone development remain to be characterized.
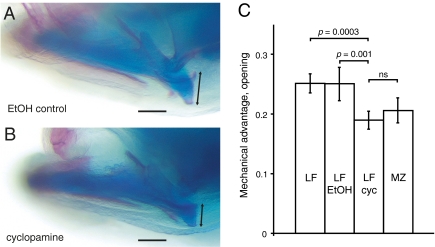
Fig. 4.
Treatment of biting species larvae with a hedgehog pathway inhibitor recapitulates a suction-feeding jaw phenotype. LF larvae were treated with (A) 0.5% ethanol (EtOH control, n = 7) or (B) 50 μM cyclopamine (cyc, n = 10) for 6 h at stage 12; RA length measured at 12 dpf (black arrow in A and B). (C) Relative to the length of the lower jaw (measured as the distance between the center of the jaw joint and the tip of the dentary), RA length (measured as the distance from the posterior tip of the posterior articulation process to the ventral tip of the retroarticular) was significantly reduced in cyclopamine treatment group compared with the control LF group but was not significantly different from MZ control larvae (n = 7). Larvae treated with ethanol were indistinguishable from untreated siblings (n = 6). P values, one-way ANOVA. (Scale bar, 100 μm.)
We also noted that treated animals exhibited bifurcated expression of Col1a1 within the developing branchiostegal rays (asterisk in Fig. S1Z), as well as aberrant expression within the dermal fin ray elements of the caudal fin (arrow in Fig. S1AA). These patterns are also consistent with craniofacial defects after cyclopamine treatment, in which fusion of the branchiostegal rays and truncation of the caudal fin ray elements were observed. These data extend previously documented roles for hedgehog signaling in dermal bone development (29, 30) and suggest that this pathway plays an important role in polarizing dermal bone development along a proximal–distal axis. It is well established that hedgehog signaling is critical for the proper patterning and polarization of several organs in various animal taxa (32–37), but here a similar role has been documented for bone development. The extent to which hedgehog-mediated outgrowth of other dermal bones (e.g., RA) has influenced species-specific differences in craniofacial shape would be a fruitful area of future investigation.
Our data suggest the presence of at least two functionally distinct alleles of Ptch1 that produce different levels of transcript in a spatially and temporally restricted manner. The long allele produces relatively higher levels of Ptch1, resulting in greater bone deposition, a longer RA, and higher MAO. The short allele has relatively lower expression, resulting in less bone and a shorter RA that increases jaw opening speed. Although the association between Ptch1 expression and bone deposition is interesting, we find no significant association between Ptch1 genotype and length or thickness of the jaw, suggesting that differences in developmental outcomes of the Ptch1 alleles are largely restricted to the RA. Whether the early effects of Ptch1-mediated bone deposition represent an adaptation for the timing and/or rate of bone development among cichlids remains an open question. Certainly the results of our expression and cyclopamine experiments support a role for hedgehog signaling in modulating bone development.
Ptch1 Alleles Contribute to Microevolutionary and Macroevolutionary Processes.
Using the SNP in highest association with MAO in Tropheops, we genotyped the Ptch1 locus in additional wild-caught individuals of four genera (Fig. 2). The significant correlation to MAO phenotype remains consistent across all taxa measured, suggesting extremely tight linkage of the SNP with the causative polymorphism. Obligate biters (Labeotropheus) and suction feeders (Cynotilapia and Metriaclima) are nearly fixed for long and short alleles, respectively. The genus Tropheops is particularly interesting, because segregating Ptch1 alleles contribute to “shallow” and “deep” ecomorphs that correlate with resource use and depth distribution (24, 38). In deep ecomorph Tropheops species, the short Ptch1 allele seems to be fixed, contributing to a morphology adapted to sifting and suction feeding of loose material. In the Tropheops populations examined from shallow water, the two alleles segregate and are associated with different morphologies within species. Individuals carrying a long allele at Ptch1 have a jaw morphology similar to that of Labeotropheus, well adapted for feeding on attached algae found at shallow depth in the lake. Individuals homozygous for the short allele of Ptch1 have a trophic morphology that more closely resembles that of the deep ecomorphs, better suited for sifting and suction feeding on loose material. Thus, allelic variation at the Ptch1 locus is associated with the continuum in lower jaw morphology both within and among Tropheops species. Although these data are consistent with recent work showing that early Sonic hedgehog signaling from the embryonic forebrain can lead to continuous craniofacial variation in mouse embryos (39) and jaw width differences in the Mexican tetra (40), our results posit a unique role for hedgehog signaling in regulating fine-scale, continuous differences in feeding morphology via bone deposition.
Overtly, the genus Labeotropheus has a highly specialized trophic apparatus, whereas the genus Metriaclima has a generalist morphology, more similar to that of the ancestral lineages that invaded Lake Malawi (10). It is therefore surprising that, according to a Ptch1 gene phylogeny including other African cichlid genera from Lake Malawi and beyond (Fig. S2), the short Ptch1 allele fixed in Metriaclima is more recently derived, whereas Labeotropheus carries the ancestral allele. The SNP haplotype that labels the short allele is found in only a subset of the rock-dwelling cichlids in Malawi. The short allele also shows signatures of recent selection, including relatively higher extended haplotype homozygosity, reduced nucleotide diversity, and significantly low values for Tajima's D (Fig. S3). These data suggest that selection has acted recently to fine-tune the RA to improve suction feeding within the rock-dwelling cichlids.
East African rift-valley cichlid radiations are characterized by rapid and repeated bentho-pelagic divergence following the colonization of a lacustrine environment by riverine ancestors (10). In Lake Malawi this divergence has led to the establishment of the genetically distinct rock- and sand-dwelling clades (41, 42). On average, rock-dwelling species possess trophic morphologies consistent with a benthic lifestyle, whereas most sand-dwellers are pelagic in terms of their feeding ecology (10). We propose that, after an initial colonization of the rock niche by cichlids with specialized biting morphologies, selection for the short allele of Ptch1 contributed to the reevolution of an ancestral-like feeding strategy and concomitant expansion of the rock-dwelling niche into the water column.
Despite the major and conserved roles the hedgehog pathway plays throughout development, we show that it continues to evolve to produce adaptive changes in craniofacial phenotype. Notably, we demonstrate that the same Ptch1 alleles underlying macroevolutionary divergence between genera also form the basis of microevolutionary adaptation among species and even within populations. Where Ptch1 alleles segregate, a range of ecologically relevant morphologies are produced within populations. Importantly, the resources used by segregating ecomorphs are distributed by depth, providing an opportunity for spatial segregation of individuals by genotype within populations, possibly contributing to character displacement and even the development of incipient species (43). Thus, diversification via adaptive polymorphism could support iterative niche partitioning in complex communities. Additionally, such adaptive polymorphism segregating within populations would permit rapid morphological evolution to take place in response to sudden environmental change. Segregating fine-tuning alleles of the kind we describe here have likely been a key component in both adaptation and speciation in the ongoing explosive radiation of Lake Malawi cichlids.
Materials and Methods
Animals.
Fish for breeding and association studies were collected directly from Lake Malawi. Fish were maintained and euthanized according to Institutional Animal Care and Use Committee guidelines. Larval fish were obtained by natural matings within facilities at Syracuse University and the University of Maryland, with developmental staging based on de Jong et al. (26). A hybrid cross between LF and MZ generated an F2 mapping population (n = 173), which was phenotyped for jaw morphology and used as the basis of QTL analysis as previously described (8, 12).
Genetic Analysis.
Draft sequence of the O. niloticus genome was assembled from Illumina sequence reads totaling 60× sequence coverage. The assembly has an N50 scaffold size of 138 kb and was aligned to stickleback genome sequence using BLAST (available at http://BouillaBase.org). Shotgun genome sequence of Malawi cichlid species (20) was aligned to the Oreochromis genome assembly using BLASTN. Other polymorphisms were identified (Table S1) and genotyped by resequencing aligned sequences using Applied Biosystems Big Dye Terminator chemistry and an ABI3730 sequencer. The marker Ptch1Loc10_SNP1 was added to the genetic map as “Ptch1” by genotyping the original F2 mapping family (12); the resulting map was used for QTL analysis. Population genetic and association analyses were performed in DnaSP and PLINK, respectively (44, 45), using polymorphisms with a minor allele frequency >0.05.
In Situ Hybridization.
Whole-mount in situ hybridization (WISH) was performed as previously described (8). Embryos used for the reported experiments were raised, staged, fixed, and processed in parallel. Sense and antisense Ptch1 riboprobes were made from cichlid clones (sequence identical for LF, accession no. JN037690, and MZ, accession no. JN037691), corresponding to exons 1–7 and 7–17. These yielded identical WISH results; data derived from the exons 1–7 riboprobe are reported. Accession numbers for Gli1 and Col1a1 probes are JN037689 and JN116727, respectively. To facilitate probe penetration, alkaline hydrolysis was used to fragment probes to ≈500 bp. A solution of 20 mL RNA probe, 12 mL H20, 4 mL 0.4 M sodium bicarbonate, and 4 mL 0.6 M sodium carbonate was incubated at 60 °C for a period based on the following: time (min) = (starting kb − desired kb)/(0.11 × starting kb × desired kb).
Pharmacological Treatment.
On the basis of previously published work in cichlids, we used cyclopamine (LC Laboratories) at a working concentration of 50 μM (46). Cyclopamine (10 mM, in ethanol) or ethanol alone (control) was added to larval fish water. Animals were treated for 6 h in the dark because cyclopamine is light sensitive. After treatment, larvae were washed several times in fish water before returning to circulating culture flasks to be grown out to 12 dpf. Cartilage and bone staining was performed as previously described (47). Larvae were photographed with a Zeiss Axiocam digital imaging system mounted to an M2 Bio stereomicroscope (Zeiss). Image processing and pixel density analysis was performed in Adobe Photoshop CS4 (Fig. S4).
Supplementary Material
Acknowledgments
We thank Matthew Conte for bioinformatics support; Claire O'Quin and Jennifer Ser for fish husbandry support; Rolf Karlstrom for advice on cyclopamine treatment in fish; Justin Marshall for the Labeotropheus photograph used in Fig. 1; and J. Todd Streelman, William Jeffery, Karen Carleton, and Kelly O'Quin for their comments on previous drafts of the manuscript.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JN037665–JN037693 and JN116727).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018456108/-/DCSupplemental.
References
- 1.Gans C, Northcutt RG. Neural crest and the origin of vertebrates: A new head. Science. 1983;220:268–273. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- 2.Darwin CR. Journal of Researches into the Geology and Natural History of the Various Countries Visited During the Voyage of H.M.S. Beagle, Under the Command of Captain FitzRoy, R.N. 2nd Ed. London: John Murray; 1845. [Google Scholar]
- 3.Grant PR, Grant BR. Unpredictable evolution in a 30-year study of Darwin's finches. Science. 2002;296:707–711. doi: 10.1126/science.1070315. [DOI] [PubMed] [Google Scholar]
- 4.Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin's finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 5.Abzhanov A, et al. The calmodulin pathway and evolution of elongated beak morphology in Darwin's finches. Nature. 2006;442:563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- 6.Wu P, Jiang TX, Suksaweang S, Widelitz RB, Chuong CM. Molecular shaping of the beak. Science. 2004;305:1465–1466. doi: 10.1126/science.1098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu P, Jiang TX, Shen JY, Widelitz RB, Chuong CM. Morphoregulation of avian beaks: Comparative mapping of growth zone activities and morphological evolution. Dev Dyn. 2006;235:1400–1412. doi: 10.1002/dvdy.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albertson RC, Streelman JT, Kocher TD, Yelick PC. Integration and evolution of the cichlid mandible: the molecular basis of alternate feeding strategies. Proc Natl Acad Sci USA. 2005;102:16287–16292. doi: 10.1073/pnas.0506649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kocher TD. Adaptive evolution and explosive speciation: The cichlid fish model. Nat Rev Genet. 2004;5:288–298. doi: 10.1038/nrg1316. [DOI] [PubMed] [Google Scholar]
- 10.Cooper WJ, et al. Bentho-pelagic divergence of cichlid feeding architecture was prodigious and consistent during multiple adaptive radiations within African rift-lakes. PLoS ONE. 2010;5:e9551. doi: 10.1371/journal.pone.0009551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helms JA, Schneider RA. Cranial skeletal biology. Nature. 2003;423:326–331. doi: 10.1038/nature01656. [DOI] [PubMed] [Google Scholar]
- 12.Albertson RC, Streelman JT, Kocher TD. Directional selection has shaped the oral jaws of Lake Malawi cichlid fishes. Proc Natl Acad Sci USA. 2003;100:5252–5257. doi: 10.1073/pnas.0930235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albertson RC, Kocher TD. Assessing morphological differences in an adaptive trait: A landmark-based morphometric approach. J Exp Zool. 2001;289:385–403. doi: 10.1002/jez.1020. [DOI] [PubMed] [Google Scholar]
- 14.Skulason S, Noakes DLG, Snorrason SS. Ontogeny of trophic morphology in four sympatric morphs of arctic charr Salvelinus alpinus in Thingvallavatn, Iceland. Biol J Linn Soc Lond. 1989;38:281–301. [Google Scholar]
- 15.Walker JA. Ecological morphology of lacustrine threespine stickleback Gasterosteus aculeatus L. (Gasterosteidae) body shape. Biol J Linn Soc Lond. 1997;61:3–50. [Google Scholar]
- 16.Adams CE, Huntingford FA. The functional significance of inherited differences in feeding morphology in a sympatric polymorphic population of Arctic charr. Evol Ecol. 2002;16:15–25. [Google Scholar]
- 17.Parsons KJ, Robinson BW. Replicated evolution of integrated plastic responses during early adaptive divergence. Evolution. 2006;60:801–813. [PubMed] [Google Scholar]
- 18.Riopel C, Robinson BW, Parsons KJ. Analyzing nested variation in the body form of Lepomid sunfishes. Environ Biol Fishes. 2008;82:409–420. [Google Scholar]
- 19.Cooper WJ, Westneat MW. Form and function of damselfish skulls: Rapid and repeated evolution into a limited number of trophic niches. BMC Evol Biol. 2009;9:24. doi: 10.1186/1471-2148-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loh YH, et al. Comparative analysis reveals signatures of differentiation amid genomic polymorphism in Lake Malawi cichlids. Genome Biol. 2008;9:R113. doi: 10.1186/gb-2008-9-7-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liem KF. Modulatory multiplicity in the functional repertoire of the feeding mechanism in cichlid fishes. J Morphol. 1978;158:323–360. doi: 10.1002/jmor.1051580305. [DOI] [PubMed] [Google Scholar]
- 22.Westneat MW. Feeding mechanics of teleost fishes (Labridae; Perciformes): A test of four-bar linkage models. J Morphol. 1990;205:269–295. doi: 10.1002/jmor.1052050304. [DOI] [PubMed] [Google Scholar]
- 23.Albertson RC, Kocher TD. Genetic architecture sets limits on transgressive segregation in hybrid cichlid fishes. Evolution. 2005;59:686–690. [PubMed] [Google Scholar]
- 24.Albertson RC. Morphological divergence predicts habitat partitioning in a Lake Malawi cichlid species complex. Copeia. 2008;2008:689–698. [Google Scholar]
- 25.Carroll SB. Evolution at two levels: On genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Jong IM, Witte F, Richardson MK. Developmental stages until hatching of the Lake Victoria cichlid Haplochromis piceatus (Teleostei: Cichlidae) J Morphol. 2009;270:519–535. doi: 10.1002/jmor.10716. [DOI] [PubMed] [Google Scholar]
- 27.Albertson RC, Yelick PC. Fgf8 haploinsufficiency results in distinct craniofacial defects in adult zebrafish. Dev Biol. 2007;306:505–515. doi: 10.1016/j.ydbio.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper WJ, Albertson RC. Quantification and variation in experimental studies of morphogenesis. Dev Biol. 2008;321:295–302. doi: 10.1016/j.ydbio.2008.06.025. [DOI] [PubMed] [Google Scholar]
- 29.Abzhanov A, Rodda SJ, McMahon AP, Tabin CJ. Regulation of skeletogenic differentiation in cranial dermal bone. Development. 2007;134:3133–3144. doi: 10.1242/dev.002709. [DOI] [PubMed] [Google Scholar]
- 30.Hammond CL, Schulte-Merker S. Two populations of endochondral osteoblasts with differential sensitivity to Hedgehog signalling. Development. 2009;136:3991–4000. doi: 10.1242/dev.042150. [DOI] [PubMed] [Google Scholar]
- 31.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chuong CM, Patel N, Lin J, Jung HS, Widelitz RB. Sonic hedgehog signaling pathway in vertebrate epithelial appendage morphogenesis: Perspectives in development and evolution. Cell Mol Life Sci. 2000;57:1672–1681. doi: 10.1007/PL00000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koyama E, et al. Polarizing activity, Sonic hedgehog, and tooth development in embryonic and postnatal mouse. Dev Dyn. 1996;206:59–72. doi: 10.1002/(SICI)1097-0177(199605)206:1<59::AID-AJA6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 34.Ingham PW, Fietz MJ. Quantitative effects of hedgehog and decapentaplegic activity on the patterning of the Drosophila wing. Curr Biol. 1995;5:432–440. doi: 10.1016/s0960-9822(95)00084-4. [DOI] [PubMed] [Google Scholar]
- 35.Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka M, et al. Distribution of polarizing activity and potential for limb formation in mouse and chick embryos and possible relationships to polydactyly. Development. 2000;127:4011–4021. doi: 10.1242/dev.127.18.4011. [DOI] [PubMed] [Google Scholar]
- 37.Harris MP, Williamson S, Fallon JF, Meinhardt H, Prum RO. Molecular evidence for an activator-inhibitor mechanism in development of embryonic feather branching. Proc Natl Acad Sci USA. 2005;102:11734–11739. doi: 10.1073/pnas.0500781102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ribbink AJ, Marsh BA, March AC, Ribbink AC, Sharp BJ. A preliminary survey of the cichlid fishes of rocky habitats in Lake Malawi. S Afr J Zool. 1983;18:149–310. [Google Scholar]
- 39.Young NM, Chong HJ, Hu D, Hallgrímsson B, Marcucio RS. Quantitative analyses link modulation of sonic hedgehog signaling to continuous variation in facial growth and shape. Development. 2010;137:3405–3409. doi: 10.1242/dev.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto Y, Byerly MS, Jackman WR, Jeffery WR. Pleiotropic functions of embryonic sonic hedgehog expression link jaw and taste bud amplification with eye loss during cavefish evolution. Dev Biol. 2009;330:200–211. doi: 10.1016/j.ydbio.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Albertson RC, Markert JA, Danley PD, Kocher TD. Phylogeny of a rapidly evolving clade: the cichlid fishes of Lake Malawi, East Africa. Proc Natl Acad Sci USA. 1999;96:5107–5110. doi: 10.1073/pnas.96.9.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Danley PD, Kocher TD. Speciation in rapidly diverging systems: Lessons from Lake Malawi. Mol Ecol. 2001;10:1075–1086. doi: 10.1046/j.1365-294x.2001.01283.x. [DOI] [PubMed] [Google Scholar]
- 43.Mayr E. Animal Species and Evolution. Cambridge, MA: Harvard Univ Press; 1963. [Google Scholar]
- 44.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 45.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fraser GJ, Bloomquist RF, Streelman JT. A periodic pattern generator for dental diversity. BMC Biol. 2008;6:32. doi: 10.1186/1741-7007-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- 48.Mims MC, Darrin Hulsey C, Fitzpatrick BM, Streelman JT. Geography disentangles introgression from ancestral polymorphism in Lake Malawi cichlids. Mol Ecol. 2010;19:940–951. doi: 10.1111/j.1365-294X.2010.04529.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.