Abstract
Malaria parasites (Plasmodium spp.) have plagued humans for millennia. Less well known are related parasites (Haemosporida), with diverse life cycles and dipteran vectors that infect other vertebrates. Understanding the evolution of parasite life histories, including switches between hosts and vectors, depends on knowledge of evolutionary relationships among parasite lineages. In particular, inferences concerning time of origin and trait evolution require correct placement of the root of the evolutionary tree. Phylogenetic reconstructions of the diversification of malaria parasites from DNA sequences have suffered from uncertainty concerning outgroup taxa, limited taxon sampling, and selection on genes used to assess relationships. As a result, inferred relationships among the Haemosporida have been unstable, and questions concerning evolutionary diversification and host switching remain unanswered. A recent phylogeny placed mammalian malaria parasites, as well as avian/reptilian Plasmodium, in a derived position relative to the avian parasite genera Leucocytozoon and Haemoproteus, implying that the ancestral forms lacked merogony in the blood and that their vectors were non-mosquito dipterans. Bayesian, outgroup-free phylogenetic reconstruction using relaxed molecular clocks with uncorrelated rates instead suggested that mammalian and avian/reptilian Plasmodium parasites, spread by mosquito vectors, are ancestral sister taxa, from which a variety of specialized parasite lineages with modified life histories have evolved.
Keywords: Bayes factors, parasite diversification, Plasmodiidae
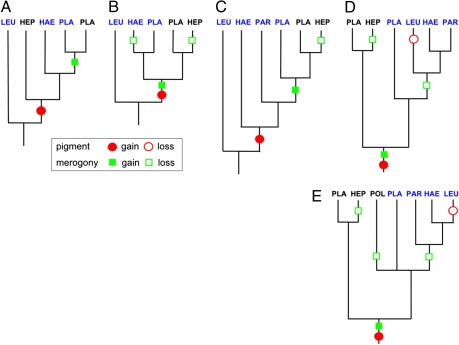
Malaria parasites [broadly Apicomplexa: Haemosporida (1, 2)] have been well sampled in primates and songbirds, but are poorly known in other vertebrate groups. Recent surveys of blood parasites in vertebrate wildlife populations, using PCR to screen hosts for infections and DNA sequencing to identify parasite lineages, have revealed a rich diversity of hemosporidian parasites (3–6), possibly comparable to the number of hosts surveyed (7, 8). It is important to reevaluate our interpretation of hemosporidian evolution as we expand sampling, to provide insight into shifts among hosts and vectors—often implicated in emerging infectious diseases—and to interpret the evolution of malaria parasite life cycles. Because choice of outgroup taxa critically influences the reconstruction of evolutionary relationships (9), it is also important to reassess assumptions about ancestral relationships and the monophyly of taxa. Premolecular reconstructions based on morphology and life history traits presumed that a monophyletic clade of Plasmodium parasites exhibited the most derived traits [e.g., asexual reproduction (merogony) in the circulating blood of the vertebrate host, production of hemozoin pigment from the metabolism of hemoglobin] (10, 11); accordingly, Leucocytozoon, a parasite of birds that lacks these traits, was placed in a basal position (Fig. 1A). Such a position for Leucocytozoon was indeed suggested in early molecular analyses based on the mitochondrial cytochrome b gene (cyt b) using genetically distant apicomplexan outgroups (Toxoplasma and Theileria), which rendered Plasmodium polyphyletic or paraphyletic (Fig. 1B) (12, 13). More comprehensive phylogenetic analyses have adopted Leucocytozoon as the outgroup taxon, that is, placed the root of the tree a priori between Leucocytozoon and all remaining taxa without independent confirmation, which results in a paraphyletic Plasmodium (Fig. 1C; SI Text).
Fig. 1.
Series of topologies for the Haemosporida in the style of Martinsen et al. (19). (A) The classic topology based on these traits places Leucocytozoon, which lacks these traits, as a sister to all other Haemosporida. (B) The topology of Perkins and Schall (13) based on cytochrome b sequences rooted with Theileria. (C) The topology of Martinsen et al. (19), based on sequences from two mitochondrial genes, one apicoplast gene, and one nuclear gene, with Leucocytozoon assigned as the outgroup taxon. (D) The data of Martinsen et al. reanalyzed under a Bayesian relaxed molecular clock model, in which Plasmodium becomes paraphyletic with respect to the other genera and Leucocytozoon is sister to the avian Haemoproteus and Parahaemoproteus clade. (E) An enlarged cytochrome b dataset analyzed under a relaxed clock model again places the root between the clades of mammalian and avian/reptilian parasites. This topology is also found under maximum likelihood optimization (Fig. 4). A minimum set of inferred character changes for hemozoin pigment (red circles) and merogony (green squares) is indicated. LEU, Leucocytozoon; PLA, Plasmodium; HEP, Hepatocystis; HAE, Haemoproteus; PAR, Parahaemoproteus; POL, Polychromophilus. Mammalian lineages are shown in black; avian/reptilian lineages, in blue.
Rooting phylogenies can be problematic when outgroup taxa are not well established, as in the case of malaria parasites (13). In many cases, the midpoint rooting method (14), which defines the root at the node between the two most divergent groups, is used and provides plausible results comparable to outgroup rooting methods (reviewed in ref. 15). However, when rates of evolution are heterogeneous across branches in a phylogeny, midpoint rooting can be confounded, as can outgroup rooting methods (9), whereas molecular clock rooting methods seem robust to moderate violations of homogeneity. Although designating an appropriate outgroup taxon is the best approach to rooting a phylogenetic tree (16), both simulated and empirical data sets indicate that molecular clock rooting methods, particularly those accounting for evolutionary rate heterogeneity, are preferred over rooting with a distant outgroup taxon (9, 17, 18).
Results and Discussion
We provide an alternative rooting of the malaria parasite phylogeny by applying a rooting method (9, 17, 18) that does not require specifying ancestors and descendants a priori. Using this approach with the data analyzed in the most recent comprehensive phylogeny (19), we find an alternative evolutionary scenario (Figs. 1D and 2A) that differs markedly from previous molecular studies (Fig. 1C); Plasmodium (avian/reptilian and mammalian) becomes paraphyletic with respect to the remaining hemosporidian genera rather than being derived, and Leucocytozoon (avian) becomes a sister to Haemoproteus and Parahaemoproteus (avian). The two major subgroups are clearly associated with vertebrate host: mammalian Plasmodium and Hepatocystis versus avian/reptilian Plasmodium and avian Leucocytozoon, Haemoproteus, and Parahaemoproteus. The chiropteran (bat) parasites Polychromophilus evidently were derived from parasites of birds or reptiles (3, 20). Increasing the taxon sampling in the ingroup and in the putative outgroup (i.e., Leucocytozoon) for the mitochondrial cyt b gene (for which large samples are available) and using the same outgroup-free rooting approach also places Leucocytozoon inside the hemosporidian phylogeny, in this case as sister to Haemoproteus, which together are sister to Parahaemoproteus (Figs. 1E and 3).
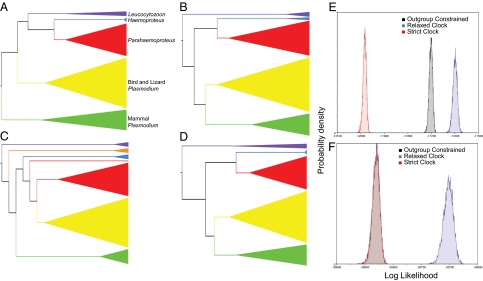
The relationships revealed by our application of an outgroup-free approach to rooting (Fig. 2A) are clearly distinguished from the outcomes of alternative rooting methods (Fig. 2 B–D), including the most recent comprehensive phylogeny (19). Midpoint rooting and the distance-based unweighted pair group method with arithmetic mean (UPGMA), both of which assume rate constancy, indicate an outgroup taxon position for Leucocytozoon (Fig. 2 B and C). Although likelihood ratio tests between alternative topologies cannot use rooted phylogenies, such as midpoint and UPGMA trees, per se, constraining the ingroup to exclude Leucocytozoon (Fig. 2D) results in a significantly worse estimate of phylogenetic relationships [Δ(−2lnL) = 148.14; P < 0.0001]. The distributions of Bayesian tree likelihoods from (i) a strict clock analysis (Fig. 2B, essentially a midpoint root; Fig. 2C, UPGMA), (ii) a relaxed clock analysis (Fig. 2A), and (iii) an outgroup-constrained analysis (Fig. 2D) show that both strict clock and outgroup-constrained analyses poorly describe the data in both datasets (Table 1).
Fig. 2.
Series of cladograms based on data analyzed by Martinsen et al. (19). (A–D) Our outgroup-free rooting (A) and three alternative topologies: midpoint rooting (B), UPGMA, in which Hepatocystis spp. form their own clade (orange) and one lizard Plasmodium lineage falls outside the others (C); and outgroup rooting (D). The size of a clade reflects relative taxon sampling. Purple indicates Leucocytozoon; blue, Haemoproteus (dove); red, bird and lizard Plasmodium; yellow, bird Parahaemoproteus; green, mammal Plasmodium. (E and F) Marginal densities of tree likelihoods of all data from Martinsen et al. (19) (E) and cytochrome b data (F).
Table 1.
Bayes factor analyses of marginal likelihoods
| Log-likelihood, lnL | SD | Δ2lnL | |
| All data | |||
| Outgroup constrained | −30,842.156 | 8.32 | −245.6 |
| Relaxed clock | −30,719.351 | 12.30 | 0.0 |
| Strict clock | −30,843.256 | 9.46 | −247.8 |
| Cytochrome b | |||
| Outgroup constrained | −11,715.948 | 9.46 | −196.0 |
| Relaxed clock | −11,617.934 | 11.29 | 0.0 |
| Strict clock | −11,997.605 | 9.36 | −759.4 |
Analyses of marginal tree likelihood distributions with Bayes factors suggest that a relaxed clock model explains the data better (higher lnL) than either a strict clock or forcing Leucocytozoon to an outgroup position.
The rapid evolution of the cyt b gene in some Leucocytozoon taxa and mammalian Plasmodium (plus Hepatocystis) (Figs. 3 and 4) represents substantial nonsynonymous nucleotide substitution and possibly adaptive amino acid changes. We have previously identified heterogeneity in protein evolution (ratio of synonymous to nonsynonymous nucleotide substitutions) in the cyt b gene of malaria parasites, particularly in the transition of Plasmodium lineages between mammalian and avian/reptilian hosts (21). High relative rates of nucleotide substitution lead to long-branch attraction (22–24) in phylogenetic hypotheses rooted with distant outgroups, which likely brought Leucocytozoon to a basal position with respect to the remaining hemosporidian parasites in previous analyses (12, 13) and resulted in previous acceptance of Leucocytozoon as the appropriate outgroup taxon.
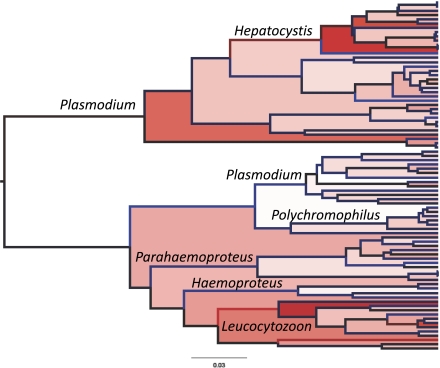
Fig. 3.
Phylogenetic tree produced using a relaxed clock with an HKY + gamma model of nucleotide substitution and uncorrelated, lognormally distributed rates. Relative rates of evolution on branches increase from blue through black to red, and also increase as shown by fills from white to red. High rates of nucleotide substitution particularly characterize mammalian Hepatocystis and avian Leucocytozoon; also see Fig. 4.
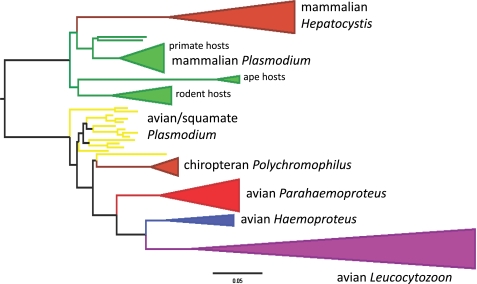
Fig. 4.
Maximum likelihood phylogram based on cytochrome b data using a GTR + gamma model of nucleotide substitution. The tree was rooted between mammal Plasmodium/Hepatocystis and all other parasite lineages. Colored lines indicate >95% bootstrap values. Breadths of cartooned clades are proportional to species sampling in this analysis. (Scale bar: 5% nucleotide substitution.)
Whereas recent phylogenetic reconstructions have Plasmodium as monophyletic (Fig. 1A), polyphyletic (Fig. 1B), or paraphyletic (Fig. 1C), our analyses show that Plasmodium as currently circumscribed is polyphyletic. The basal split in the phylogeny of contemporary malaria lineages is between mammalian and avian/reptilian Plasmodium, each of which is more closely related to other genera of Haemosporida than to each other (Figs. 1E and 2A). Accordingly, the taxon Plasmodium will need to be redefined as a result of the new phylogeny, and character evolution in malaria parasites will need to be reinterpreted by morphological taxonomists and molecular systematists working closely together (25).
The evolution of host specialization, vector specialization, and life cycle traits in the Haemosporida remain poorly characterized. However, a properly rooted phylogeny that indicates the directions of evolutionary transitions is a first step toward understanding host and vector switching leading to the emergence of new infectious diseases. Our placement of the root of the malaria parasite phylogeny indicates that merogony (three inferred losses) and hemozoin pigment (one inferred loss) are neither derived characters nor evolutionarily conservative (Fig. 1E). The direction of vertebrate (secondary, i.e., asexual phase) host switching can be inferred in the case of Polychromophilus as being from avian/reptilian to mammalian hosts. Assuming that mosquitoes (Culicidae) are the ancestral primary (i.e., sexual phase) vectors of contemporary Haemosporida, lineages of parasites have switched to Culicoides midges (Hepatocystis and Parahaemoproteus), Simulium blackflies (Leucocytozoon), and Hippoboscoidea louse flies (Haemoproteus) and bat flies (Polychromophilus).
The diversification of contemporary Haemosporida evidently has occurred recently compared with the evolution of host and vector lineages, likely within the last 20 million years (26), and the long history of evolution in this group, inferred from the presence of parasite stages in dipteran vectors preserved in mid-Tertiary and Cretaceous ambers (27, 28) is lost. Nevertheless, phylogenetic reconstruction of the history of contemporary members provides an appropriate context for understanding evolutionary transitions associated with switching to new vectors and hosts, and thus the emergence of new infectious diseases. Extended sampling of Haemosporida in nature, particularly of the parasites of mammals, combined with comparative genomic analyses associated with host switching, should greatly improve our understanding of the evolution of pathogenic organisms.
Methods
We used sequence data from four genes analyzed by Martinsen et al. (19) for as many as 60 parasite lineages: mitochondrial cytochrome b (607 nt), cytochrome oxidase I (998 nt), apicoplast caseinolytic protease C (523 nt), and nuclear argininosuccinate lysase (206 nt). Incomplete data were treated as missing values in a concatenated data set. We used a relaxed molecular clock in BEAST (29) [two runs each; GTR + I + G (four rate categories); estimated base frequencies; Yule tree prior; 10,000,000 generations, sampling every 1,000th tree with a 10% burn-in] and generated consensus data from 18,000 trees. We also downloaded additional avian Leucocytozoon, Hepatocystis, Haemoproteus, and Parahaemoproteus and mammalian Plasmodium and Polychromophilus (3) cyt b sequences from GenBank (1,125 nt), which were combined with cyt b data from Martinsen et al. (19) in a maximum likelihood analysis [RAxML (30), using GTR + G; 100 bootstrap iterations] and in BEAST runs [as above or using HKY + gamma (four rate categories), Yule tree prior, 10,000,000 generations, sampling every 5,000th tree with 10% burn-in (i.e., 1,800 trees)]. The estimated sample size was >200 in all BEAST runs. For each of the foregoing data sets, we also conducted BEAST runs using a strict clock and a relaxed clock with Leucocytozoon and all remaining taxa constrained to be monophyletic. We then compared the likelihood distributions from the relaxed clock, the strict clock, and the outgroup-constrained analyses using BayesFactors (1,000 bootstrap replicates) in Tracer version 1.5 (31). We tested two alternative topologies, one with Leucocytozoon taxa as a sister to other avian hemosporidians (our rooting) and one with Leucocytozoon taxa unconstrained, using the Shimodaira–Hasegawa test (32), with resampling estimated log-likelihood approximation, 1,000 bootstrap pseudoreplicates, and PAUP* (33).
Supplementary Material
Acknowledgments
We thank C. P. Brooks, B. Counterman, S. Edwards, R. K. Outlaw, S. S. Renner, S. Rich, and V. W. Walstrom for their comments on the manuscript. This work was supported by National Science Foundation Grant DEB-0089226 (to R.E.R.), an Alexander von Humboldt Research Fellowship (to R.E.R.), and research funds from Mississippi State University (to D.C.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
See Commentary on page 12973.
This article contains supporting information online www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109153108/-/DCSupplemental.
References
- 1.Garnham PCC. Malaria Parasites and Other Haemosporidia. Oxford: Blackwell Scientific; 1966. [Google Scholar]
- 2.Valkiunas G. Avian Malaria Parasites and Other Haemosporidia. Boca Raton, FL: CRC Press; 2005. [Google Scholar]
- 3.Duval L, et al. Multiple host-switching of Haemosporidia parasites in bats. Malar J. 2007;6:157. doi: 10.1186/1475-2875-6-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chasar A, et al. Prevalence and diversity patterns of avian blood parasites in degraded African rainforest habitats. Mol Ecol. 2009;18:4121–4133. doi: 10.1111/j.1365-294X.2009.04346.x. [DOI] [PubMed] [Google Scholar]
- 5.Beadell JS, et al. Host associations and evolutionary relationships of avian blood parasites from West Africa. Int J Parasitol. 2009;39:257–266. doi: 10.1016/j.ijpara.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishtiaq F, et al. Prevalence and diversity of avian hematozoan parasites in Asia: A regional survey. J Wildl Dis. 2007;43:382–398. doi: 10.7589/0090-3558-43.3.382. [DOI] [PubMed] [Google Scholar]
- 7.Bensch S, Hellgren O, Pérez-Tris J. MalAvi: A public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour. 2009;9:1353–1358. doi: 10.1111/j.1755-0998.2009.02692.x. [DOI] [PubMed] [Google Scholar]
- 8.Ricklefs RE, Fallon SM. Diversification and host switching in avian malaria parasites. Proc Biol Sci. 2002;269:885–892. doi: 10.1098/rspb.2001.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huelsenbeck JP, Bollback JP, Levine AM. Inferring the root of a phylogenetic tree. Syst Biol. 2002;51:32–43. doi: 10.1080/106351502753475862. [DOI] [PubMed] [Google Scholar]
- 10.Esposito A, et al. FRET imaging of hemoglobin concentration in Plasmodium falciparum–infected red cells. PLoS ONE. 2008;3:e3780. doi: 10.1371/journal.pone.0003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenthal PJ, Meshnick SR. Hemoglobin catabolism and iron utilization by malaria parasites. Mol Biochem Parasitol. 1996;83:131–139. doi: 10.1016/s0166-6851(96)02763-6. [DOI] [PubMed] [Google Scholar]
- 12.Escalante AA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc Natl Acad Sci USA. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perkins SL, Schall JJ. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol. 2002;88:972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 14.Farris JS. Estimating phylogenetic trees from distance matrices. Am Nat. 1972;106:645–668. [Google Scholar]
- 15.Hess PN, De Moraes Russo CA. An empirical test of the midpoint rooting method. Biol J Linn Soc. 2007;92:669–674. doi: 10.1111/j.1095-8312.2007.00864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maddison WP, Donoghue MJ, Maddison DR. Outgroup analysis and parsimony. Syst Zool. 1984;33:88–103. [Google Scholar]
- 17.Boykin LM, Kubatko LS, Lowrey TK. Comparison of methods for rooting phylogenetic trees: A case study using Orcuttieae (Poaceae: Chloridoideae) Mol Phylogenet Evol. 2010;54:687–700. doi: 10.1016/j.ympev.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 18.Renner SS, Grimm GW, Schneeweiss GM, Stuessy TF, Ricklefs RE. Rooting and dating maples (Acer) with an uncorrelated-rates molecular clock: Implications for North American/Asian disjunctions. Syst Biol. 2008;57:795–808. doi: 10.1080/10635150802422282. [DOI] [PubMed] [Google Scholar]
- 19.Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): Evolution of life-history traits and host switches. Mol Phylogenet Evol. 2008;47:261–273. doi: 10.1016/j.ympev.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Megali A, Yannic G, Christe P. Disease in the dark: Molecular characterization of Polychromophilus murinus in temperate zone bats revealed a worldwide distribution of this malaria-like disease. Mol Ecol. 2011;20:1039–1048. doi: 10.1111/j.1365-294X.2010.04905.x. [DOI] [PubMed] [Google Scholar]
- 21.Outlaw DC, Ricklefs RE. Comparative gene evolution in haemosporidian (Apicomplexa) parasites of birds and mammals. Mol Biol Evol. 2010;27:537–542. doi: 10.1093/molbev/msp283. [DOI] [PubMed] [Google Scholar]
- 22.Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–193. doi: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- 23.Felsenstein J. Inferring Phylogenies. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- 24.Kolaczkowski B, Thornton JW. Long-branch attraction bias and inconsistency in Bayesian phylogenetics. PLoS ONE. 2009;4:e7891. doi: 10.1371/journal.pone.0007891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valkiūnas G, et al. Linkage between mitochondrial cytochrome b lineages and morphospecies of two avian malaria parasites, with a description of Plasmodium (Novyella) ashfordi sp. nov. Parasitol Res. 2007;100:1311–1322. doi: 10.1007/s00436-006-0409-3. [DOI] [PubMed] [Google Scholar]
- 26.Ricklefs RE, Outlaw DC. A molecular clock for malaria parasites. Science. 2010;329:226–229. doi: 10.1126/science.1188954. [DOI] [PubMed] [Google Scholar]
- 27.Poinar G., Jr Plasmodium dominicana n. sp. (Plasmodiidae: Haemospororida) from Tertiary Dominican amber. Syst Parasitol. 2005;61:47–52. doi: 10.1007/s11230-004-6354-6. [DOI] [PubMed] [Google Scholar]
- 28.Poinar G, Jr, Telford SR., Jr Paleohaemoproteus burmacis gen. n., sp. n. (Haemospororida: Plasmodiidae) from an Early Cretaceous biting midge (Diptera: Ceratopogonidae) Parasitology. 2005;131:79–84. doi: 10.1017/s0031182005007298. [DOI] [PubMed] [Google Scholar]
- 29.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 31.Rambaut A, Drummond AJ. Tracer version 1.4. 2007. Available from http://beast.bio.ed.ac.uk/Tracer.
- 32.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 33.Swofford D. PAUP* 4.0: Phylogenetic Analysis Using Parsimony *and Other Methods. Sutherland, MA: Sinauer Associates; 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.