Abstract
Homing endonucleases mobilize their own genes by generating double-strand breaks at individual target sites within potential host DNA. Because of their high specificity, these proteins are used for “genome editing” in higher eukaryotes. However, alteration of homing endonuclease specificity is quite challenging. Here we describe the identification and phylogenetic analysis of over 200 naturally occurring LAGLIDADG homing endonucleases (LHEs). Biochemical and structural characterization of endonucleases from one clade within the phylogenetic tree demonstrates strong conservation of protein structure contrasted against highly diverged DNA target sites and indicates that a significant fraction of these proteins are sufficiently stable and active to serve as engineering scaffolds. This information was exploited to create a targeting enzyme to disrupt the endogenous monoamine oxidase B gene in human cells. The ubiquitous presence and diversity of LHEs described in this study may facilitate the creation of many tailored nucleases for genome editing.
Keywords: gene targeting, gene therapy, protein engineering
Several types of highly specific DNA recognition and cleavage enzymes, including homing endonucleases (HEs), zinc finger nucleases (ZFNs), and transcription activator-like (TAL) effector nucleases, are being developed for targeted gene modification, ranging from gene disruption to corrective gene therapy (1–5). Regardless of the identity of the protein scaffold, site-specific gene modification generally requires the creation of individually tailored site-specific endonucleases that generate double-strand breaks (DSBs) at unique chromosomal targets. These lesions induce intrinsic DNA repair responses in the targeted cells (primarily homologous recombination and nonhomologous end joining) that lead to DNA sequence alterations at the target (3, 5).
Homing endonucleases are highly specific DNA cleaving enzymes that drive the genetic mobilization of their own reading frames (3, 4, 6). Their catalytic activity is usually tightly coupled to recognition of their cognate DNA target sites, which allows them to be used for site-specific genome modification (7). Among the HE families that have been identified so far, members of LAGLIDADG homing endonuclease (LHE) family, which are primarily encoded in archaea and in organellar DNA in green algae and fungi, display the highest overall DNA recognition specificity (8, 9). These proteins possess one or two LAGLIDADG catalytic motifs per protein chain and function as homodimers or monomers, respectively. To date, four naturally occurring LHEs (I-SceI, I-CreI, I-MsoI, and I-AniI) and one chimeric enzyme (DmoCre) have been engineered to alter their sequence recognition specificity (3, 4, 8–10), but only I-CreI has been used to modify endogenous chromosomal targets (11, 12). Successful gene targeting required extensive alteration of its DNA contact points and specificity, at up to two-thirds of the base-pair positions in its target site.
Recent analyses of metagenomic and microbial sequence databases have hinted at the presence of large numbers of uncharacterized LHEs, as illustrated by the discovery of multiple endonuclease reading frames inserted at several positions within the ribosomal protein S3 gene in ascomycete fungi (13). To expand upon this concept, we analyzed available genome sequence databases for unique LHEs. Over 200 such endonucleases were identified and subjected to a phylogenetic analysis that illustrated the evolutionary diversity of monomeric LHE scaffolds. This analysis implies that such a diverse and growing collection of naturally occurring endonucleases might be exploited to create unique genome editing enzymes. To further test this concept, we focused on a single LHE subfamily and demonstrated that a significant fraction of its proteins are well behaved and cleave a predictable, specific DNA target site. Members of this endonuclease subfamily display high overall identity across their amino acid sequences, but act upon a diverse set of DNA sequences. We determined the DNA-bound crystal structure of two representative enzymes in order to assess the conservation of their protein folds and DNA recognition mechanisms and then engineered one of those enzymes in order to cleave and disrupt the human monoamine oxidase B (MAO-B) gene. This study implies that systematic characterization of sufficient numbers of LHE scaffolds might allow their full potential to be realized for genome editing applications.
Results
Global Analyses of Single-Chain LAGLIDADG Enzymes Reveals Distinct Subfamilies.
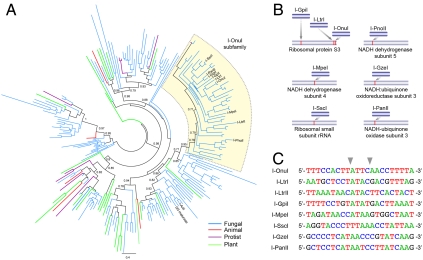
The recent discovery of I-OnuI and I-LtrI, which display 47% amino acid identity and yet specifically cleave distinct DNA target sites (13), prompted us to survey and characterize homologues of these proteins. Using structure-based alignment methods, we identified 211 putative single-chain LHEs (Fig. 1A and Fig. S1A). Phylogenetic analyses of these sequences revealed significant diversity. Interestingly, endonucleases encoded within fungal mitochondrial genomes constitute a significant proportion of the monomeric enzymes, with fewer examples found among plant, animal, and protozoan genomes.
Fig. 1.
Diversity of LHE genes and their sequence recognition specificities. (A) Shown is a phylogenetic analysis of 211 single-chain LHEs as identified by structure-based alignments. Branches are color-coded according to the taxonomic source from which the LHE was identified, as indicated. Groups of LHEs are colored the same if all members of that subfamily are derived from the same taxonomic group. Black lines indicate that members of a subfamily are derived from more than one taxonomic grouping. The I-OnuI subfamily is highlighted in yellow, with individual LHEs named (e.g., I-OnuI). The phylogenetic tree was generated by PhyML (14), and approximate likelihood-ratio test values greater than 0.7 are indicated on major nodes. A version of the tree with branches labeled by accession numbers and partial sequences of all the LHEs identified are provided as Fig. S1A and Dataset S1, respectively. (B) Schematic of homing sites recognized by the I-OnuI subfamily. (C) Target sequences for I-OnuI, I-LtrI, and I-LtrII were identified in previous studies (13, 15). Recognition sequences for the other LHEs were predicted through comparative sequence alignments of each host gene to related species lacking an embedded endonuclease. Cleavage activity against each predicted site was verified using yeast surface-displayed enzyme in both in vitro and flow cytometric cleavage assays (see SI Text for detail). Sequence analysis of cleaved products showed that all of the homologues generated 3′, 4-base overhangs by hydrolyzing the phosphodiester bonds between the base-pair positions ± 2 and ± 3 (indicated by gray arrows).
In the subset of LHEs that included the previously characterized enzymes I-OnuI, I-LtrI, and I-LtrII (13, 15), many additional putative endonucleases reside within a wide variety of different host genes (see the I-OnuI subfamily in Fig. 1 A and B and protein alignment in Fig. S1B). This suggests that members of the I-OnuI subfamily can target highly diverse DNA sequences. To test this hypothesis, and to determine how often LHE genes identified solely on the basis of sequence homology actually encode active endonucleases, we subcloned and characterized I-OnuI, I-LtrI, and 11 putative endonucleases. Eight of the putative enzymes were efficiently expressed on yeast cell surface, and six of those enzymes displayed robust cleavage of predictable DNA target sites (that correspond to the LHEs’ intron insertion sites within their host genes, as described in SI Materials and Methods). Subsequent sequencing of individual cleaved DNA products demonstrated that all the enzymes generate 3′, 4-base overhangs that typify the LAGLIDADG family and that the target sites for this subfamily are widely diverged (Fig. 1C).
Target Sequence Recognition by I-OnuI and I-LtrI.
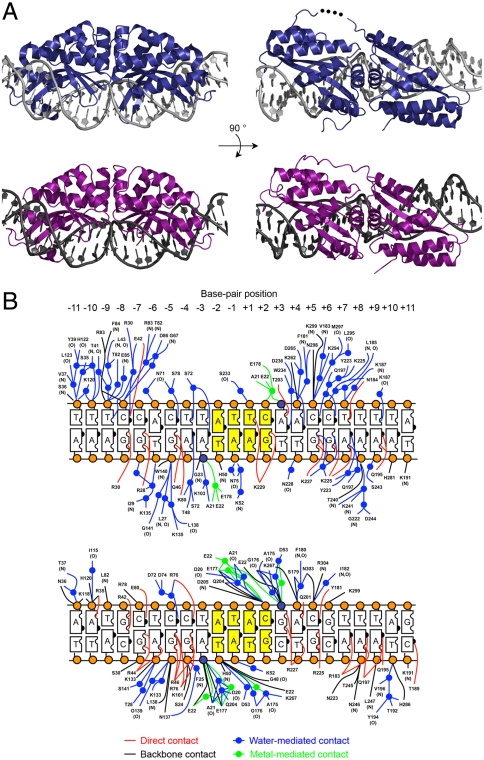
To visualize the molecular contacts that facilitate recognition of different DNA sequences by otherwise very closely related proteins, we solved the crystal structures of I-OnuI and I-LtrI bound to their DNA targets at 2.4-Å and 2.7-Å resolution, respectively. These enzymes displayed somewhat low sequence identities to previously well-characterized LHEs, including I-AniI and I-CreI (< 25%), but displayed very similar LAGLIDADG folds relative to both of those enzymes as well as to each other (Fig. 2A and Fig. S2A). Both proteins make contacts with approximately one-half of the nucleotide bases and backbone phosphate groups of their target sequences, via a mixture of direct and water-mediated contacts, as observed in previous LHE crystal structures (16–18) (Fig. 2B). Superposition of the two structures yields a rmsd across 284 superimposed α-carbons of approximately 1.3 Å, as well as similar DNA backbone conformations. The DNA substrate was uncleaved in the I-OnuI complex (solved in the presence of magnesium at pH 4.5), whereas both DNA strands were cleaved by I-LtrI (solved in the presence of manganese and magnesium at pH 6.5). Nonetheless, the active sites of these proteins were closely superposed, with the exception of the exact positions of bound metal ions, the scissile phosphates, and the side-chain rotamer of a metal-coordinating glutamate residue (E178 in I-OnuI and E177 in I-LtrI) (Fig. S2B).
Fig. 2.
Structure determination of I-OnuI and I-LtrI. (A) Crystal structures of I-OnuI (Upper, dark blue) and I-LtrI (Lower, purple) bound to their physiological target sites. Residues 156–158 in the middle of linker between the two pseudosymmetric half domains of I-OnuI were disordered (represented as black dots). The loop region (residues 236–244) between the third and fourth β-sheets of the C-terminal half domain of I-LtrI could not be assigned due to poor electron density. (B) Schematic of I-OnuI (Upper) and I-LtrI (Lower) DNA contacts. The two scissile phosphates and the other backbone phosphates are depicted as dark blue and orange spheres, respectively. The central four base pairs (positions ± 1 and ± 2) are colored in yellow. Residue numbers of I-LtrI crystal shown here are shifted from the numbers assigned in the deposited PDB file, in order to align the residue numbers of the first LAGLIDADG motif to the corresponding numbers of I-OnuI crystal. E29 in the original PDB file is labeled as E22.
Outside of their active sites, the two protein–DNA interfaces are quite dissimilar. Although 12 of 22 base pairs are identical between the two target sites, only one contact between a side chain and a nucleotide contact (corresponding to glutamine 195 and the adenine ring at base-pair position +9 in I-OnuI) is observed in both structures (Fig. 2B). These results suggest that even two closely related LHEs such as I-OnuI and I-LtrI rapidly evolve unique, diverged surfaces to recognize corresponding DNA target sites, while maintaining conserved protein folds and catalytic mechanisms.
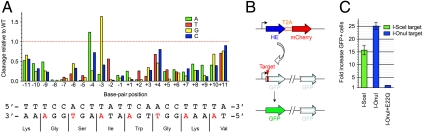
We next determined the specificity profile of I-OnuI, by measuring the enzyme’s relative cleavage activity on a series of DNA targets containing single base-pair substitutions relative to its WT target. This experiment was performed using a previously described cleavage assay involving display of the endonuclease on the surface of yeast, followed by staining with fluorescently labeled DNA substrates (19, 20). Similar to other LHEs (19–22), I-OnuI displayed significantly reduced cleavage activities for most of the single base-pair substitutions across its target site (Fig. 3A). The direct side-chain contacts, and those mediated by bridging water molecules, both appear to contribute to recognition specificity at many base-pair positions. This assay probably provides a conservative estimate of the enzyme’s true specificity, because the DNA substrates are physically tethered near the protein (potentially suppressing the effect of DNA substitutions that slightly reduce binding affinity) (20).
Fig. 3.
Evaluation of specificity and activity of I-OnuI. (A) Cleavage activity of I-OnuI for each single base-pair substituted target site from its physiological target was assayed by tethering fluorescence-labeled substrates to the I-OnuI protein expressed on the surface of yeast. Each bar represents relative cleavage activity for a target site containing a single base-pair substitution from the WT I-OnuI target. A, green; T, red; G, yellow; C, blue. The bottom strand encodes a host gene product, and wobble positions in the host gene reading frame are colored in red. (B) Schematic representation of the plasmids used in an episomal gene conversion assay. A homing endonuclease (HE) gene was linked to the mCherry gene through a 2A peptide sequence from T2A, leading to a coexpression of the two genes separated by the ribosomal skipping mechanism (23). The episomal DR-GFP reporter harbored two nonfunctional GFP gene (24): One was interrupted by a LHE target site and a stop codon, and the other was truncated. Double-strand breaks induced by a HE promote the conversion between the two nonfunctional GFP genes on the episomal DR-GFP reporter, resulting in the restoration of the GFP expression. (C) Gene conversion activity was assayed using the episomal DR-GFP reporter containing a target site for a LHE. Each bar represents an increase in a fraction of GFP positive cells by transient expression of a LHE compared to the background observed by transfection with the DR-GFP reporter alone. Errors refer to ± SD of three independent experiments.
Because relatively little selective pressure acts upon an HE to maintain its protein fold and activity after successful invasion of their host genes (6), naturally occurring HEs often exhibit compromised activity, particularly for in vivo applications (25). We therefore measured the ability of I-OnuI to induce homologous recombination in cultured human cells, by using an episomal DR-GFP reporter that harbors two nonfunctional GFP genes (24). In this system, one reporter gene is interrupted by a LHE target site and a stop codon, whereas the other is truncated; an LHE-induced double-strand break of the target site promotes recombination between the nonfunctional GFP genes and restores the fluorescent signal (Fig. 3B). In these experiments, the signal generated by transfection of HEK 293T cells with only the reporter plasmid was 0.3 to 0.8% gene conversion frequency (Fig. 3C). In contrast, cotransfection with an I-OnuI expression plasmid and a reporter plasmid containing the I-OnuI target site increased the fraction of GFP positive cells by 22- to 27-fold in individual experiments (corresponding to gene conversion frequencies of 15 to 19% of the total cell population, under conditions where approximately 80% of the cells were transfected). An inactivating point mutation in the active site of I-OnuI (E22Q), corresponding to the metal-binding residue in the enzyme’s first LAGLIDADG motif, resulted in dramatic reduction in the GFP recombination frequency (Fig. 3C), as was observed in previous mutational studies of LHEs (26–28). These results indicated that I-OnuI is sufficiently active for use in genome editing applications. Its level of gene conversion activity was comparable to positive controls using the I-SceI LHE and its cognate target.
Engineering and Endogenous Gene Disruption.
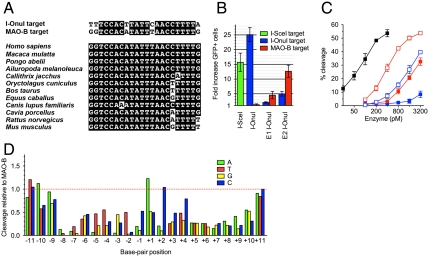
LHE variants that are either computationally engineered or selected through directed evolution for altered DNA cleavage specificity may display compromised activity and/or broadened specificity that might require substantial effort to ameliorate (10–12, 29, 30). To evaluate these effects using an enzyme that recognizes a target site that is closely related to an endogenous human gene target, we engineered I-OnuI to cleave a DNA sequence that is found in the third exon of the human MAO-B gene and that differs from the WT I-OnuI target site at only five base-pair positions (Fig. 4A). MAO-B is one of two monoamine oxidases localized on the mitochondrial outer membrane, where it oxidizes neurotransmitters and dietary amines and produces hydrogen peroxide as a by-product (a known oxidative cytotoxin). This protein is associated with and being studied as a potential therapeutic target for a wide variety of neurodegenerative disorders including Parkinson disease (PD) (31). Pharmacological MAO-B inhibitors appear to slow the progress of PD symptoms through a neuroprotective activity, but the disease-modifying effect and action mechanism of the inhibitors has been controversial (32–36). The ability to generate tissue-specific disruption or modifications of the MAO-B gene might therefore be a valuable tool for future clinical research. The target sequence in MAO-B is completely conserved among some primates and is slightly diverged in the corresponding regions of other mammalian genomes (Fig. 4A).
Fig. 4.
Redesign of I-OnuI to target the human monoamine oxidase B gene. (A) Sequence alignment of the I-OnuI and MAO-B target sites (Upper) or the human MAO-B target and the corresponding sequences coded in other mammalian genomes (Lower). (B) An episomal GFP gene conversion assay was carried out similar to those shown in Fig. 3C. Error bars refer to ± SD of three independent experiments. E1 I-OnuI was selected from directed evolution of I-OnuI in bacteria, and E2 I-OnuI was generated by an addition of E178D in E1 I-OnuI. (C) In vitro cleavage activity was assayed using 32P-labeled DNA substrates. Error bars refer to ± SD of three independent experiments. The I-OnuI target is shown as closed squares and the MAO-B target as open squares; I-OnuI, black; E1 I-OnuI, blue; E2 I-OnuI, red. (D) Cleavage activity of E2 I-OnuI for a target sequence containing a single base-pair substitution from the MAO-B target was tested similarly to that shown in Fig. 3A. A, green; T, red; G, yellow; C, blue.
Directed evolution of I-OnuI endonuclease (see SI Results and Fig. S3 for detail) identified an enzyme variant termed “E1 I-OnuI” that preferentially induced gene conversion in HEK 293T cells on an episomal DR-GFP reporter containing the MAO-B target (Fig. 4B). E1 I-OnuI contained eight amino acid substitutions, of which all except one are located within the protein–DNA interface (Fig. S4A). Addition of an E178D substitution into the active site of E1 I-OnuI, creating a construct termed “E2 I-OnuI,” increased the fraction of GFP positive cells for both of the I-OnuI and MAO-B targets by approximately 3-fold (E178 is one of two residues that coordinate divalent metal ions in the active site: see Fig. S2B), suggesting this substitution primarily enhances the enzyme’s catalysis. Western blotting verified that the engineered enzymes were as stable as the parental enzyme in the transfected cells (Fig. S4C).
Electrophoretic mobility shift assays demonstrated that wild-type I-OnuI preferentially bound its physiological target with a very low dissociation constant (190 ± 15 pM). The E1 and E2 I-OnuI proteins displayed similar affinity for both the WT and MAO-B targets (Table S1); however, these enzymes significantly discriminated between the two target sites in cleavage reactions (Fig. 4C). The relative cleavage activities assayed in vitro correlated well with the GFP gene conversion frequencies that were measured using the DR-GFP reporter. For example, E2 I-OnuI induced GFP gene conversion on the MAO-B target approximately 3-fold more efficiently than E1 I-OnuI and displayed a very similar level of in vitro cleavage activity for the MAO-B target at approximately 4-fold lower enzyme concentrations.
We subsequently determined the DNA sequence specificity profile of E2 I-OnuI across the MAO-B target. Overall, the profile of E2 I-OnuI was very similar to that of the native I-OnuI (Fig. 3A), but specificity was slightly reduced at positions -11, -10, -9, -5, +1, +2, and +11 and appeared to have increased relative to that of native I-OnuI at position -3. The positions of attenuated specificity correlate well with the positions of mismatches between the native I-OnuI target and the MAO-B target (-11, -10, -4, +2, and +11). This suggests that altered specificity as a result of amino acid substitution at the protein–DNA interface is confined to the regions of the altered residues.
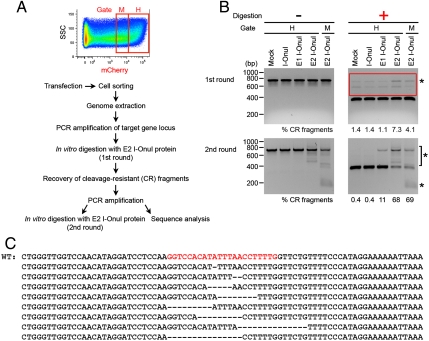
We next tested whether the I-OnuI variants could induce mutagenesis of the endogenous MAO-B gene locus in HEK 293T cells. We transfected the cells with an expression plasmid encoding the I-OnuI variant and sorted the cell population expressing the endonuclease [visualized by coupled expression of mCherry (Fig. 5A)]. The mCherry and the nuclease genes are linked by a 2A peptide sequence from Thosea asigna virus (T2A) (Fig. 3B) and are thereby cotranslated as separate peptide chains through a ribosomal skip mechanism (23). The sorting gates were set to collect approximately the top 25% (gate H) and the following 25% (gate M) of mCherry positive cells (Fig. 5A).
Fig. 5.
Targeted mutagenesis of the endogenous MAO-B gene in human tissue culture cells. (A) Experimental procedure for detection of mutations at the endogenous MAO-B gene locus that are induced by transiently expressed I-OnuI variants. The sorting gates H and M were set to collect approximately the top 25% and the following 25% of mCherry positive cells. The mCherry is cotranslated with I-OnuI variants as a separate protein (Fig. 3B). (B) The impaired, chromosomal MAO-B target was detected by in vitro digestion with E2 I-OnuI recombinant protein, following PCR amplification of the surrounding sequence. Asterisks indicate cleavage-resistant (CR) fragments that are significantly or selectively observed in PCR amplicons from E1 or E2 I-OnuI-transfected cells. DNAs larger than the cleaved fragments in the first round of digestion (included in a red box; Upper Right) were recovered, reamplified, and subjected to the second round of digestion (Lower Panel). (C) CR fragments from E2 I-OnuI-transfected cells (collected in the sorting gate H) were analyzed by sequencing. Small deletions were found within the endogenous MAO-B target site. The intact genome sequence is shown on the top (WT), and the MAO-B target site is in red.
In the absence of a donor DNA template with homology to a break site, DSBs in human cells are primarily repaired by mutagenic pathways involving nonhomologous end joining (NHEJ), which can frequently lead to small insertions or deletions (indels) (3, 37). To detect the indels accumulated at the MAO-B target locus, the genomic DNA was extracted from the sorted cells and used as a template to amplify a DNA sequence that spanned the endogenous MAO-B target. The resulting approximately 700 base-pair fragments were then incubated in vitro with purified E2 I-OnuI recombinant protein in order to cleave intact MAO-B target site. Similar experiments previously allowed us to detect indels induced by the Y2 I-AniI variant at an integrated target in the human genome (38). We observed that a significantly higher fraction (approximately 4 to 7%) of PCR amplicons from E2 I-OnuI-transfected cells was not cleaved by the recombinant endonuclease than those from mock-treated cells (Fig. 5B, Upper Right), suggesting the presence of indels at the target site resulted in cleavage resistance.
To ensure that the CR fragments were not simply due to incomplete in vitro cleavage by E2 I-OnuI, rather than actual indels, the MAO-B target locus was recovered and reamplified from genomic DNA, followed by a second round of digestion. Various-sized PCR fragments were amplified selectively from E2 I-OnuI-transfected cells (Fig. 5B, Lower Left), suggesting that relatively large indels (approximately 100 to 500 base pairs) were induced at the endogenous MAO-B target site of E2 I-OnuI-transfected cells with low frequency, because these fragments were visible only after PCR amplification from the predigested fragments.
The PCR products that were indistinguishable in size from the DNA band containing the intact MAO-B target were then subjected to sequence analyses. Short deletions that were randomly distributed within the MAO-B target were readily identified in the sequences obtained from E2 I-OnuI-transfected cells (8 clones out of 23, sorted in gate H, Fig. 5C), whereas neither indels nor base substitutions were observed in the clones sequenced from mock-treated cells. The second round of in vitro digestion showed that the fraction of cleavage-resistant fragments were enriched in the PCR amplicons from E1 and E2 I-OnuI-transfected cells (Fig. 5B, Lower Right). The mutation frequencies at the endogenous MAO-B target site of the cells transfected with I-OnuI variants were estimated from the results of two rounds of in vitro digestion (Table S2), which indicated that E1 I-OnuI and E2 I-OnuI significantly increased the mutation frequency over the background calculated using mock-treated cells. Taken together, these results indicated the I-OnuI variants indeed induced targeted mutagenesis of the endogenous MAO-B gene locus.
Undesired, off-target cleavage events induced by engineered endonucleases could lead to gene disruption and/or a variety of additional undesirable mutagenic events including carcinogenesis. To test whether off-target cleavage by the WT or E2 I-OnuI enzyme was predictable and measurable on the basis of sequence homology to a desired chromosomal target site, we conducted a BLAST search for DNA sequences in the human genome that are similar to the central 18 base-pair sequence of the MAO-B target site. We concentrated on these positions because these endonucleases (particularly E2 I-OnuI) were already known to relatively tolerate single base-pair mismatches at the most distal base-pair positions in their respective targets during catalysis (base-pair positions ± 10 and ± 11; see Figs. 3A and 4D). In the list of the off-target sites shown in Table S3, four potentially cleavable chromosomal loci were investigated by the same assays used to detect indels at the endogenous MAO-B locus. None of these sites were located within protein-coding regions. E2 I-OnuI increased the intensity of CR fragments at off-target sites #1 and #2, while WT I-OnuI accumulated the CR fragment at the off-target site #2 (Fig. S5). In contrast, no CR fragment induced by either enzyme was significantly detected at the off-target sites #6 and #7 over the slightly high background in the single round of digestion with E2 I-OnuI protein, although an additional in vitro digestion of the reamplified CR fragments is required to detect infrequent indels (as performed for the MAO-B locus). These results suggest that potential off-target cleavage sites can be predicted by a sequence homology search of genome sequence, and the presence of off-target cleavage product caused by an engineered endonuclease can be assayed.
Discussion
While many engineered ZFNs have been used successfully to modify chromosomal target loci in a variety of multicellular eukaryotic organisms (5, 39), to date only the homodimeric I-CreI enzyme has been engineered to modify endogenous chromosomal loci (11, 12). However, the properties of monomeric LHEs argue for their continued development as genome-modifying enzymes: They are encoded by particularly short open reading frames that are less than 1,000 base pairs in length, and their cleavage specificities are often very high (18–22). In addition, site-specific DNA nicking enzymes can be generated from monomeric LHEs (28, 40). One such enzyme, derived from I-AniI, was shown to promote homologous recombination while reducing both cellular toxicity and NHEJ-induced mutagenesis compared to the parental scaffold (38).
The analysis described in this paper indicates that a significant fraction of putative monomeric LHEs, chosen strictly on the basis of sequence similarity to previously characterized endonucleases, may have retained sufficient stability and activity to serve as protein scaffolds for engineering and future gene targeting. For those enzymes that have accumulated mutations that reduce their stability or activity, random mutagenesis might be employed to rescue compromised function, as illustrated by prior studies of the WT I-AniI and DmoCre hybrid enzyme (10, 41). Because the endonucleases within the I-OnuI subfamily appear to have recently diverged during their search for and invasion of a variety of “homing” sites, it is possible that many of them have retained most of their key residues for DNA binding and enzymatic activity.
The concept of tapping the reservoir of natural diversity within the LHE family has been recently described (42) as an attractive alternative to extensive protein engineering. In that study, the tendency of LHEs (in particular, those associated with inteins) to tolerate base substitutions in their DNA target sites that correspond to degenerate or “wobble” positions in their host genes (which lead to synonymous or neutral mutations in the host gene product) was documented. This correlation, if generalizable, might further facilitate homing endonuclease engineering, by allowing the prediction of base-pair mismatches in DNA target sites that are naturally tolerated by wild-type LHEs. The specificity profile of I-OnuI partially supports this analysis (Fig. 3A); slightly reduced fidelity is observed at three wobble positions (base pairs -6, -3, and +4), relative to that at the immediately neighboring positions in its target site. However, this trend is clearly rather weak: Several individual base-pair substitutions at these positions are still strongly disfavored by the wild-type enzyme, and one of the least specifically recognized positions in the central region of the target site is actually located at a nonwobble position in the host gene (at base pair -4).
Although we have focused on characterization of only the I-OnuI subfamily in this study, identification of target sequences for additional LHEs will allow us to evaluate sequence recognition diversity within each subfamily. We anticipate that characterization of a wide range of monomeric LHEs will accelerate their continued development and application for genome editing, particularly when combined with protein chimerization and DNA shuffling approaches, eventually allowing coverage of DNA sequence space by engineered homing endonucleases at a much increased density and success rate.
Materials and Methods
See detailed SI Materials and Methods. Putative single-chain LAGLIDADG homing endonuclease sequences were collected and aligned against the structure of I-AniI (16), using Cn3D (http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml). Target sequences for individual LHEs were predicted through comparison of the LHE-harboring host gene to related genes lacking an endonuclease and verified using a DNA binding and cleavage assay conducted on enzyme displayed on yeast surface (19). Verification and sequencing of cleaved DNA products were subsequently performed in vitro using free solubilized enzyme and linearized DNA plasmids containing the appropriate target site. I-OnuI was both subcloned in a GST fusion expression vector (pGEX6P-3) and purified to homogeneity using glutathione affinity chromatography; I-LtrI was purified using metal affinity chromatography as previously described (7). Measurements of DNA binding affinity and cleavage were performed using radiolabeled 34 base-pair synthetic DNA substrates that contain each enzymes’ natural target sequence. Crystals of I-OnuI and I-LtrI were obtained in the presence of their cognate DNA targets. Statistics for the crystallographic data are shown in Table S4. Mutagenesis and selection of I-OnuI variants that cleave and disrupt the human MAO-B gene were conducted using protocols for selection described in ref. 41.
Supplementary Material
Acknowledgments.
X-ray data were collected at the Advanced Light Source synchrotron facility at the Lawrence Berkeley National Laboratory on beamlines 5.0.1, 5.0.2, and 8.2.1 with the assistance of multiple staff. We thank Michael Certo (Seattle Children’s Research Institute, Seattle, WA) for providing I-SceI expression plasmid used in episomal GFP reporter gene conversion assays and additional members of the Stoddard and Scharenberg labs for invaluable advice and assistance. This work was supported by the National Institutes of Health Grants R01 GM49857 and RL1 CA133833 (to B.L.S.) and RL1 CA133832 (to A.M.S.), the Gates Foundation Grand Challenge Program (to A.M.S. and B.L.S.), the Canadian Institutes of Health Research (MOP 977800) and pilot project funding from the Northwest Genome Engineering Consortium (to D.R.E.), postdoctoral support from the Interdisciplinary Training in Genome Engineering Program (to R.T., A.R.L., and A.N.-S.M.) and support from the Japan Society for the Promotion of Science (to R.T.).
Footnotes
Conflict of interest statement: B.L.S. and A.M.S. are founders of a biotechnology company (Precision Genome Engineering) that conducts engineering and selection studies on homing endonucleases for gene targeting.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factor amplitudes corresponding to the DNA-bound structures of I-OnuI and I-LtrI have been deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank, www.rcsb.org/pdb (RCSB ID codes 3QQY and 3R7P, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107719108/-/DCSupplemental.
References
- 1.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li T, et al. TAL nucleases (TALNs): Hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2010;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcaida MJ, Munoz IG, Blanco FJ, Prieto J, Montoya G. Homing endonucleases: from basics to therapeutic applications. Cell Mol Life Sci. 2010;67:727–748. doi: 10.1007/s00018-009-0188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoddard BL. Homing endonucleases: From microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 6.Stoddard BL. Homing endonuclease structure and function. Q Rev Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 7.Arnould S, et al. The I-CreI meganuclease and its engineered derivatives: Applications from cell modification to gene therapy. Protein Eng Des Sel. 2010;24:27–31. doi: 10.1093/protein/gzq083. [DOI] [PubMed] [Google Scholar]
- 8.Paques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: Perspectives for gene therapy. Curr Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 9.Stoddard BL, Scharenberg AM, Monnat RJ., Jr . Advances in engineering homing endonucleases for gene targeting: Ten years after structures. In: Bertolotti R, Ozawa K, editors. Progress in Gene Therapy: Autologous and Cancer Stem Cell Gene Therapy. World Scientific eBooks; 2007. pp. 135–167. [Google Scholar]
- 10.Grizot S, et al. Generation of redesigned homing endonucleases comprising DNA-binding domains derived from two different scaffolds. Nucleic Acids Res. 2010;38:2006–2018. doi: 10.1093/nar/gkp1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao H, et al. Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 2010;61:176–187. doi: 10.1111/j.1365-313X.2009.04041.x. [DOI] [PubMed] [Google Scholar]
- 12.Grizot S, et al. Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 2009;37:5405–5419. doi: 10.1093/nar/gkp548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sethuraman J, Majer A, Friedrich NC, Edgell DR, Hausner G. Genes within genes: Multiple LAGLIDADG homing endonucleases target the ribosomal protein S3 gene encoded within an rnl group I intron of Ophiostoma and related taxa. Mol Biol Evol. 2009;26:2299–2315. doi: 10.1093/molbev/msp145. [DOI] [PubMed] [Google Scholar]
- 14.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 15.Mullineux ST, Costa M, Bassi GS, Michel F, Hausner G. A group II intron encodes a functional LAGLIDADG homing endonuclease and self-splices under moderate temperature and ionic conditions. RNA. 2010;16:1818–1831. doi: 10.1261/rna.2184010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolduc JM, et al. Structural and biochemical analyses of DNA and RNA binding by a bifunctional homing endonuclease and group I intron splicing factor. Genes Dev. 2003;17:2875–2888. doi: 10.1101/gad.1109003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moure CM, Gimble FS, Quiocho FA. The crystal structure of the gene targeting homing endonuclease I-SceI reveals the origins of its target site specificity. J Mol Biol. 2003;334:685–695. doi: 10.1016/j.jmb.2003.09.068. [DOI] [PubMed] [Google Scholar]
- 18.Scalley-Kim M, McConnell-Smith A, Stoddard BL. Coevolution of a homing endonuclease and its host target sequence. J Mol Biol. 2007;372:1305–1319. doi: 10.1016/j.jmb.2007.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarjour J, et al. High-resolution profiling of homing endonuclease binding and catalytic specificity using yeast surface display. Nucleic Acids Res. 2009;37:6871–6880. doi: 10.1093/nar/gkp726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thyme SB, et al. Exploitation of binding energy for catalysis and design. Nature. 2009;461:1300–1304. doi: 10.1038/nature08508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Argast GM, Stephens KM, Emond MJ, Monnat RJ., Jr I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J Mol Biol. 1998;280:345–353. doi: 10.1006/jmbi.1998.1886. [DOI] [PubMed] [Google Scholar]
- 22.Chevalier B, Turmel M, Lemieux C, Monnat RJ, Jr, Stoddard BL. Flexible DNA target site recognition by divergent homing endonuclease isoschizomers I-CreI and I-MsoI. J Mol Biol. 2003;329:253–269. doi: 10.1016/s0022-2836(03)00447-9. [DOI] [PubMed] [Google Scholar]
- 23.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 24.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burt A, Koufopanou V. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr Opin Genet Dev. 2004;14:609–615. doi: 10.1016/j.gde.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Zhao H. A highly sensitive selection method for directed evolution of homing endonucleases. Nucleic Acids Res. 2005;33:e154. doi: 10.1093/nar/gni148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chevalier B, et al. Metal-dependent DNA cleavage mechanism of the I-CreI LAGLIDADG homing endonuclease. Biochemistry. 2004;43:14015–14026. doi: 10.1021/bi048970c. [DOI] [PubMed] [Google Scholar]
- 28.McConnell Smith A, et al. Generation of a nicking enzyme that stimulates site-specific gene conversion from the I-AniI LAGLIDADG homing endonuclease. Proc Natl Acad Sci USA. 2009;106:5099–5104. doi: 10.1073/pnas.0810588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashworth J, et al. Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs. Nucleic Acids Res. 2010;38:5601–5608. doi: 10.1093/nar/gkq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redondo P, et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- 31.Youdim MB, Edmondson D, Tipton KF. The therapeutic potential of monoamine oxidase inhibitors. Nat Rev Neurosci. 2006;7:295–309. doi: 10.1038/nrn1883. [DOI] [PubMed] [Google Scholar]
- 32.Deftereos SN, Andronis CA. Discordant effects of rasagiline doses in Parkinson disease. Nat Rev Neurol. 2010;6 doi: 10.1038/nrneurol.2010.2-c1. 10.1038/nrneurol.2010.2-c1. [DOI] [PubMed] [Google Scholar]
- 33.Hauser RA. Early pharmacologic treatment in Parkinson’s disease. Am J Manag Care. 2010;16:S100–S107. [PubMed] [Google Scholar]
- 34.Olanow CW, et al. A double-blind, delayed-start trial of rasagiline in Parkinson’s disease. N Engl J Med. 2009;361:1268–1278. doi: 10.1056/NEJMoa0809335. [DOI] [PubMed] [Google Scholar]
- 35.Sampaio C, Ferreira JJ. Parkinson disease: ADAGIO trial hints that rasagiline slows disease progression. Nat Rev Neurol. 2010;6:126–128. doi: 10.1038/nrneurol.2010.2. [DOI] [PubMed] [Google Scholar]
- 36.Youdim MB. Rasagiline in Parkinson’s disease. N Engl J Med. 2010;362:657–658. doi: 10.1056/NEJMc0910491. [DOI] [PubMed] [Google Scholar]
- 37.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metzger MJ, McConnell-Smith A, Stoddard BL, Miller AD. Single-strand nicks induce homologous recombination with less toxicity than double-strand breaks using an AAV vector template. Nucleic Acids Res. 2011;39:926–935. doi: 10.1093/nar/gkq826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le Provost F, et al. Zinc finger nuclease technology heralds a new era in mammalian transgenesis. Trends Biotechnol. 2010;28:134–141. doi: 10.1016/j.tibtech.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Niu Y, Tenney K, Li H, Gimble FS. Engineering variants of the I-SceI homing endonuclease with strand-specific and site-specific DNA-nicking activity. J Mol Biol. 2008;382:188–202. doi: 10.1016/j.jmb.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeuchi R, Certo M, Caprara MG, Scharenberg AM, Stoddard BL. Optimization of in vivo activity of a bifunctional homing endonuclease and maturase reverses evolutionary degradation. Nucleic Acids Res. 2009;37:877–890. doi: 10.1093/nar/gkn1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barzel A, et al. Native homing endonucleases can target conserved genes in humans and in animal models. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr242. 10.1093/nar/gkr242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.