Abstract
5-Hydroxymethylcytosine (5-hmC) is a newly discovered DNA base in mammalian cells that is believed to be another important epigenetic modification. Here we report the use of a methylation-insensitive restriction enzyme TaqαI coupled with selective chemical labeling of 5-hmC in a Combined Glycosylation Restriction Analysis (CGRA) to detect 5-hmC in TCGA sequences. This method, differentiates fully versus hemi-hydroxymethylated cytosine in the CpG dinucleotide, adds a new tool to facilitate biological studies of 5-hmC.
Keywords: 5-Hydroxymethylcytosine, Selective chemical labeling, Restriction enzyme assay
In addition to the four standard DNA bases, base modifications can add extra epigenetic information to the DNA that is not encoded in the sequence. For example, in mammalian genome, 5-methylcytosine (5-mC), which constitutes about 1% of total nucleotide and mainly occurs in the CpG dinucleotide, has a profound effect on gene regulation and dysregulation.1–3 In 2009, two groups independently reported the existence of 5-hydroxymethylcytosine (5-hmC) in mammalian genomic DNA.4, 5 Meanwhile the TET family enzymes, which are methylcytosine dioxygenases, have been identified to oxidize 5-mC to 5-hmC, and play roles in embryonic stem (ES) cell regulation and myelopoiesis.5–8 These studies alter our perception of how DNA methylation impacts mammalian cells by suggesting that 5-hmC might also play an important role in epigenetic regulation.
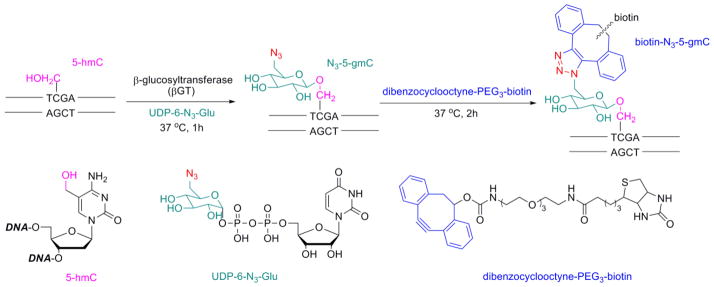
To further reveal the function of 5-hmC, sensitive detection and sequencing methods are required, as the widely used sequencing methods for 5-mC, such as bisulfite sequencing and methylation-sensitive restriction digestion, cannot discrimination between the two cytosine modifications.9–12 Commercially available anti-5-hmC antibodies are unlikely to provide high-resolution sequencing of 5-hmC. Recently, we have developed a chemical labeling method to selectively label 5-hmC with an azide-modified glucose by β-glucosyltransferase (βGT). A subsequent coupling of the azide group with an alkyne-biotin probe allows detection of 5-hmC in genomic DNA (Fig. 1).13 With this method, 5-hmC-containing genomic DNA fragments can be enriched and sequenced to provide the genomic distribution of this modification.13 However, a high-throughput method for single-base resolution detection of 5-hmC has yet to be achieved.
Figure 1.
The βGT-catalyzed formation of N3-5-gmC and the subsequent click chemistry to yield biotin-N3-5-gmC on the TCGA site in duplex DNA. Modification on only one strand is shown.
The use of methylation-sensitive restriction enzymes is a classic approach to the study of DNA methylation at specific loci.14 However, due to the similar size of the methyl and hydroxymethyl groups, 5-mC and 5-hmC are indistinguishable to most restriction enzymes.10, 11, 15, 16 We anticipated that after adding the bulky glucose group or a subsequent biotin attachment to 5-hmC, the glucosylated base would have different properties from 5-mC.17 For example, the resulting 5-N3-gmC or biotin-5-N3-gmC in a duplex DNA may be able to block digestion from the methylation-insensitive restriction enzyme, which can digest both 5-hmC- and 5-mC-containing DNA. While we are developing this method, two companies, Zymo Research and New England Biolabs, have launched products based on this combined glycosylation restriction analysis (CGRA). They utilize βGT to transfer a regular glucose to 5-hmC and show that it can block the methylation-insensitive restriction enzyme MspI, which has a recognized sequence of C^CGG.18 Although the use of MspI in CGRA can detect the presence of 5-hmC on CCGG site, it has several limitations: i) MspI is also blocked if the outer C is 5-mC or 5-hmC, regardless of the cytosine modification status of the inner C, which limits the use of this approach on many CCGG sites where the outer C methylated;15 ii) it cannot tell whether 5-hmC occurs on only one strand or both strands of the CpG dinucleotide (see below). Herein, we show that TaqαI, another methylation-insensitive restriction enzyme that recognizes and cuts T^CGA, can also be used in CGRA when coupled with our chemical labeling method. This new approach can differentiate fully versus hemi-hydroxymethylated states in the CpG dinucleotide.
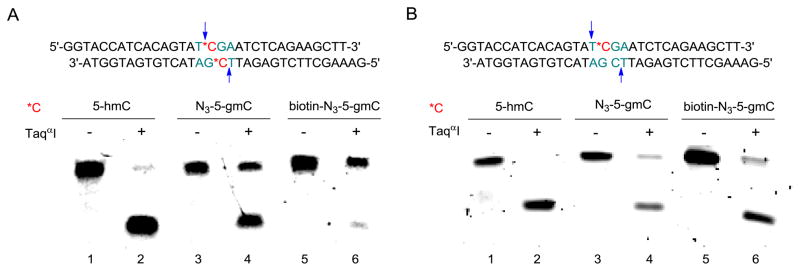
We first synthesized a 32-mer double strand DNA bearing T*CGA (*C = 5-hmC) on both strands (Fig. 2A). Instead of glucose, we employed βGT to transfer our chemically modified 6-N3-glucose, onto 5-hmC (Fig. 1). Then we subjected the modified DNA to TaqαI-mediated digestion. As expected, TaqαI can completely cut unmodified 5-hmC, but only partially for N3-5-gmC. (Fig. 2A, lane 1-4).17 The relatively high tolerance of TaqαI to the cytosine modification requires more bulky modifications in order to achieve a satisfactory difference in CGRA. The presence of azide group on the glucose allows us to add further modification using click chemistry.19–21 We therefore coupled N3-5-gmC with dibenzocyclooctyne-modified biotin using copper-free click chemistry to introduce a sterically bulky dibenzocyclooctyne moiety (Fig. 1).22, 23 To our delight, when we applied the biotin-N3-5-gmC modification into TaqαI digestion, it showed an almost complete blocking effect (Fig. 2A, lane 5–6).
Figure 2.
TaqαI-mediated digestion of 5-hmC-, N3-5-gmC-, and biotin-N3-5-gmC-containing DNA with the sequences showing on top. *C indicates the modified position; arrows indicate TaqαI cutting sites. (A), Digestion of fully-modified DNA. (B), Digestion of hemi-modified DNA. The 32-mer dsDNA (1 pmol) was digested with 100 U of TaqαI (New England BioLabs) for 1 hr at 65 °C. Samples were analyzed by 16% PAGE/Urea gel and visualized using SYBR Green I staining (Lumiprobe).
Due to the semi-conservative DNA replication, besides a full methylation state, hemi-methylation state also exists in mammalian genome. The conversion of 5-mC to 5-hmC suggests that fully and hemi-hydroxymethylation states may also exist in the mammalian genome. If this is the case, developing a method to distinguish between these two states will be very important to understanding the formation of 5-hmC and conversion process between 5-mC and 5-hmC. Since the blocking efficiency of TaqαI is largely dependent on the size of the modification group on the hydroxyl group, we wondered if TaqαI can behave differently over fully and hemi-hyroxymethylation states. We prepared the same 32-mer double strand DNA with hemi-hyroxymethylation, performed the same labeling procedure, and subjected to TaqαI digestion (Fig. 2B). While the hemi-5-hmC can be cut, hemi-N3-5-glocose cannot block digestion as well as the fully-modified one (Fig. 2B, lane 1-4). Even with the bulkier group, biotin-N3-5-gmC, present, the majority of DNA was still digested (Fig. 2B, lane 5-6). Thus, TaqαI digests the hemi-modified sequence but is blocked by the fully-modified one with biotin-N3-5-gmC. This noticeable difference of TaqαI in response to fully- and hemi-hydroxymethylation states after modification provides a method to distinguish these two states on TCGA sites.
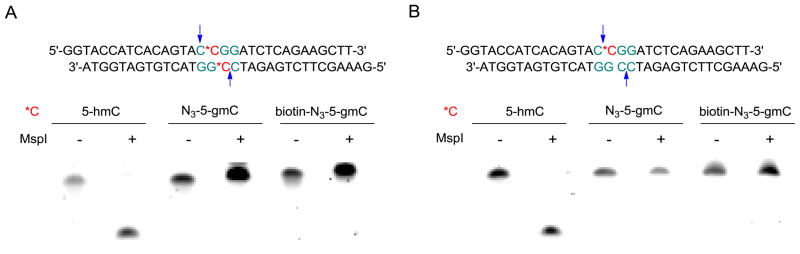
To further investigate if the difference in digestion between fully and hemi-hydroxymethylation states is universal in restriction enzymes, we replaced the T*CGA site with C*CGG (*C = 5-hmC) in the previous 32-mer duplex DNA and performed the same assays on MspI (Fig. 3). Both fully and hemi-5-hmC can be cut by MspI completely before modification (Fig. 3A, lane 1-2 and Fig. 3B, lane 1-2). For a fully 5-hmC site, both N3-gmC and biotin-N3-5-gmC can block MspI digestion completely as expected (Fig. 3A, lane 3-6), suggesting that MspI is more sensitive towards the steric hindrance of cytosine modification of the CpG dinucleotide; the presence of a glucose group on the inner C is enough for the protection from digestion. For the hemi-5-hmC site, it gave the same results as the fully-modified one: both the N3-5-gmC and biotin-N3-5-gmC can be protected from digestion (Fig. 3B, lane 3-6). This result is in accordance with our assumption that MspI is much easier to block and that it cannot distinguish the fully-modified 5-hmC site from hemi-5-hmC site as TaqαI.
Figure 3.
MspI digestion of 5-hmC-, N3-5-gmC-, and biotin-N3-5-gmC-containing DNA with the sequences showing on top. *C indicates the modified position; arrows indicate MspI cutting sites. (A), Digestion of fully-modified DNA. (B), Digestion of hemi-modified DNA. The 32-mer dsDNA (1 pmol) was digested with 100 U of MspI (New England BioLabs) for 1 hr at 37 °C. Samples were analyzed by 16% PAGE/Urea gel and visualized using SYBR Green I staining (Lumiprobe).
In conclusion, we discovered the formerly known methyl-insensitive restriction enzyme TaqαI can be blocked by a biotin-N3-5-gmC modification in order to detect the existence of 5-hmC on TCGA sites in CGRA. Furthermore, TaqαI is unique in that it is almost completely inhibited by the fully-modified biotin-N3-gmC site while cutting most of the hemi-modified site, therefore may be used to distinguish fully versus hemi-hydroxymethylation sites on specific loci. Given the possible importance of 5-hmC in gene regulation, the development of this detection method will greatly facilitate future epigenetic studies. Work to employ this assay to discover 5-hmC in real biological samples is currently underway in our laboratory.
Acknowledgments
This work was partially supported by the National Institutes of Health (GM 071440 to C.H.). We thank Sarah Frank Reichard, M.A. for help with editing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goll MG, Bestor TH. Annu Rev Biochem. 2005;74:481. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 2.Reik W. Nature. 2007;447:425. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 3.Gal-Yam EN, Saito Y, Egger G, Jones PA. Annu Rev Med. 2008;59:267. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 4.Kriaucionis S, Heintz N. Science. 2009;324:929. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Science. 2009;324:930. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Nature. 2010;466:1129. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP, Rao A. Nature. 2010;468:839. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A. Cell Stem Cell. 2011;8:200. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin SG, Kadam S, Pfeifer GP. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nestor C, Ruzov A, Meehan RR, Dunican DS. BioTechniques. 2010;48:317. doi: 10.2144/000113403. [DOI] [PubMed] [Google Scholar]
- 12.Zheng Y, Cohen-Karni D, Xu D, Chin HG, Wilson G, Pradhan S, Roberts RJ. Nucleic Acids Res. 2010;38:5527. doi: 10.1093/nar/gkq327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song C-X, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen C-H, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Nat Biotechnol. 2011;29:68. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singer-Sam J, Grant M, LeBon JM, Okuyama K, Chapman V, Monk M, Riggs AD. Mol Cell Biol. 1990;10:4987. doi: 10.1128/mcb.10.9.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tardy-Planechaud S, Fujimoto J, Lin SS, Sowers LC. Nucleic Acids Res. 1997;25:553. doi: 10.1093/nar/25.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szwagierczak A, Brachmann A, Schmidt CS, Bultmann S, Leonhardt H, Spada F. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang LH, Farnet CM, Ehrlich KC, Ehrlich M. Nucleic Acids Res. 1982;10:1579. doi: 10.1093/nar/10.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis T, Vaisvila R. J Vis Exp. 2011;48 doi: 10.3791/2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem, Int Ed. 2002;41:2596. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 20.Sletten EM, Bertozzi CR. Angew Chem, Int Ed. 2009;48:6974. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Speers AE, Cravatt BF. Chem Biol. 2004;11:535. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Ning X, Guo J, Wolfert MA, Boons GJ. Angew Chem, Int Ed. 2008;47:2253. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jewett JC, Bertozzi CR. Chem Soc Rev. 2010;39:1272. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]