Abstract
Cdc42, a member of the Rho GTPase family, is a fundamental regulator of the actin cytoskeleton during cell migration. To generate a sensor for Cdc42 activation, we employed a multi-pronged approach, utilizing cysteine labeling and expressed protein ligation, to incorporate the environment sensitive fluorophore 4-N,N-dimethylamino-1,8-naphthalimide (4-DMN) into the GTPase binding domain of the WASP protein. These constructs bind only the active, GTP-bound conformation of Cdc42 to produce a fluorescence signal. Studies with a panel of five sensor analogs revealed a derivative that exhibits a 32-fold increase in fluorescence intensity in the presence of activated Cdc42 compared to incubation with the inactive GDP-bound form of the protein. We demonstrate that this sensor can be exploited to monitor Cdc42 nucleotide exchange and GTPase activity in a continuous, fluorescence assay.
Keywords: Biosensors, Protein design, Expressed protein ligation, Cysteine labeling, Solvatochromic fluorophores
Fluorescent probes have been used to monitor a variety of dynamic signaling interactions, including protein-protein interactions, enzyme activation, and changes in protein conformation.1, 2 In addition to enabling biochemical studies in real-time, these tools have been utilized in high-thoughput assays to screen for new small molecule drugs and inhibitors.3, 4 These methods generally rely on a change in a fluorescent read out, such as FRET, anisotropy, fluorescence lifetime, quantum yield, or emission wavelength.5 Solvatochromic fluorophores, which have emissive properties that are dependent on the nature of the solvent environment, are ideal for measuring these interactions because they are capable of providing information on even subtle changes in protein conformation.2 While many such fluorophores are commercially available, we have been particularly interested in the development and application of those derived from the dimethylaminonaphthalimide family.6–9 Specifically, 4-N,N-dimethylamino-1,8-naphthalimide (4-DMN) has recently been demonstrated to be a powerful probe of protein binding interactions.10 When the fluorophore is in an unbound, water-exposed state, the quantum yields of fluorescence are exceedingly low. However, upon exposure to a hydrophobic environment, such as that at the interface of a protein-protein interaction, increases in fluorescence can exceed 100-fold. Thus, when the fluorophore is appropriately positioned in a sensor protein, it behaves as an “off-on” switch to indicate a binding event. The 4-DMN chromophore can be readily incorporated into peptides and proteins either as an amino acid building block10 or through cysteine labeling with maleimide or α-bromoacetamide derivatives.11 Herein, we exploit the unique properties of this fluorophore to generate a sensor for the Rho GTPase Cdc42 (Figure 1).
Figure 1.
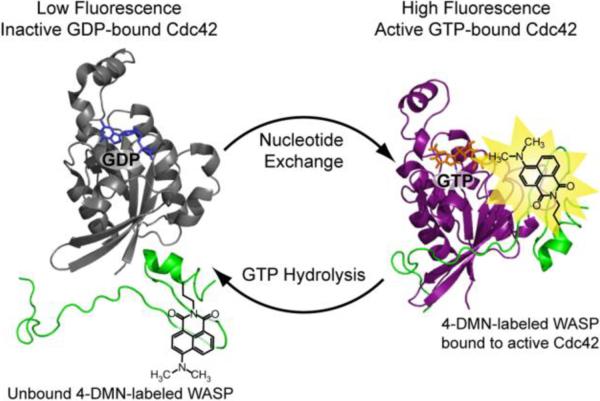
4-DMN-Based sensor for active GTP-bound Cdc42. Labeling of the WASP fragment with 4-DMN generates a sensor that discriminates between the inactive and active forms of Cdc42 through a fluorescence increase. Image was modified from Protein Data Bank files 2WMN and 1CEE.
Rho GTPases are small proteins central to the regulation of the actin cytoskeleton and cell migration. Alternating between an inactive GDP-bound and active GTP-bound state, these proteins function as molecular switches through transient interactions with effector proteins.12 One prominent member of the Rho GTPases is Cdc42, which is responsible for the reorganization of actin filaments into parallel bundles for filopodial protrusions. Additionally, Cdc42 activation generates cell polarity, a critical step in cell migration.13 Due to the fundamental role of Cdc42 in migratory processes, methods to monitor the real-time activity of the protein in vitro and in cells are essential.
Although FRET-based sensors have been developed for cellular applications, these methods use recombinant Cdc4214 or require overexpression of the protein.15 However, a particularly elegant sensor, generated by incorporating an environment sensitive fluorophore onto a protein fragment known to bind only the activated conformation of Cdc42, is capable of probing endogenous levels of wild-type Cdc42. This sensor, which was derived from the Cdc42/Rac interactive binding (CRIB) domain of the Wiscott-Aldrich syndrome protein (WASP), was modified with a merocyanine dye and exhibited a 3-fold increase in fluorescence intensity in the presence of activated Cdc42 compared to incubation with the inactive form of the protein.16 Due to the long excitation and emission wavelengths and high extinction coefficient of the merocyanine fluorophore, this sensor has been successfully used in live cell imaging studies and has been applied in combination with FRET-based sensors of the RhoA and Rac GTPases to interrogate activity at cell protrusions.17 To extend these studies and to exploit the high environment sensitivity of the 4-DMN fluorophore, we developed a complementary sensor for Cdc42 with 4-DMN and used it to monitor in vitro Cdc42 activity. We anticipated that the 4-DMN fluorophore would provide advantages for in vitro studies due to excellent signal-to-noise ratios and could be readily adapted for high-throughput screening.
The Cdc42 sensor was based on the CRIB domain of the WASP protein (residues 230–277) and incorporates the 4-DMN fluorophore in place of phenylalanine at position 271, a site previously reported to undergo changes in solvent environment upon interaction with Cdc42(GTP) due to exposure to hydrophobic interactions at the binding interface.16, 18 To increase heterologous expression levels and protein stability, the GB1 protein19 was fused to the N-terminus of the sensor, and to facilitate purification, a hexahistidine tag was incorporated at the C-terminus. Because the length of the linker tethering the fluorophore to the protein can significantly affect the fluorescent properties of the sensor,11 we tested a panel of five analogs, which append the 4-DMN fluorophore to the sensor with varying linker lengths, ranging from no linker to chains between 4 and 9 Å. We developed two complementary approaches for the site-specific incorporation of 4-DMN into the WASP fragment (Figure 2). In the first strategy, we examined a cysteine-labeling method using previously reported 4-DMN-modified α-bromoacetamide and maleimide reagents,11 and in an alternative approach, we exploited protein semisynthesis to incorporate the 4-N,N-dimethylamino-1,8-naphthalimido-alanine amino acid (4-DMNA), which eliminates the need for a linker.10 Because this amino acid derivative is similar in size to tryptophan, this route introduces a more conservative mutation, and potential structural perturbations are minimized. We then compared the fluorescence intensities of the sensors in the presence of inactive GDP-bound and active GTP-γS-bound Cdc42.
Figure 2.
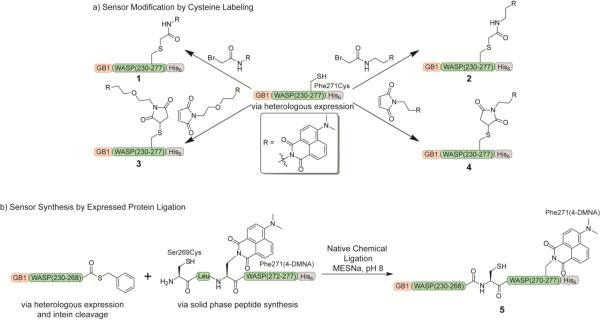
Approaches for the synthesis of 4-DMN-labeled Cdc42 sensors. a) Generation of the sensor by cysteine labeling with 4-DMN-modified α-bromoacetamides and maleimides. The GB1-WASP-His6 protein was obtained through heterologous expression in E. coli, and Cys271 was labeled with 4-DMN reagents of varying linker length. b) Incorporation of the 4-DMN amino acid into the sensor through expressed protein ligation. The N-terminal portion of the sensor was expressed in E. coli as a fusion to an intein, allowing cleavage with sodium 2-mercaptoethanesulfonate (MESNa) to yield the corresponding thioester, and a peptide containing the 4-DMNA amino acid was generated by Fmoc-based solid phase peptide synthesis. Reaction of the recombinant and peptide fragments in the native chemical ligation reaction yielded the full-length sensor.
With the 4-DMN labeling strategy, we expressed the GB1-WASP protein with a Phe271Cys mutation to enable selective labeling of the protein sensor with 4-DMN thiol-reactive reagents (Figure 2a). Following expression in E. coli and purification by Ni-NTA affinity chromatography, the protein was labeled through an overnight incubation with a 20-fold excess of the cysteine-modifying reagent in the presence of TCEP. Purification of the labeling reactions from excess fluorophore by desalting yielded 1, 2, 3, and 4. Labeling efficiencies for all derivatives routinely reached around 60–70% (Figure S1).
The second approach to incorporation of the 4-DMN chromophore utilized expressed protein ligation, in which the N-terminal portion of the protein was prepared as a thioester, and the C-terminal fragment of the sensor containing the 4-DMNA amino acid was synthesized by Fmoc-based solid phase peptide synthesis (Figure 2b). The recombinant fragment (GB1-WASP(230–268)) was expressed as a fusion to intein and chitin binding domains to enable on-resin cleavage with sodium 2-mercaptoethanesulfonate to yield the corresponding thioester. Native chemical ligation between this thioester and the N-terminal cysteine of the synthetic peptide furnished the full-length sensor at about 60% conversion relative to unligated protein (Figure S2). The sensor was then purified from the unligated protein by Ni-NTA affinity chromatography, and excess peptide was eliminated by size exclusion chromatography. In addition to incorporation of the 4-DMNA residue, this approach offers the ability to integrate other functional handles into the C-terminal region of the sensor.
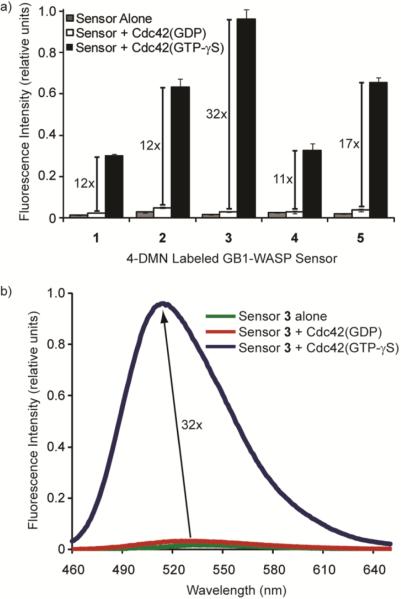
We next examined the fluorescent responses of the 4-DMN-based sensors to Cdc42 (Figures 3, S3). We expressed Cdc42 heterologously in E. coli, and loaded the purified protein with either GDP or GTP-γS, a non-hydrolyzable analog of GTP. Incubation of each 4-DMN sensor with the inactive GDP-bound form of the protein resulted in a negligible 1.1 – 1.9-fold increase in fluorescence over that of the sensor alone. However, dramatic increases in fluorescence, ranging from 11- to 32-fold, were observed in the presence of the active GTP-γS-bound Cdc42 compared to incubation with the inactive form of the protein (Figure 3a). Thus, the sensor readily discriminates between the two states of the protein and effectively acts as a signal of Cdc42 activation. Concurrent with the increase in fluorescence intensity was a hypsochromic shift in the emission maximum from 534 nm in the absence of Cdc42 to 514 nm in the presence of the activated protein.
Figure 3.
Fluorescence properties of the 4-DMN-based sensor for activated Cdc42. a) The relative fluorescence intensity of each sensor derivative (1 – 5) alone and in the presence of Cdc42(GDP) and Cdc42(GTP-γS) is plotted. The fold differences between fluorescence with active and inactive Cdc42 are noted. b) Fluorescence spectra of sensor 3 alone and after incubation with Cdc42(GDP) and Cdc42(GTP-γS). Measurements were taken with 150 nM sensor and 700 nM Cdc42 at 25 °C.
Whereas the fluorescence of all five sensors was similarly low in the presence of the inactive GDP-bound protein, deviations in fluorescence intensities appeared upon Cdc42 activation. Of all the derivatives, probe 3 exhibited the most promising properties with a 32-fold increase in emission intensity and the strongest fluorescence signal upon binding activated Cdc42 (Figure 3b). In this construct, the long 9-Å linker provides the greatest flexibility for the 4-DMN fluorophore, likely allowing it to better embed within the hydrophobic interface of the two proteins. Importantly, because signal from the sensor alone or in the presence of Cdc42(GDP) is extremely low, background fluorescence is virtually non-existent. This switch-like property of the 4-DMN sensor is particularly useful because significant fluorescence differences can be observed without removing excess, unbound sensor before analysis. Additionally, we examined a double mutant of the sensor (His246Asp, His249Asp), which cannot bind Cdc42. This labeled mutant did not demonstrate fluorescence increases, even in the presence of Cdc42(GTP-γS), confirming that the fluorescence differences with 1 – 5 resulted from the sensor recognizing activated Cdc42 (Figure S4).
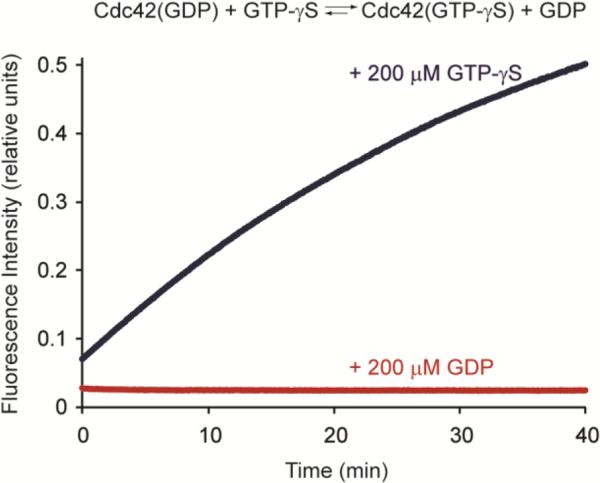
After demonstrating that the sensors exhibit excellent fluorescent responses to activated Cdc42, we explored the use of these tools to monitor nucleotide exchange and GTPase activity in real-time. We envisioned that this would provide a convenient method to measure the in vitro activity of Cdc42 and interactions with guanine nucleotide exchange factors (GEFs), which activate Cdc42 by exchanging GDP for GTP, and GTPase activating proteins (GAPs), which accelerate the rate of GTP hydrolysis. In the case of nucleotide exchange, the addition of GTP-γS to a solution of Cdc42 pre-equilibrated with GDP resulted in a continuous increase in the fluorescence of 3, while addition of GDP did not change the signal (Figure 4). Thus, the nucleotide exchange promoted by GEFs can be easily monitored, allowing convenient characterization of these regulating proteins or rapid screens of GEF inhibitors. Alternatively, the probe can be used to monitor GTP hydrolysis. For this assay, Cdc42 is pre-loaded with GTP, and in the presence of the sensor, a decrease in fluorescence is observed as the nucleotide is hydrolyzed (Figure S5). However, because the WASP fragment has been shown to inhibit the intrinsic rate of GTP hydrolysis,20 this application may be best suited for comparing relative rates of hydrolysis. The tool provides advantages over current approaches for evaluating Cdc42 activity, which include discontinuous radioactive filter binding approaches,21 because dynamic activity can be monitored directly in a continuous fashion with a simple fluorescence plate reader. Additionally, with a high signal-to-noise ratio, the probe exhibits great sensitivity and can be readily adapted for high-throughput screening of small molecule modulators of Cdc42 or associated regulatory proteins.
Figure 4.
Application of the sensor to monitor Cdc42 nucleotide exchange. Cdc42 was pre-equilibrated with GDP, and the fluorescence of sensor 3 was monitored immediately upon the addition of GTP-γS or GDP. Measurements were taken with 150 nM 4-DMN sensor 3 and 700 nM Cdc42 at 30 °C.
Finally, we measured the dissociation constant of the 4-DMN sensor with GTP-γS-bound Cdc42 to demonstrate the utility of the probe in quantifying bimolecular interactions and to ensure that incorporation of the fluorophore does not significantly alter binding. To eliminate complicating effects resulting from the presence of unmodified GB1-WASP in samples obtained through the cysteine-labeling approach, the sensor generated by expressed protein ligation was initially employed for these studies. Fluorescence spectra from titrations in which Cdc42(GTP-γS) was added to 5 were fit to a 1:1 binding model, which yielded an observed KD of 385 ± 9 nM (Figure S6). Similarly, the observed KD for sensor 3 was 341 ± 22 nM. These values agree well with the dissociation constants of similar WASP fragments, which lie within the mid-nanomolar range.22
In summary, we have developed a powerful probe for detecting the activated form of Cdc42 by incorporation of the 4-DMN solvatochromic fluorophore into the WASP binding domain. Cysteine labeling of the WASP fragment or protein semisynthesis furnishes a sensor that discriminates between the inactive and active conformations of Cdc42 through dramatic increases in fluorescence. These remarkable properties can be exploited to monitor the nucleotide exchange and GTPase activities of Cdc42 as well as the influence of regulatory proteins. By offering a large dynamic range and high sensitivity, this 4-DMN-based sensor promises to be valuable in the study of Cdc42 and represents another example of the versatility of the 4-DMN fluorophore as a probe of important protein binding interactions.
Supplementary Material
Acknowledgments
This research was supported by the NIH Cell Migration Consortium (GM064346). B.N.G. and G.S.L. were supported by the NIGMS Biotechnology Training Grant (T32-GM08334). We thank Professor Martin Schwartz of the University of Virginia for the gift of the plasmid encoding Cdc42.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Fan C, Plaxco KW, Heeger AJ. Trends Biotechnol. 2005;23:186. doi: 10.1016/j.tibtech.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Loving GS, Sainlos M, Imperiali B. Trends Biotechnol. 2010;28:73. doi: 10.1016/j.tibtech.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noble JE, Ganju P, Cass AE. Anal. Chem. 2003;75:2042. doi: 10.1021/ac025838e. [DOI] [PubMed] [Google Scholar]
- 4.Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. Nat. Chem. Biol. 2007;3:466. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 5.Lakowicz JR. Principles of fluorescence spectroscopy. 3rd ed. Springer; New York: 2006. Chapter 19. [Google Scholar]
- 6.Vázquez ME, Rothman DM, Imperiali B. Org. Biomol. Chem. 2004;2:1965. doi: 10.1039/b408001g. [DOI] [PubMed] [Google Scholar]
- 7.Sainlos M, Imperiali B. Nat. Protoc. 2007;2:3201. doi: 10.1038/nprot.2007.442. [DOI] [PubMed] [Google Scholar]
- 8.Sainlos M, Tigaret C, Poujol C, Olivier NB, Bard L, Breillat C, Thiolon K, Choquet D, Imperiali B. Nat. Chem. Biol. 2011;7:81. doi: 10.1038/nchembio.498. [DOI] [PubMed] [Google Scholar]
- 9.Vázquez ME, Blanco JB, Imperiali B. J. Am. Chem. Soc. 2005;127:1300. doi: 10.1021/ja0449168. [DOI] [PubMed] [Google Scholar]
- 10.Loving G, Imperiali B. J. Am. Chem. Soc. 2008;130:13630. doi: 10.1021/ja804754y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loving G, Imperiali B. Bioconjugate Chem. 2009;20:2133. doi: 10.1021/bc900319z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaffe AB, Hall A. Annu. Rev. Cell Dev. Biol. 2005;21:247. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 13.Nobes CD, Hall A. J. Cell Biol. 1999;144:1235. doi: 10.1083/jcb.144.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh RE, Kurokawa K, Ohba Y, Yoshizaki H, Mochizuki N, Matsuda M. Mol. Cell Biol. 2002;22:6582. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seth A, Otomo T, Yin HL, Rosen MK. Biochemistry. 2003;42:3997. doi: 10.1021/bi026881z. [DOI] [PubMed] [Google Scholar]
- 16.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Science. 2004;305:1615. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 17.Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Nature. 2009;461:99. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdul-Manan N, Aghazadeh B, Liu GA, Majumdar A, Ouerfelli O, Siminovitch KA, Rosen MK. Nature. 1999;399:379. doi: 10.1038/20726. [DOI] [PubMed] [Google Scholar]
- 19.Bao WJ, Gao YG, Chang YG, Zhang TY, Lin XJ, Yan XZ, Hu HY. Protein Expression Purif. 2006;47:599. doi: 10.1016/j.pep.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, Wang ZX, Zheng Y. J. Biol. Chem. 1997;272:21999. doi: 10.1074/jbc.272.35.21999. [DOI] [PubMed] [Google Scholar]
- 21.Self AJ, Hall A. Methods Enzymol. 1995;256:67. doi: 10.1016/0076-6879(95)56010-6. [DOI] [PubMed] [Google Scholar]
- 22.Rudolph MG, Bayer P, Abo A, Kuhlmann J, Vetter IR, Wittinghofer A. J. Biol. Chem. 1998;273:18067. doi: 10.1074/jbc.273.29.18067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.