Summary
The classification of follicular thyroid neoplasms requires surgical resection for histologic evaluation of malignancy. As variable clinical behavior exists, genomic expression profiling may lead to the identification of novel markers that facilitate better biological classification. We performed for the first time gene expression analysis on clinically aggressive and non-aggressive follicular carcinomas (FCs) from patients for whom long-term follow-up data were available. We examined matched fresh/frozen tissue from 15 histopathologically diagnosed FCs (7 patients with documented distant metastasis and/or death from disease and 8 patients without recurrence). For categorical comparison, we analyzed 4 follicular adenomas (FAs). The biological control comprised 11 normal thyroid tissue specimens. High-quality RNA was extracted from the tissues, labeled, and hybridized to an Affymetrix oligonucleotide microarray (HG-U133A). With the exceptions of 1 FA and 1 FC, unsupervised hierarchical cluster analysis revealed 2 distinct groups—one containing normal thyroid tissue and FAs and another containing FCs. We identified 421 genes that were differentially expressed between histologically normal thyroid tissues and all follicular neoplasms (P < 0.01; fold-change >2), 94 genes that distinguished FCs from FAs (including PBP and CKS2), and 4 genes that distinguished aggressive FCs from non-aggressive FCs (NID2, TM7SF2, TRIM2 and GLTSCR2). Comparative genomic groupings identified differentially expressed genes that may lead to better classification of follicular thyroid neoplasms. Such genes may be used in future prospective validation studies to establish clinically useful and complementary diagnostic markers.
Keywords: Follicular neoplasms, follicular carcinoma, gene expression, Affymetrix
1. Introduction
Fine needle aspiration (FNA) for the primary evaluation of follicular thyroid neoplasms is generally non-definitive in differentiating follicular adenomas (FAs) from follicular carcinomas (FCs) [1–3]. Accordingly, the majority of follicular thyroid neoplasms must be surgically excised so they can be histopathologically evaluated and definitively classified as either benign or malignant; consequently, many patients with benign neoplasms unnecessarily undergo surgery. Although conventional histomorphologic assessment remains central to the management of patients with follicular thyroid neoplasms, the ability to definitively predict the biology of a given differentiated follicular tumor remains elusive due to the variable clinical behavior of these lesions. Not uncommonly, FCs behave non-aggressively and do not recur, and in rare instances, FAs metastasize [4]. Therefore, ancillary techniques that augment existing means of biologically classifying follicular thyroid neoplasms and specifically predicting their biologic behavior are needed.
Gene expression profiling provides a powerful tool for surveying genetic alterations and identifying novel markers of potential practical significance in the assessment of thyroid tumorigenesis [3]. Previous studies have used gene expression profiling to differentiate non-neoplastic thyroid tissue from malignant thyroid neoplasms, focusing mainly on papillary thyroid carcinomas [5–9]. However, few gene expression studies have focused on patients with follicular thyroid neoplasms, and fewer studies have investigated potential correlations between genetic profiles and clinical outcome [6, 10–14]. Owing to the protracted course, previous genomic studies of differentiated thyroid tumors have lacked clinical endpoints of disease progression focusing on tumors by histologic classification which is known to have limitations. We contend that if molecular signatures identified on the basis of the differential expression of genes can be used to predict the biologic behavior of follicular thyroid neoplasms, then such identified genes can be used to complement the pathologic diagnosis and/or prognostic assessment of follicular thyroid neoplasms at the initial assessment with FNA.
To investigate the impact of gene expression profiling on the biological classification of follicular thyroid neoplasms, we used the Affymetrix microarray platform (HG-U133A) to identify differentially expressed genes in a spectrum of follicular carcinomas with known aggressive biologic behavior with metastases versus indolent tumors with long term follow-up.
2. Materials and methods
2.1. Tumor samples
Following approval of this study by The University of Texas MD Anderson Cancer Center’s Institutional Review Board, we searched the clinical database and identified 7 patients with histologically confirmed FC with documented metastases for whom flash-frozen tissue specimens were available (clinically aggressive FC). We also identified a comparative group of 8 matched patients with FCs with lymphovascular invasion and no evidence of recurrence or metastases with at least 5 years of follow-up (clinically non-aggressive), who were also treated at MD Anderson Cancer Center during the same period, 1993 to 2003, and for whom flash-frozen tissue were available. The tissues had been stored at −80°C until their use. We also included fresh-frozen tissue from 4 FAs and 11 non-neoplastic thyroid tissues (3 non-pathologic thyroids and 8 histologically normal thyroid parenchyma from resected tumor specimens) were also included for comparison. All tissues were histologically re-reviewed; tumors were classified using current World Health Organization criteria [15] and staged using the 7th edition of the American Joint Committee on Cancer staging criteria [16]. The nuclei were carefully reviewed in order to exclude cases of follicular variant of papillary thyroid carcinoma.
2.2. RNA extraction and microarray experiments
High-quality mRNA was extracted from the tissues using standard guanidinium thiocyanate-phenol-chloroform extraction method (TRIzol reagent, Invitrogen, Inc. Carlsbad, California). The extracted mRNA was converted to biotin-labeled cRNA, which was then hybridized to HG-U133A Affymetrix oligonucleotide microarrays (Santa Clara, CA). The arrays were washed and scanned according to standard Affymetrix protocol (www.affymetrix.com).
2.3. Gene expression by Rt-PCR
To support array findings, gene expression levels were performed by real-time rt-PCR from cDNA for CKS2, and PBP with GAPDH as the endogenous control. Using the recommended Applied Biosystems (Forest city, CA) protocols, mRNA was converted to cDNA and Taqman gene expression assays were run using manufactured primers and probes (CKS2 (Hs01048812_g1), PBP (Hs00831506_g1), and GAPDH). Forty nanograms of cDNA were used in each reaction which were performed in triplicate for 40 cycles. Relative expression comparisons were calculated from normal thyroid tissue as the calibrator.
2.4. Bioinformatics
The microarray data were processed using the position-dependent nearest-neighbor model [17]. To ensure data quality, we inspected the array images and checked the control genes’ 5′-end-to-3′-end ratios. We then converted the gene expressions to log scale for downstream analysis.
For the unsupervised cluster analysis, we filtered out genes that were undetected or unchanged across the samples. Genes with a mean log-expression level >8.13 and a standard deviation (SD) of the log-expression level >0.33, passed through the filtering. The SD threshold of 0.33 was determined using the following analysis. Each gene on the Affymetrix microarray platform was interrogated by an 11-probe set. We divided each probe set into 2 subsets containing 5 and 6 probes, respectively. Theoretically, because the 2 subsets targeted the same gene, the expression profile would display high correlation across all samples. However, the expression profiles obtained from the 2 subsets may not always correlate because some genes may not be expressed in the samples or may not change beyond the noise level. When the SD is < 0.33, the range of correlation coefficients quickly expands, indicating that the profiles of the same genes from the probe subsets are often inconsistent. Consequently, we considered genes with an SD < 0.33 to be uninformative and excluded them from the cluster analysis.
2.5. Comparative gene expression analysis
The gene expression profiles of normal thyroid and follicular thyroid tumor tissues were compared as follows: Normal thyroid parenchyma versus neoplasms (FA and FC); FA versus all FCs; and clinically non-aggressive FC versus clinically aggressive FC. To identify significant differentially expressed genes, we used t-test of log-expression values to compute the P values across the more than 22,000 probe sets. Genes that had P values <0.01 and fold-changes >2 were considered significantly differentially expressed. To correct for multiple tests, we used permutation (permuting the cancer type labels only) and repeated the analysis. In each comparison, the rate of false discovery was < 20% (data not shown).
Genes identified as differentially expressed were classified by their biological functions using Ingenuity Pathways Analysis Software (Ingenuity Systems, Redwood City, CA) and compared between groups.
3. Results
3.1. Follicular thyroid carcinomas
Clinicopathological data for the 15 patients (10 men and 5 women) with histologically classified FCs are summarized in Table 1. The patients’ mean age was 60.1 years (range, 23–79 years) and the mean tumor size was 6.3 cm (range, 2.0–16.0 cm). Four patients presented with stage I disease, 3 with stage II disease, 2 with stage III disease, 2 with stage IVa disease, and 4 with stage IVc disease. FCs were segregated into biologically aggressive, defined by the presence of distant metastases or death from disease (7 FCs), and non-aggressive without evidence of recurrence (8 FCs).
Table 1.
Clinicopathological data for the 15 patients with follicular thyroid carcinomas analyzed
| Patient | Sex | Age, y | Tumor size, cm | Metastases | Disease stage | Follow- up time, mo | Patient status |
|---|---|---|---|---|---|---|---|
| 1 | M | 36 | 6.0 | None | I | 65 | ANED |
| 2 | F | 45 | 6.5 | None | III | 105 | ANED |
| 3 | M | 57 | 2.0 | None | I | 93 | ANED |
| 4a | F | 69 | 2.5 | None | II | 120 | DNED |
| 5 | F | 79 | 2.0 | None | I | 84 | ANED |
| 6d | M | 23 | 3.2 | None | I | 161 | ANED |
| 7d | M | 68 | 4.0 | None | II | 94 | ANED |
| 8d | F | 69 | 4.0 | None | II | 158 | ANED |
| 9 | M | 77 | 5.5 | Boneb | IVc | 89 | ANED |
| 10 | M | 47 | 11.0 | Bone, lung, CNS | IVa | 26 | DWD |
| 11 | M | 49 | 15.0 | Lymph nodesb | IVa | 212 | DWD |
| 12c | F | 65 | 6.0 | Bone, lungb | IVc | 33 | DWD |
| 13 | M | 66 | 16.0 | Bone, lung, liver gingivab | IVc | 9 | DWD |
| 14 | M | 78 | 2.6 | Bone, kidneyb | IVc | 71 | DWD |
| 15c | M | 73 | 8.5 | Bone, lung | III | 36 | DWD |
Abbreviations: ANED–alive, no evidence of disease; CNS–central nervous system; DNED–dead, no evidence of disease; DWD–dead with disease; mo–months; y–years.
PPARg expression elevated consistent with the PAX8/PPARg translocation (t(2;3)(q13;25).
Metastases were present at initial diagnosis.
Poorly differentiated features present.
Hurthle cell phenotype.
3.1.1. Non-aggressive follicular carcinoma
Eight patients (4 men and 4 women) had histologically documented FC (three showed Hurthle cell features) with lymphovascular invasion and no evidence of recurrence or metastasis during follow-up. The patients’ mean age was 55.8 years (range, 23–79 years) and the median tumor size was 3.6 cm (range, 2.0–6.0 cm). Four patients had stage I disease, 3 patients had stage II disease, and 1 patient had stage III disease. One patient died without evidence of disease 120 months after initial presentation; the other 7 patients were alive without evidence of recurrence or metastases at a mean of 110 months after initial presentation (range, 65–161 months).
3.1.2. Aggressive follicular carcinoma
Seven patients (6 men and 1 woman) had clinically aggressive FCs; two tumors had a poorly differentiated component with solid growth, necrosis and increased mitoses. The patients’ mean age was 65 years (range, 47–78 years). The mean tumor size was 9.2 cm (range, 2.6–16.0 cm). Metastatic sites included the bone (6 patients), lung (4 patients), lymph node (1 patient), and other sites (3 patients; Table 1). One patient had stage III disease, 2 patients had stage IVa disease, and 4 patients had stage IVc disease. Six patients died of disease; the mean time to death after initial presentation was 64 months (range, 9–212 months). At the time of the current study, 1 patient was living with no evidence of disease 89 months following the resection of a single bone metastasis that had been identified at initial presentation.
3.1.3. Comparison of non-aggressive and aggressive follicular carcinoma
The clinicopathologic parameters shown in Table 1 were analyzed between the patients with aggressive FC and the patients with non-aggressive FC. Single-parameter statistical analysis using Fisher’s exact test revealed that tumor size (P = .04, ≤ 4.0 cm versus >4.0 cm) and disease stage (P = .004) were significant predictors of disease progression. However, disease stage was most influenced by metastases at presentation and large size alone (≥6.0 cm) did not always equal aggressive disease.
3.2. Follicular thyroid adenomas
For comparative purpose, we included 4 patients (1 man and 3 women) who had macrofollicular (1 patient) or microfollicular (3 patients) FAs. The patients’ mean age was 55 years (range, 34–74 years). The mean tumor size was 4.2 cm (range, 2.5–5.2 cm). At the time of the current study, all patients were alive with no evidence of recurrence at a mean follow-up time of 83 months (range, 73–95 months).
3.3. Gene expression profiling
A total of 1181 genes met the gene expression criteria (mean log-expression level > 8.13, SD > 0.33) and were thus considered to be informative. Unsupervised hierarchical clustering using these genes are shown in Figure 1.
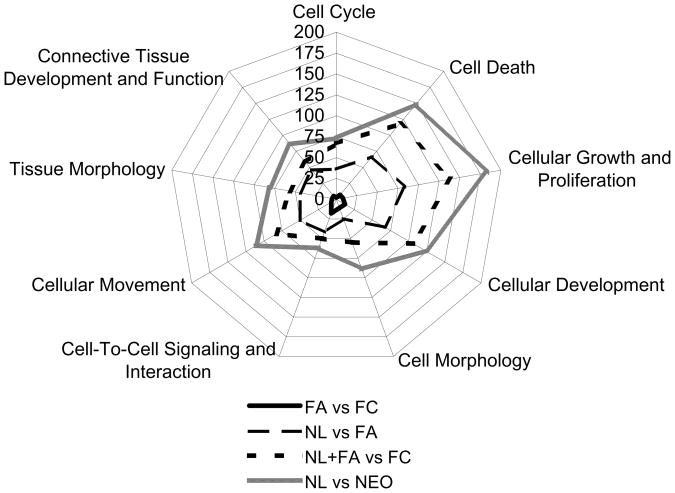
Fig. 1. Differential gene expression by cellular function pathways.
Major cellular pathways and the corresponding numbers of differentially expressed genes between follicular adenomas (FAs), follicular carcinoma (FCs), and normal thyroid tissue (NL). NL versus neoplasms (NEO; all FAs and FCs) differentially expressed 421 genes, with similar pathways to NL versus FA (203 differentially expressed genes) and benign (NL and FA) versus FC (302 differentially expressed genes). FAs and FCs had similar gene expression profiles, with only 94 genes differentially expressed between the two groups.
3.3.1. Normal thyroid tissue vs. follicular thyroid neoplasms
To broadly identify differentially expressed genes associated with follicular tumorigenesis, we compared normal thyroid parenchyma from follicular neoplasms (FAs and FCs) and identified 421 genes that were differentially expressed between normal thyroid tissues and FAs and FCs. Of these 421 genes, 152 were overexpressed and 269 were underexpressed in the follicular thyroid neoplasms compared to normal thyroid tissue. Most of the differentially expressed genes were involved in cellular growth and proliferation (183 genes), death (149 genes), development (123 genes), migration (111 genes), and/or differentiation (89 genes)(Figure 1). The genes with the highest fold-change between the normal thyroid parenchyma and the FA and FC parenchyma were related to the synthesis and maintenance of stromal proteins (decorin [DCN], trefoil factor 3 [TFF3]) and growth factors (insulin growth factor [IGF] and epidermal growth factor [EGF]).
Unsupervised hierarchical clustering revealed that 10 of the 11 normal thyroid tissues were tightly grouped and segregated from all FAs and FCs (Figure 2). The remaining normal thyroid tissue specimen, which was harvested from grossly normal thyroid tissue in a lobectomy specimen for an adenoma, was clustered with the adenomas.
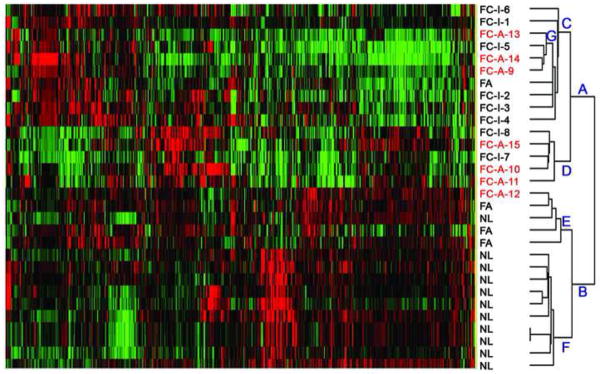
Fig. 2.
Hierarchical clustering of thyroid samples including normal thyroid tissue (NL), follicular adenomas (FAs), and follicular carcinomas (FCs) by 1181 genes. Branches are designated A to G. The 2 major divisions are (A) comprised of all FCs and 1 FA, and (B) comprised of all NL and 3 of the 4 FAs. Subdivisions within branch (B) are (F) 10 of 11 NL samples further clustered tightly at the bottom adjacent to (E), 3 FAs and the remaining NL. FCs with metastases (FC-A; indicated in red) clustered in 2 regions in subdivision (D) and within branch (C) in cluster (G) admixed with indolent FCs. Sample numbers correspond to the patient numbers in Table 1.
3.3.2. Normal thyroid tissue vs. follicular adenomas
To identify genes that may be associated with benign tumor development, we compared normal thyroid tissue to FAs. Two hundred and three genes were differentially expressed comprised of 89 upregulated and 114 downregulated genes between normal thyroid tissue and FAs. The functional classification of these differentially expressed genes is shown in Figure 1.
Three of the 4 FAs were clustered in close proximity to the normal tissues in the hierarchical clustering (Figure 2). One FA grouped within the clinically non-aggressive FCs; a re-evaluation of the FA’s pathologic features confirmed the original diagnosis.
3.3.3. Normal thyroid tissue and follicular adenomas vs. follicular carcinoma
To identify genes associated with malignancy, we compared FCs to combined normal thyroid tissues and FAs. This led to the identification of 302 differentially expressed genes; 75 were upregulated and 227 were downregulated in the FC group compared to the combined normal tissue plus FA samples (Figure 3).
Fig. 3.
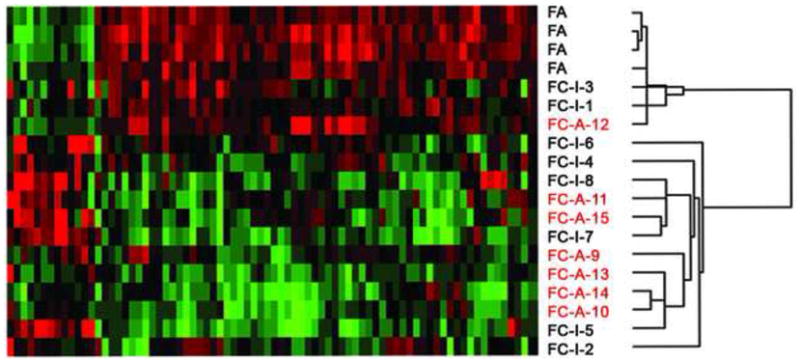
Clustering of the 94 genes that differentiate follicular adenoma (FA; benign lesions) vs. follicular carcinoma (FC; malignant lesions). FCs with metastases (FC-A; indicated in red) clustered. Sample numbers correspond to patient numbers in Table 1. FC-I, indolent/non-aggressive follicular carcinoma.
Cluster analysis showed 14 of the 15 FCs were clustered together and were distinct from normal thyroid tissue and FA (Figure 2). One clinically aggressive FCs was grouped with the FAs; compared with other FC samples. One FA was clustered with the clinically non-aggressive FCs and had a similar profile was grouped with 2 adjacent non-aggressive FCs (Figure 1).
3.3.4. Follicular adenomas vs. follicular carcinomas
To further characterize genes potentially associated with histopathologic follicular malignancy, we compared the gene expression patterns of FAs with those of FCs and found that 94 genes were significantly differentially expressed between FAs and all FCs. These comprised genes commonly involved in small molecule biochemistry (18 genes), growth and proliferation (8 genes), genomic regulation (7 genes), and differentiation (6 genes)(Figure 1). Of these 94 genes, 70 genes were relatively lost in FC compared to FAs including genes associated with tumorigenesis: sorbin and SK3 domain containing 2 (SORBS2/argBP2), prostatic binding protein (PBP/phosphatidylethanolamine binding protein 1 [PEBP1]), chromobox homolog 7 (CBX7), and catenin (cadherin-associated protein - delta1, CTNND1) (Table 2). Twenty-four genes showed higher expression in FCs than in FAs and included growth differentiation factor 15 (GDF-15), CDC28 protein kinase regulatory subunit 2 (CKS2), lysosomal-associated membrane protein 3 (LAMP3), among others (Table 2). Thyroid-associated genes especially thyroid peroxidase (TPO) and thyroglobulin (TG) were also found in general to have lower but variable expression in FCs in comparison to FAs, and this was irrespective of the presence or absence of poor differentiation.
Table 2.
Genes differentially expressed in follicular adenomas (A) compared to follicular carcinomas (C).
| Gene Symbol | Gene Name | P-value | Fold-change (A/C) |
|---|---|---|---|
| TPO | thyroid peroxidase | 0.0040 | 22.64 |
| CLIC3 | chloride intracellular channel 3 | 0.0072 | 16.96 |
| FCGBP | Fc fragment of IgG binding protein | 0.0083 | 9.94 |
| SORBS2 | sorbin and SH3 domain containing 2 | 0.0010 | 8.34 |
| SNRPN /// SNURF | small nuclear ribonucleoprotein polypeptide N /// SNRPN upstream reading frame | 0.0031 | 5.73 |
| TG | thyroglobulin | 0.0014 | 5.60 |
| HSPA5 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | 0.0015 | 5.10 |
| SDF2L1 | stromal cell-derived factor 2-like 1 | 0.0067 | 4.97 |
| FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 0.0017 | 4.60 |
| CAPG | capping protein (actin filament), gelsolin-like | 0.0013 | 4.23 |
| PDIA6 | protein disulfide isomerase family A, member 6 | 0.0004 | 4.20 |
| ARMET | arginine-rich, mutated in early stage tumors | 0.0005 | 4.12 |
| RRBP1 | ribosome binding protein 1 homolog 180kDa (dog) | 0.0063 | 4.01 |
| MPPED2 | metallophosphoesterase domain containing 2 | 0.0086 | 3.95 |
| HYOU1 | hypoxia up-regulated 1 | 0.0016 | 3.87 |
| FKBP2 | FK506 binding protein 2, 13kDa | 0.0066 | 3.87 |
| PBP | prostatic binding protein | 0.0053 | 3.85 |
| ATP1A1 | ATPase, Na+/K+ transporting, alpha 1 polypeptide | 0.0017 | 3.82 |
| C20orf3 | chromosome 20 open reading frame 3 | 0.0045 | 3.81 |
| PPIB | peptidylprolyl isomerase B (cyclophilin B) | 0.0037 | 3.67 |
| CPE | carboxypeptidase E | 0.0019 | 3.56 |
| TNS1 | tensin 1 /// tensin 1 | 0.0023 | 3.05 |
|
| |||
| PCSK1N | proprotein convertase subtilisin/kexin type 1 inhibitor | 0.0021 | −3.03 |
| C3 | complement component 3 | 0.0009 | −3.05 |
| CKS2 | CDC28 protein kinase regulatory subunit 2 | 0.0066 | −3.67 |
| NELL2 | NEL-like 2 (chicken) /// NEL-like 2 (chicken) | 0.0005 | −3.69 |
| HLA-DPA1 | major histocompatibility complex, class II, DP alpha 1 | 0.0027 | −5.09 |
| GDF15 | growth differentiation factor 15 | 0.0006 | −8.02 |
Hierarchical cluster analysis distinctly separated the FAs along with 2 indolent FCs and the 1 aggressive FC with low gene variability from the remaining aggressive FCs, which formed 2 subclusters (Figure 3).
3.3.5. Non-aggressive follicular carcinomas vs. aggressive follicular carcinomas
To identify genes that may be associated with biological aggressiveness within histologically diagnosed carcinomas, we compared aggressive FCs (FCs with metastases) with non-aggressive FCs (FCs without recurrence). Four genes were differentially expressed between the 2 groups; 3 genes—nidogen 2 (NID2), transmembrane 7 superfamily member 2 (TM7SF2), and tripartite motif-containing 2 (TRIM2)—had higher expression in the non-aggressive FCs than in the aggressive FCs and 1 gene—glioma tumor suppressor candidate region gene 2 (GLTSCR2)—had higher expression in aggressive FCs than in non-aggressive FCs (Table 3).
Table 3.
Genes differentially expressed between follicular carcinomas, non-aggressive (I) versus metastatic (M)
| Gene Symbol | Gene Name | P-value | Fold- change (I/M) |
|---|---|---|---|
| NID2 | nidogen 2 (osteonidogen) | 0.0099 | 6.40 |
| TM7SF2 | transmembrane 7 superfamily member 2 | 0.0056 | 2.59 |
| TRIM2 | tripartite motif-containing 2 | 0.0008 | 2.15 |
| GLTSCR2 | glioma tumor suppressor candidate region gene 2 | 0.0092 | −2.40 |
Aggressive FC aggregated in two subgroups within the larger subdivision of FC; one of these subgroups with 5 FC had 3 aggressive tumors while the second subgroup showed that three of the four tightly associated FC were aggressive (Figure 2).
3.3.6 Quantitative gene expression by real-time rt-PCR
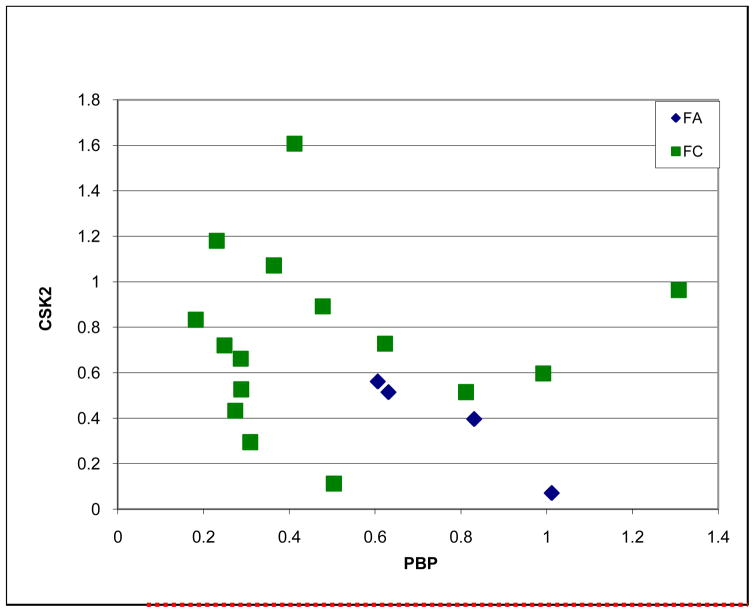
The analysis showed higher expression of PBP in adenoma versus carcinoma and in general paradoxically higher levels of CSK2 in FC versus FA (P<0.03, Figure 4).
Fig. 4.
Quantitative gene expression by real-time rt-PCR of PBP and CSK2 genes in follicular adenomas (FA) and follicular carcinomas (FC) (T-test, P<0.03).
4. Discussion
In the current study, we identified a set of genes that differentiated histologically normal thyroid parenchyma from follicular thyroid neoplasms and certain genes that, with few exceptions, segregated FAs from FCs. Among these samples, 1 normal thyroid parenchyma from a patient with an FC segregated proximal to FAs, which suggests that genomic modifications within the thyroid parenchyma may underlie a field effect. We also found that tumors within the defined histological categories were generally closer to each other in their genomic profiling, a finding that lends additional support for the use of histopathological evaluation.
However, we observed 1 aggressive FC that clustered with the FAs. Although, an isolated instance, it is tempting to speculate that follicular thyroid neoplasms share certain fundamental genes and that variable genes associated with biological progression in this particular FC were either underrepresented in our platform or were expressed at levels below the defined significant expression threshold (> 2.0 fold change). However, the finding may also point to the inherent limitations of using genomic and informatics tools to assess individual tumor variation and intertumoral heterogeneity for the classification of follicular thyroid neoplasms [18].
In the current study, 1 FA was grouped with the non-aggressive FCs, suggesting that similar biologic characteristics are shared and reflect a common clinical behavior despite their morphologic classification. In this context, if FC is a potential transformation of some FAs as a result of additional acquired alterations, one may expect that some histologically diagnosed FAs may progress to FCs if left unresected. Conceptually, FCs may arise de novo or evolve from certain FAs [4]. In the latter case, FA carrying RAS mutations (10%) may be prone to the evolution to carcinoma. Paradoxically, some of the histologically malignant tumors may behave in a benign fashion as in the majority of FC. Although these examples underscore the inherent limitations of using phenotyping to assess the biology of follicular thyroid neoplasms, they also highlight the potential advantages of integrating pathologic criteria with submicroscopic biological analysis.
Although limited, our results show that within FCs, individual tumors manifest variable biologic outcomes and that better biological assessment can be further achieved using complimentary predictors. As an example, a minimally invasive FC (FC14) was molecularly most closely associated with a widely invasive metastatic tumor (Table 1, Figure 2). Whether this tumor would have pursued an aggressive course had it been left unresected is a matter of speculation. Moreover, from a diagnostic perspective, the validation of the array findings by quantitative rt-PCT analysis supports the potential use of molecular analysis for future characterization of thyroid neoplasms into benign and malignant from small specimens including FNA biopsy material. In the future, integrating the pathologic review of the FNA specimen and the molecular assessment of a limited set of genes could optimize the evaluation of indeterminate thyroid nodules, thereby reducing the uncertainty in assigning patients to surgery or clinical observation [13]. However, prospective diagnostic validation studies of gene expression in FNA samples will be critical.
In the current study, we identified 76 genes that had also been identified in previous studies of gene alterations in follicular neoplasms [6, 12–14, 19, 20]. Twenty of these genes were also identified by Weber et al. in a study of benign and malignant follicular neoplasms [13]. An additional 20 of 105 (19.0%) genes were also reported by Barden et al. to be differentially expressed in follicular neoplasms [6]. In other smaller studies, 17 of 54 (31.5%) unique genes reported by Arora et al. to identify borderline follicular neoplasms [20], 12 of 131 genes that differentiated FA from FC by Zhao et al. [14], and 2 of the 13 genes (15.4%) found to differentiate minimally invasive by Lubitz et al. were also identified in our study [12]. However, the differences in these genes, in our study, were predominantly identified to discriminate between normal and follicular neoplasms and were not specific to the segregation of FA from FCs as in these prior studies.
Variation in array platforms and the heterogeneity of tumors being compared contributes to the complexity of comparative analysis across studies. Moreover, the interobserver inconsistency in histologic classification (e.g., follicular variant of papillary thyroid carcinoma versus widely or minimally invasive FC,) and the absence of long-term follow-up data preclude any definitive understanding of the true biologic potential of these entities. Careful consideration of these factors and validation will be necessary to determine the true value of these differentially expressed genes’ diagnostic potential and/or prognostic significance.
In conclusion, gene expression profiling can be used to identify molecular events that are differentially activated or suppressed between benign and malignant follicular neoplasms and between follicular neoplasms and normal thyroid tissues. Selected genes can also be used to segregate non-aggressive or aggressive FCs. Combining genetic profiling with morphologic evaluation may facilitate a more accurate classification of benign and malignant follicular thyroid neoplasms. However, genomic platforms must be optimized to accurately characterize these tumors. In addition, the decreased expression of potential tumor or metastasis suppressor genes suggests that the molecular pathways contributing to aggressive FCs should be investigated further.
Acknowledgments
Grant Support: This work was supported in part by the Kenneth D. Müller Professorship, a National Cancer Institute Specialized Program of Research Excellence (SPORE) grant in Head and Neck Cancer (Award Number P50CA097007), and National Cancer Institute Cancer Center Support Grant CA16672. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baloch ZW, LiVolsi VA, Asa SL, et al. Diagnostic terminology and morphologic criteria for cytologic diagnosis of thyroid lesions: a synopsis of the National Cancer Institute Thyroid Fine-Needle Aspiration State of the Science Conference. Diagn Cytopathol. 2008;36:425–37. doi: 10.1002/dc.20830. [DOI] [PubMed] [Google Scholar]
- 2.Williams MD, Suliburk JW, Staerkel GA, et al. Clinical significance of distinguishing between follicular lesion and follicular neoplasm in thyroid fine-needle aspiration biopsy. Ann Surg Oncol. 2009;16:3146–53. doi: 10.1245/s10434-009-0666-3. [DOI] [PubMed] [Google Scholar]
- 3.Baloch ZW, Fleisher S, LiVolsi VA, Gupta PK. Diagnosis of “follicular neoplasm”: a gray zone in thyroid fine-needle aspiration cytology. Diagn Cytopathol. 2002;26:41–4. doi: 10.1002/dc.10043. [DOI] [PubMed] [Google Scholar]
- 4.Nikiforova MN, Nikiforov YE. Molecular genetics of thyroid cancer: implications for diagnosis, treatment and prognosis. Expert Rev Mol Diagn. 2008;8:83–95. doi: 10.1586/14737159.8.1.83. [DOI] [PubMed] [Google Scholar]
- 5.Jarzab B, Wiench M, Fujarewicz K, et al. Gene expression profile of papillary thyroid cancer: sources of variability and diagnostic implications. Cancer Res. 2005;65:1587–97. doi: 10.1158/0008-5472.CAN-04-3078. [DOI] [PubMed] [Google Scholar]
- 6.Barden CB, Shister KW, Zhu B, et al. Classification of follicular thyroid tumors by molecular signature: results of gene profiling. Clin Cancer Res. 2003;9:1792–800. [PubMed] [Google Scholar]
- 7.Huang Y, Prasad M, Lemon WJ, et al. Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. Proc Natl Acad Sci U S A. 2001;98:15044–9. doi: 10.1073/pnas.251547398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazzanti C, Zeiger MA, Costouros NG, et al. Using gene expression profiling to differentiate benign versus malignant thyroid tumors. Cancer Res. 2004;64:2898–903. doi: 10.1158/0008-5472.can-03-3811. [DOI] [PubMed] [Google Scholar]
- 9.Finley DJ, Arora N, Zhu B, Gallagher L, Fahey TJ., 3rd Molecular profiling distinguishes papillary carcinoma from benign thyroid nodules. J Clin Endocrinol Metab. 2004;89:3214–23. doi: 10.1210/jc.2003-031811. [DOI] [PubMed] [Google Scholar]
- 10.Giordano TJ, Au AY, Kuick R, et al. Delineation, functional validation, and bioinformatic evaluation of gene expression in thyroid follicular carcinomas with the PAX8-PPARG translocation. Clin Cancer Res. 2006;12:1983–93. doi: 10.1158/1078-0432.CCR-05-2039. [DOI] [PubMed] [Google Scholar]
- 11.Aldred MA, Huang Y, Liyanarachchi S, et al. Papillary and follicular thyroid carcinomas show distinctly different microarray expression profiles and can be distinguished by a minimum of five genes. J Clin Oncol. 2004;22:3531–9. doi: 10.1200/JCO.2004.08.127. [DOI] [PubMed] [Google Scholar]
- 12.Lubitz CC, Gallagher LA, Finley DJ, Zhu B, Fahey TJ., 3rd Molecular analysis of minimally invasive follicular carcinomas by gene profiling. Surgery. 2005;138:1042–8. doi: 10.1016/j.surg.2005.09.009. discussion 8–9. [DOI] [PubMed] [Google Scholar]
- 13.Weber F, Shen L, Aldred MA, et al. Genetic classification of benign and malignant thyroid follicular neoplasia based on a three-gene combination. J Clin Endocrinol Metab. 2005;90:2512–21. doi: 10.1210/jc.2004-2028. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J, Leonard C, Gemsenjager E, et al. Differentiation of human follicular thyroid adenomas from carcinomas by gene expression profiling. Oncol Rep. 2008;19:329–37. [PubMed] [Google Scholar]
- 15.DeLellis RA, Lloyd RV, Heitz PU, Eng C, editors. Pathology & Genetics Tumours of Endocrine Organs. IARC Press; Lyon: 2004. World Health Organization Classification of Tumours. [Google Scholar]
- 16.Edge SB, Byrd DR, Compton C, Fritz AG, Green FL, Trotti A, editors. AJCC Cancer Staging Manual. 7. Chicago: Springer; 2010. Thyroid; pp. 87–96. [Google Scholar]
- 17.Zhang L, Miles MF, Aldape KD. A model of molecular interactions on short oligonucleotide microarrays. Nat Biotechnol. 2003;21:818–21. doi: 10.1038/nbt836. [DOI] [PubMed] [Google Scholar]
- 18.Weigelt B, Peterse JL, van’t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 19.Griffith OL, Melck A, Jones SJ, Wiseman SM. Meta-analysis and meta-review of thyroid cancer gene expression profiling studies identifies important diagnostic biomarkers. J Clin Oncol. 2006;24:5043–51. doi: 10.1200/JCO.2006.06.7330. [DOI] [PubMed] [Google Scholar]
- 20.Arora N, Scognamiglio T, Lubitz CC, et al. Identification of borderline thyroid tumors by gene expression array analysis. Cancer. 2009 doi: 10.1002/cncr.24616. [DOI] [PubMed] [Google Scholar]