Abstract
Thyroid hormone receptor interacting protein 6 (TRIP6), also known as zyxin-related protein-1 (ZRP-1), is an adaptor protein that belongs to the zyxin family of LIM proteins. TRIP6 is primarily localized in the cytosol or focal adhesion plaques, and may associate with the actin cytoskeleton. Additionally, it is capable of shuttling to the nucleus to serve as a transcriptional coregulator. Structural and functional analyses have revealed that through multidomain-mediated protein-protein interactions, TRIP6 serves as a platform for the recruitment of a wide variety of signaling molecules involved in diverse cellular responses, such as actin cytoskeletal reorganization, cell adhesion and migration, antiapoptotic signaling, osteoclast sealing zone formation and transcriptional control. Although the physiological functions of TRIP6 remain largely unknown, it has been implicated in cancer progression and telomere protection. Together, these studies suggest that TRIP6 plays multifunctional roles in different cellular responses, and thus may represent a novel target for therapeutic intervention.
Keywords: TRIP6, cell motility, apoptosis, transcription, LIM, PDZ
1. Introduction
Thyroid Hormone Receptor Interacting Protein 6 (TRIP6), also known as zyxin-related protein-1 (ZRP-1), is an adaptor protein that belongs to the zyxin family of LIM proteins [1–3]. These proteins, including zyxin, TRIP6, lipoma preferred partner (LPP), LIMD1, and Ajuba, contain a nuclear export signal (NES) in the N-terminal region and three LIM domains in the carboxyl-terminus [4]. The LIM domains, named by the initials of homeodomain proteins Lin-11, Isl-1 and Mec-3, contain two tandemly repeated zinc fingers that mediate protein-protein interactions [5]. These LIM proteins are primarily localized in the cytosol or focal adhesion plaques, and may associate with the actin cytoskeleton. They are also capable of shuttling between the cytoplasmic and nuclear compartments [6].
TRIP6 was originally discovered as a binding partner of the nuclear thyroid hormone receptor in a yeast two-hybrid screen in 1995 [3]. Subsequently, it has been characterized as an adaptor protein that associates with a wide variety of proteins to modulate diverse cellular responses, notably actin cytoskeletal reorganization, cell adhesion and migration, antiapoptotic signaling, transcriptional control, osteoclastic bone resorption, host-pathogen interactions and telomere protection (Table 1). In this review, we aim to update our current understanding of the multiple roles of TRIP6 in mediating these functions and the signal transduction mechanisms through which it acts, with an eye towards its biological relevance and potential roles in cancer progression.
Table 1.
Binding partners of TRIP6
| Binding Partner | Binding Domain | Functional significance | Refs. |
|---|---|---|---|
| Cell surface receptors | |||
| LPA2 | LIM2-3 | Promoting LPA-induced ERK/AKT/NF-κB activation, cell migration and antiapoptosis | [7–9, 21] |
| Fas/CD95 | LIM3 | Antagonizing Fas-induced apoptosis but promoting its effect on NF-κB activation and cell invasion | [9] |
| Nuclear receptors | |||
| (TRβ, RXR, GR) | LIM2-3 | Transcriptional regulation | [3,10] |
| Transcription factors | |||
| (p65, v-Rel, c-Fos) | LIM1-2 | Transcriptional coactivation | [10–12] |
| Bacterial proteins | |||
| (Opa, SrfH) | LIM | Host-pathogen interactions | [13–14] |
| Cytoskeletal regulators and adaptors | |||
| p130cas, CasL | LIM1-2 | Regulation of cell motility | [15] |
| supervillin | LIM | Modulation of focal adhesions | [16] |
| endoglin | LIM | Actin cytoskeletal reorganization | [32] |
| tropomyosin 4 | LIM | Osteoclastic bone resorption | [41] |
| Crk | pYXXP | Promoting cell migration | [7] |
| MAGI-1b | PDZ-binding motif | Promoting cell invasion | [22] |
| NHERF2 | PDZ-binding motif | Promoting LPA2-mediated ERK/AKT activation and antiapoptosis | [21] |
| Scrib | PDZ-binding motif | [23] | |
| RIL | LIM2-3 | [19] | |
| Kinases/Phosphatases | |||
| c-Src | Phosphorylating TRIP6 at Y55 to promote TRIP6-mediated cell motility and osteoclastic bone resorption | [7] | |
| RIP2 | LIM | ERK and NF-κB activation | [17] |
| AMPK | LIM | Phosphorylating TRIP6 and promoting transcriptional coactivator function of TRIP6 | [18] |
| PTPL1/hPTP1E/FAP-1 | LIM3&PDZ-binding motif | Dephosphorylating pY55 of TRIP6 to terminate its effect on cell motility | [37] |
| Shelterin Complex | |||
| (POT1) | LIM | Telomere protection | [20] |
2. The molecular structure and binding partners of TRIP6
2.1. NES, proline-rich domains, and SH2-binding motif
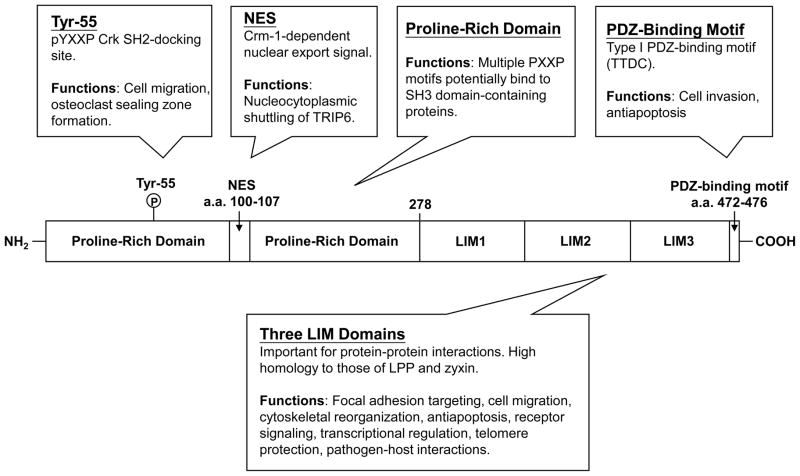
The structure of TRIP6 contains multiple domains, allowing it to interact with a wide variety of proteins involved in diverse cellular responses (Fig. 1). The N-terminal half of TRIP6 is characterized by a Crm-1-dependent leucine-rich NES (a.a. 100-107) and multiple proline-rich PXXP motifs, presumably allowing it to bind to SH3 domain-containing proteins [4]. TRIP6 also contains a YXXP motif (a.a. 55-58) that, when phosphorylated at Tyr-55 by c-Src, serves as a docking site for the SH2 domain of the adaptor protein Crk [7].
Figure 1. Schematic representation of TRIP6.
The N-terminal half of TRIP6 contains a proline-rich region with multiple PXXP motifs, a Crm-1-dependent nuclear export signal, and a phosphotyrosine-55-directed pYXXP motif that serves as a docking site for Crk SH2 domain. The carboxyl-terminal half of TRIP6 contains three tandem LIM domains and a PDZ-binding motif. These multiple elements allow TRIP6 to serve as an adaptor for a wide variety of partners with diverse functional outcomes.
2.2. LIM domains
The carboxyl-terminal half of TRIP6 contains three LIM domains in tandem that interact with a wide variety of proteins, such as cell surface receptors (LPA2, Fas/CD95), nuclear receptors (thyroid hormone receptor β [TRβ], retinoid X receptor [RXR], glucocorticoid receptor [GR]), transcription factors (v-Rel, NF-κB p65, c-Fos), pathogenic bacterial proteins (gonococcal opacity-associated protein [Opa], salmonella secreted effector SrfH), proteins involved in actin reorganization (p130cas, CasL, supervillin, endoglin), kinases (c-Src, receptor-interacting protein 2 [RIP2], AMP-activated protein kinase [AMPK]), adaptor proteins (reversion-induced LIM [RIL]), and shelterin complex proteins (protection of telomeres 1 [POT1]) [3, 8–20]. The LIM domains of TRIP6 share a high degree of sequence homology with those of LPP and zyxin, with an overall identity of 71.9% to LPP and 61.5% to zyxin. As such, it is not surprising that these LIM proteins, TRIP6 and LPP in particular, share some of the same binding partners.
2.3. PDZ-binding motif
The last four amino acid residues (TTDC) of TRIP6, known as a class I PDZ-binding motif, mediates the interaction with a number of PDZ proteins, including PTPL1 and its mouse homologue PTP-BL (PDZ2), Scrib (PDZ3), NHERF2 (Na+/H+ exchanger regulatory factor 2) (PDZ2), and MAGI-1b (membrane-associated guanylate kinase with an inverted domain structure-1b) (PDZ5) [1, 19, 21–23]. In addition to the PDZ-binding motif, the adjacent LIM3 domain is also required for TRIP6 binding to PTPL1 [1].
PDZ domains, named by the initials of three proteins that contain such domains (PSD-95, the Drosophila discs-large tumor suppressor protein DlgA, and the tight junction protein ZO-1), are composed of 80-90 amino acids that bind to the specific short peptide motif found in the carboxyl-terminus or internal region of a variety of target proteins [24–28]. Many PDZ proteins function as scaffolds to facilitate the assembly of multiprotein complexes or to help target protein anchoring to the actin cytoskeleton. Thus, it appears that the LIM domain-containing TRIP6 cooperates with its interacting PDZ proteins to coordinate the rapid assembly of macromolecular complexes, thereby regulating actin dynamics and other signaling events in a more efficient manner.
3. The roles of TRIP6 in actin cytoskeletal reorganization, adhesion turnover and cell motility
3.1. TRIP6 is involved in focal adhesion assembly and actin cytoskeletal reorganization
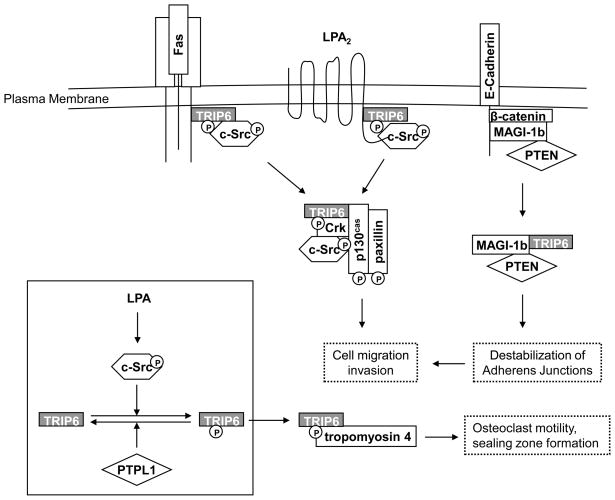
Accumulating data have pointed to a critical role for TRIP6 in cytoskeletal reorganization, focal adhesion assembly/disassembly, and migration by serving as a platform for the recruitment of a number of proteins involved in these dynamic processes (Fig. 2). Notably, TRIP6 associates directly or indirectly with the components of focal complexes, including paxillin, p130cas, focal adhesion kinase (FAK), and c-Src, upon stimulation with lysophosphatidic acid (LPA) [8]. TRIP6 and its close family members, LPP and zyxin, are targeted to the “integrin adhesome” through a myosin II-dependent recruitment [29]. Both TRIP6 and LPP, but not zyxin, are capable of interacting with supervillin at large focal adhesions [16]. Supervillin is a peripheral membrane protein that is known to bind to myosin II and F-actin to reorganize the actin cytoskeleton [30, 31]. By binding to TRIP6, supervillin promotes the loss of focal adhesion structure and function [16]. TRIP6 has also been shown to cooperate with endoglin, a component of the TGF-β receptor complex, to regulate actin cytoskeletal reorganization [32].
Figure 2. TRIP6 promotes cell motility, invasion and osteoclast sealing zone formation through c-Src-mediated phosphorylation and the interaction with a number of cell-surface receptors and signaling molecules involved in these dynamic processes.
TRIP6 binds to the juxtamembrane domains of the Fas/CD95 and LPA2 receptors upon stimulation with FasL and LPA, respectively. This induces phosphorylation of TRIP6 at Tyr-55 by c-Src, an event reversed by PTPL1, and triggers its association with Crk and various regulators of actin remodeling, leading to an increase in cell motility and promoting osteoclast sealing zone formation. TRIP6 also competes with β-catenin for binding to the MAGI-1b/PTEN signalsome. As a result, this destabilizes adherens junctions and increases cell invasiveness.
3.2. The function of TRIP6 in adhesion turnover and cell motility is regulated by c-Src kinase and PTPL1 phosphatase
The Src family kinases play a fundamental role in adhesion turnover and cell migration via the phosphorylation of a number of focal adhesion molecules, such as paxillin and p130cas [33, 34]. The c-Src-dependent tyrosine phosphorylation of p130cas at multiple YXXP motifs mediates its coupling to the SH2 domain of Crk [35, 36]. Likewise, TRIP6 contains a YXXP motif (a.a. 55-58), which is a unique feature that is not present in LPP or zyxin [7]. When the Tyr-55 residue is phosphorylated by c-Src, this allows its binding to Crk. Although TRIP6 itself can interact with p130cas directly through its LIM1-2 domains, phosphorylation of Tyr-55 is also important for TRIP6 to form a stable complex with p130cas. Ultimately, these c-Src-directed macromolecular complexes coordinate to promote adhesion turnover and cell migration.
However, it should be noted that, as TRIP6 is involved in both assembly and disassembly of focal adhesions, the effect of TRIP6 on promoting or reducing cell motility may depend on the cellular context. It has been reported that overexpression of TRIP6 slows cell migration in 10T1/2 fibroblasts [15]. Moreover, in c-Src/Yes/Fyn-null mouse embryonic fibroblasts (MEFs) where TRIP6 cannot be phosphorylated at Tyr-55, overexpression of TRIP6 does not promote adhesion turnover; instead, it increases the formation of larger focal adhesions and slows LPA-induced morphological changes [7]. Unlike the wild-type TRIP6, the TRIP6-Y55F mutant failed to promote Fas/CD95-mediated cell migration in glioblastoma cells [9]. Together, these findings suggest that the levels of activated c-Src may play a critical role in determining whether TRIP6 promotes or slows cell migration in different cell types.
In contrast to c-Src, the protein tyrosine phosphatase PTPL1 plays a counteracting role in regulating TRIP6 function in cell motility [37]. Human PTPL1, also known as hPTP1E, PTPN13 or Fas-associated phosphatase-1 (FAP-1), and its mouse homologue, PTP-BL, are cytosolic tyrosine phosphatases that have been shown to bind to a number of molecules involved in actin dynamics [38]. Following prolonged LPA stimulation, PTPL1 binds to TRIP6 and dephosphorylates the phosphotyrosine-55 residue of TRIP6, resulting in the inhibition of both the binding of TRIP6 to Crk and the turnover of TRIP6 at focal adhesion sites. This serves as a negative feedback regulatory mechanism to terminate the function of TRIP6 in promoting LPA-induced morphological changes and cell migration [37].
3.3. c-Src-mediated Tyr-55 phosphorylation regulates TRIP6 function in promoting LPA2- and Fas/CD95-mediated cell migration, as well as osteoclastic bone resorption
Given its adaptor role in the assembly of multiprotein complexes involved in cell motility, TRIP6 is also capable of linking cell surface receptors, including the LPA2 and Fas/CD95 receptors, to downstream signaling pathways that regulate cell motility [8, 9]. LPA is a growth factor-like phospholipid that mediates cell proliferation, survival, and migration [39]. Upon LPA stimulation, TRIP6 binds to the LPA2 receptor, but not to other LPA receptor subtypes, through a LIM domain-mediated interaction with the CXXC motif of the LPA2 receptor, which is present in the juxtamembrane region of its carboxyl-terminal tail [8, 21]. This binding promotes the targeting of TRIP6 to focal adhesions and its association with actin stress fibers. Consequently, TRIP6 enhances LPA-induced cell migration in ovarian cancer cells that express high levels of the LPA2 receptor.
Analogously, TRIP6 also promotes Fas ligand (FasL)-mediated glioma cell invasion through a LIM3-mediated interaction with the juxtamembrane domain of Fas/CD95 [9]. Fas/CD95 is traditionally viewed as a proapoptotic death receptor; however, Fas/CD95 engagement can induce invasion in apoptosis-resistant tumor cells via the activation of the Src/Yes and phosphatidylinositol-3-kinase (PI3K)/AKT signaling pathways [40]. Stimulation with Fas ligand induces the activation of c-Src, which in turn mediates the tyrosine phosphorylation of TRIP6 at Tyr-55 [9]. This phosphorylation is required for TRIP6 to promote Fas/CD95-mediated cell migration in apoptosis-resistant glioblastoma cells.
c-Src-mediated phosphorylation of TRIP6 not only regulates its function in cell motility, but is also involved in LPA-stimulated osteoclast sealing zone formation [41]. Upon LPA stimulation, TRIP6 is recruited to the sealing zone through its association with tropomyosin 4, an actin-binding protein that regulates osteoclast motility, sealing zone dimensions, and bone resorptive capacity. These signaling events are regulated by Tyr-55 phosphorylation of TRIP6, whereas treatment with LPA2 receptor antagonists, knockdown of TRIP6, or overexpression of the TRIP6-Y55F mutant impairs these effects. These results demonstrate that c-Src-mediated phosphorylation of TRIP6 promotes osteoclastic bone resorption.
3.4. TRIP6 promotes cell invasion via an interaction with the MAGI-1b PDZ scaffold protein
It has been shown that TRIP6 promotes invasiveness and impairs cell-cell aggregation in epithelial MDCK cells, which is in part regulated through binding to the MAGI-1b PDZ protein [22]. MAGI-1b is a scaffold protein that plays a critical role in the maintenance of cell-cell contacts by recruiting PTEN to the cadherin/β-catenin complex [42]. As both TRIP6 and β-catenin bind to the fifth PDZ domain of MAGI-1b, it is possible that TRIP6 competes with β-catenin for the binding to the MAGI-1b/PTEN signalosome, thereby destabilizing the adherens junctional complexes and promoting cell motility [22]. TRIP6 also binds to another PDZ protein, Scrib, which is involved in cell-cell adhesion and cell polarity; however, the functional significance of this interaction remains to be elucidated [23].
3.5. TRIP6 is involved in host-pathogen interactions
It has been shown that the Salmonella typhimurium-secreted effector protein SrfH promotes phagocyte motility and systemic spread of infection in the gut via a direct interaction with TRIP6 in the host cells [14]. TRIP6 also associates with the Opa proteins of Neisseria gonorrhoeae, which are a family of outer membrane proteins involved in gonococcal adherence to and invasion of human cells [13]. Although the molecular basis for these interactions is still elusive, these data suggest that TRIP6 may be involved in the interplay between pathogenic bacteria and host cells and therefore plays a role in the pathogenesis of certain infectious diseases.
4. The roles of TRIP6 in antiapoptotic signaling and NF-κB activation
4.1. TRIP6 regulates LPA2 receptor-mediated antiapoptotic signaling and antagonizes Fas/CD95-induced apoptosis
We are just now beginning to understand that, beyond its central role in cell motility, TRIP6 can also serve as an adaptor for the assembly of multiprotein complexes involved in prosurvival signaling pathways. Notably, TRIP6 positively regulates LPA-induced antiapoptotic effects through its binding to the LPA2 receptor and antagonizes Fas/CD95-initiated apoptosis via its interaction with Fas/CD95 [9, 21].
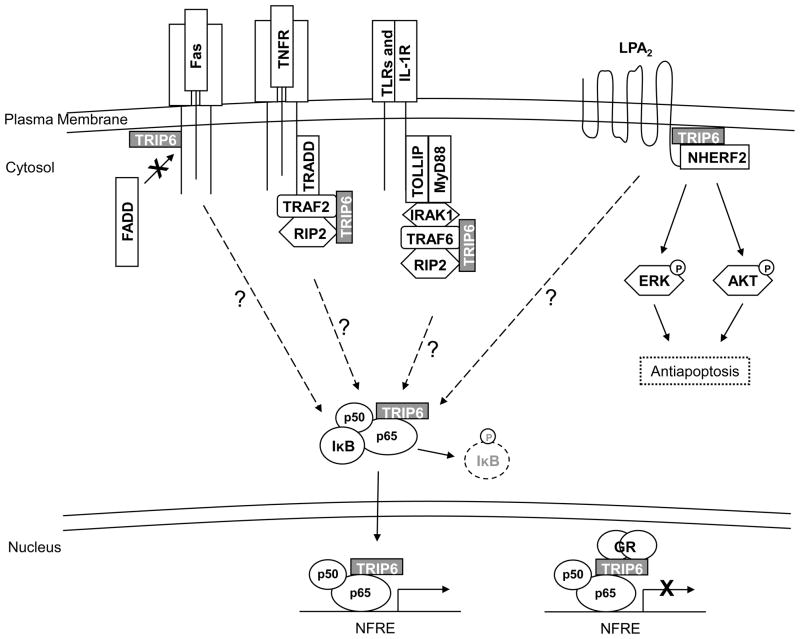
The LPA2 receptor mediates LPA-induced antiapoptotic responses through the activation of the Ras/ERK, PI3K/AKT and NF-κB signaling pathways [43]. While the LPA2 receptor can couple to the heterotrimeric G proteins to elicit these signals, activation of the full antiapoptotic responses also requires its binding to other elements, such as TRIP6 and NHERF2 [21]. The carboxyl-terminal tail of the LPA2 receptor contains a unique CXXC motif that interacts with the LIM2-3 domains of TRIP6 and a PDZ-binding motif responsible for NHERF2 binding [21, 44, 45]. Additionally, TRIP6 itself can associate with NHERF2 directly through a PDZ-mediated interaction [21]. Together, they form a ternary complex to coordinately regulate LPA2 receptor-mediated ERK and AKT activation, rendering cells resistant to chemotherapeutic agent-induced apoptosis [21] (Fig. 3).
Figure 3. TRIP6 regulates antiapoptotic responses and NF-κB signaling at multiple levels.
TRIP6 forms a ternary complex with the LPA2 receptor and NHERF2 upon LPA stimulation. Together, they coordinately regulate antiapoptotic signaling through the activation of ERK and AKT pathways. In addition to binding to Fas/CD95 and the LPA2 receptor directly, TRIP6 also associates with multiple elements of signal transduction complexes recruited by TNFR, TLRs, and IL-1R. These interactions ultimately result in the degradation of IκB and allow NF-κB to shuttle to the nucleus where TRIP6 binds to and serves as a coactivator of NF-κB p65 to activate promoters containing NF-κB responsive elements (NFREs). On the other hand, upon treatment with gluococorticoid, TRIP6 recruits glucocorticoid receptor (GR) to NF-κB at target gene promoters. This results in the transrepression of NF-κB.
4.2. TRIP6 regulates NF-κB signaling pathway at multiple different levels
The NF-κB transcription factor mediates resistance to apoptosis by promoting the gene transcription of a variety of antiapoptotic proteins [46]. In unstimulated cells, NF-κB forms an inactive complex with IκB in the cytosol. Upon stimulation with various signals on the cell surface, the IKK complex is activated. Phosphorylation of IκB by IKK leads to the ubiquitination and proteosomal degradation of IκB. This dissociates NF-κB from the cytosolic NF-κB/IκB complex, allowing it to translocate to the nucleus to activate the expression of target genes [47]. Mounting evidence suggests that TRIP6 regulates NF-κB activity at multiple levels (Fig. 3). It has been shown that the LIM domains of TRIP6 interact with the CARD (caspase activation and recruitment domain) and kinase domain of RIP2 [17]. RIP2 is a member of the RIP kinase family that is critically involved in inflammation and innate and adaptive immune responses triggered by TNF, IL-1, Nod1, and the Toll-like receptors (TLRs) [48, 49]. RIP2 regulates NF-κB signaling through TNF receptor associated factor (TRAF)-mediated recruitment to the receptor signaling complex. It has been shown that TRIP6 facilitates RIP2-mediated NF-κB activation following IL-1, Nod1, and TLR2 stimulation [17]. TRIP6 also potentiates RIP2- and Nod1-mediated ERK activation. Intriguingly, TRIP6 has also been found to associate with IL-1 receptor (IL-1R), Nod1, and TLR2, as well as a number of the signaling components involved in NF-κB activation, including TRAF2, TRAF6, MyD88, Tollip, and IRAK1 [17]. These observations suggest that TRIP6 is present in the macromolecular complexes involved in NF-κB activation. However, whether TRIP6 binds to these molecules directly or indirectly and how TRIP6 might facilitate the assembly of these signaling complexes have not yet been determined.
TRIP6 can also bind to NF-κB p65 directly to regulate its nuclear translocation and transcriptional activity upon LPA stimulation or Fas/CD95 activation [9]. This serves as an alternate mechanism by which TRIP6 promotes the LPA-mediated antiapoptotic effect as well as resistance to Fas/CD95-induced apoptosis.
5. The roles of TRIP6 in transcriptional control and other nuclear functions
5.1. TRIP6 serves as a cofactor for a number of transcription factors
Similar to other zyxin family members of LIM proteins, TRIP6 contains a leucine-rich NES between amino acid residues 100-107, which is responsible for its primarily cytosolic localization. However, mutation/deletion of the NES or treatment with leptomycin B, an inhibitor of Crm1-dependent nuclear export, results in its nuclear localization, suggesting that TRIP6 may play a role in the nucleus [4].
Nuclear TRIP6 does not bind directly to DNA, but it does contain two transactivation domains within the carboxyl-terminal LIM domain region and the N-terminal region that overlaps with the NES [4]. Indeed, TRIP6 binds to and serves as a transcriptional coactivator for a number of transcription factors, including the v-Rel oncoprotein, the NF-κB p65 subunit, and a number of c-Fos family members [10–12]. It has been reported that the transcriptional coactivator properties of TRIP6 can be enhanced through binding to AMPK and that AMPK phosphorylates TRIP6 in the N-terminal region in vitro [18]. However, the specific phosphorylation sites as well as the mechanism for this regulation have not yet been determined.
5.2. TRIP6 is a transcriptional cofactor of the nuclear glucocorticoid receptor
Accumulating evidence suggests that TRIP6 is involved in transcriptional regulation mediated by various nuclear hormone receptors. TRIP6 was originally discovered as a binding partner of thyroid hormone receptor β (TRβ) and retinoid X receptor (RXR); however, the functional significance of these interactions has not been characterized. Later, it was found to associate with glucocorticoid receptor (GR) through its LIM2 and LIM3 domains [10]. GR mediates the anti-inflammatory effect of glucocorticoids through the transrepression of transcription factors NF-κB and AP-1 [50, 51]. Normally, nuclear TRIP6 is recruited to the promoters of target genes via binding to NF-κB p65 or the c-Fos, but not the c-Jun, component of the AP-1 complex, and functions as a transcriptional coactivator [10, 12]. Upon treatment with glucocorticoids, TRIP6 recruits GR to NF-κB and AP-1 complexes at target gene promoters. This forms a basis for GR-mediated transrepression of NF-κB and AP-1, which is responsible for most of the anti-inflammatory effects of glucocorticoids [10]. Intriguingly, it has been shown that nuclear TRIP6 can increase GR-mediated transcription, but it is also required for the transrepression of GR by NF-κB and AP-1 [52]. Thus, TRIP6 serves as a platform to bridge GR and the NF-κB and AP-1 transcription factors, and therefore integrates activating or repressing signals at the promoters of target genes.
5.3. TRIP6 is implicated in telomere protection through interaction with the shelterin complex
In addition to serving as a transcriptional cofactor, the nuclear TRIP6 has been shown to bind to POT1, a single stranded telomeric overhang binding protein that regulates telomere length and protects chromosome ends [20]. POT1 is a component of the shelterin complex, which contains six proteins essential for proper telomere function [53]. Both TRIP6 and its structurally related LPP form complexes with POT1, TRF2 and TIN2 of the shelterin complex. Depletion of TRIP6 leads to the induction of telomere dysfunction induced foci (TIFs), suggesting that TRIP6 is involved in telomere protection [20]. In the future, additional work will be needed to define the precise roles of these LIM proteins in the regulation of shelterin functions and telomere protection.
6. Concluding remarks
Over the past 15 years, numerous studies have uncovered the diverse functions of TRIP6 in cell motility, antiapoptotic signaling, transcriptional control and other cellular responses. Through multidomain-mediated protein-protein interactions and phosphorylation modification, TRIP6 cooperates with a variety of PDZ proteins, LIM-binding proteins, and other interacting partners to modulate multiple signaling pathways from the cell surface to the nucleus. As TRIP6 is widely involved in cell motility, antiapoptotic signaling, as well as many other cellular responses, dysregulation of TRIP6 may lead to pathologic consequences. Notably, TRIP6 has been implicated in tumor progression. It has been shown that TRIP6 is dramatically overexpressed in glioblastomas and colon cancers where c-Src kinase activity is often aberrantly elevated [9, 22]. It is likely that TRIP6 contributes significantly to tumor invasiveness and resistance to apoptotic agents in these tumors, and therefore it may represent a novel therapeutic target for cancer treatment. Although these studies have shed new insights into the understanding of the biochemical and cellular properties of TRIP6, the physiological functions of TRIP6 remain largely unknown. In the near future, more work needs to be undertaken to clarify the molecular basis by which TRIP6 regulates these diverse pathways through protein-protein interactions and the biological relevance of these interactions. In the end, the knowledge obtained from these studies may provide new directions to specifically target TRIP6 for therapeutic intervention in the diseases such as cancers, inflammatory and infectious diseases, and disorders of bone remodeling.
Acknowledgments
We would like to acknowledge the multiple investigators who have made important contributions to advance our knowledge of the functions of TRIP6 summarized in this article. This work was supported by the National Institute of Health Grant NS066332 (F.-T. L).
Footnotes
The authors declare that they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Murthy KK, Clark K, Fortin Y, Shen SH, Banville D. J Biol Chem. 1999;274:20679–20687. doi: 10.1074/jbc.274.29.20679. [DOI] [PubMed] [Google Scholar]
- 2.Yi J, Beckerle MC. Genomics. 1998;49:314–316. doi: 10.1006/geno.1998.5248. [DOI] [PubMed] [Google Scholar]
- 3.Lee JW, Choi HS, Gyuris J, Brent R, Moore DD. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Gilmore TD. Biochim Biophys Acta. 2001;1538:260–272. doi: 10.1016/s0167-4889(01)00077-5. [DOI] [PubMed] [Google Scholar]
- 5.Bach I. Mech Dev. 2000;91:5–17. doi: 10.1016/s0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Gilmore TD. Biochim Biophys Acta. 2003;1593:115–120. doi: 10.1016/s0167-4889(02)00349-x. [DOI] [PubMed] [Google Scholar]
- 7.Lai YJ, Chen CS, Lin WC, Lin FT. Mol Cell Biol. 2005;25:5859–5868. doi: 10.1128/MCB.25.14.5859-5868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu J, Lai YJ, Lin WC, Lin FT. J Biol Chem. 2004;279:10459–10468. doi: 10.1074/jbc.M311891200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai YJ, Lin VT, Zheng Y, Benveniste EN, Lin FT. Mol Cell Biol. 2010;30:5582–5596. doi: 10.1128/MCB.00134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kassel O, Schneider S, Heilbock C, Litfin M, Gottlicher M, Herrlich P. Genes Dev. 2004;18:2518–2528. doi: 10.1101/gad.322404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao MK, Wang Y, Murphy K, Yi J, Beckerle MC, Gilmore TD. Gene Expr. 1999;8:207–217. [PMC free article] [PubMed] [Google Scholar]
- 12.Diefenbacher M, Sekula S, Heilbock C, Maier JV, Litfin M, van Dam H, Castellazzi M, Herrlich P, Kassel O. Mol Endocrinol. 2008;22:1767–1780. doi: 10.1210/me.2007-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams JM, Chen GC, Zhu L, Rest RF. Mol Microbiol. 1998;27:171–186. doi: 10.1046/j.1365-2958.1998.00670.x. [DOI] [PubMed] [Google Scholar]
- 14.Worley MJ, Nieman GS, Geddes K, Heffron F. Proc Natl Acad Sci U S A. 2006;103:17915–17920. doi: 10.1073/pnas.0604054103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yi J, Kloeker S, Jensen CC, Bockholt S, Honda H, Hirai H, Beckerle MC. J Biol Chem. 2002;277:9580–9589. doi: 10.1074/jbc.M106922200. [DOI] [PubMed] [Google Scholar]
- 16.Takizawa N, Smith TC, Nebl T, Crowley JL, Palmieri SJ, Lifshitz LM, Ehrhardt AG, Hoffman LM, Beckerle MC, Luna EJ. J Cell Biol. 2006;174:447–458. doi: 10.1083/jcb.200512051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, Bin LH, Li F, Liu Y, Chen D, Zhai Z, Shu HB. J Cell Sci. 2005;118:555–563. doi: 10.1242/jcs.01641. [DOI] [PubMed] [Google Scholar]
- 18.Solaz-Fuster MC, Gimeno-Alcaniz JV, Casado M, Sanz P. Cell Signal. 2006 doi: 10.1016/j.cellsig.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 19.Cuppen E, van Ham M, Wansink DG, de Leeuw A, Wieringa B, Hendriks W. Eur J Cell Biol. 2000;79:283–293. doi: 10.1078/S0171-9335(04)70031-X. [DOI] [PubMed] [Google Scholar]
- 20.Sheppard SA, Loayza D. Aging (Albany NY) 2010;2:432–444. doi: 10.18632/aging.100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.SE, Lai YJ, Tsukahara R, Chen CS, Fujiwara Y, Yue J, Yu JH, Guo H, Kihara A, Tigyi G, Lin FT. J Biol Chem. 2009;284:14558–14571. doi: 10.1074/jbc.M900185200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chastre E, Abdessamad M, Kruglov A, Bruyneel E, Bracke M, Di Gioia Y, Beckerle MC, van Roy F, Kotelevets L. Faseb J. 2009;23:916–928. doi: 10.1096/fj.08-106344. [DOI] [PubMed] [Google Scholar]
- 23.Petit MM, Crombez KR, Vervenne HB, Weyns N, Van de Ven WJ. FEBS Lett. 2005;579:5061–5068. doi: 10.1016/j.febslet.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Cho KO, Hunt CA, Kennedy MB. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 25.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 26.Fanning AS, Anderson JM. Curr Biol. 1996;6:1385–1388. doi: 10.1016/s0960-9822(96)00737-3. [DOI] [PubMed] [Google Scholar]
- 27.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 28.Sato T, Irie S, Kitada S, Reed JC. Science. 1995;268:411–415. doi: 10.1126/science.7536343. [DOI] [PubMed] [Google Scholar]
- 29.Schiller HB, Friedel CC, Boulegue C, Fassler R. EMBO Rep. 2011;12:259–266. doi: 10.1038/embor.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nebl T, Pestonjamasp KN, Leszyk JD, Crowley JL, Oh SW, Luna EJ. J Biol Chem. 2002;277:43399–43409. doi: 10.1074/jbc.M205386200. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y, Takizawa N, Crowley JL, Oh SW, Gatto CL, Kambara T, Sato O, Li XD, Ikebe M, Luna EJ. J Biol Chem. 2003;278:46094–46106. doi: 10.1074/jbc.M305311200. [DOI] [PubMed] [Google Scholar]
- 32.Sanz-Rodriguez F, Guerrero-Esteo M, Botella LM, Banville D, Vary CP, Bernabeu C. J Biol Chem. 2004;279:32858–32868. doi: 10.1074/jbc.M400843200. [DOI] [PubMed] [Google Scholar]
- 33.Panetti TS. Front Biosci. 2002;7:d143–150. doi: 10.2741/A771. [DOI] [PubMed] [Google Scholar]
- 34.Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. Nat Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 35.Burnham MR, Harte MT, Richardson A, Parsons JT, Bouton AH. Oncogene. 1996;12:2467–2472. [PubMed] [Google Scholar]
- 36.Klemke RL, Leng J, Molander R, Brooks PC, Vuori K, Cheresh DA. J Cell Biol. 1998;140:961–972. doi: 10.1083/jcb.140.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai YJ, Lin WC, Lin FT. J Biol Chem. 2007;282:24381–24387. doi: 10.1074/jbc.M701499200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erdmann KS. Eur J Biochem. 2003;270:4789–4798. doi: 10.1046/j.1432-1033.2003.03895.x. [DOI] [PubMed] [Google Scholar]
- 39.Moolenaar WH. Trends Cell Biol. 1994;4:213–219. doi: 10.1016/0962-8924(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 40.Kleber S, Sancho-Martinez I, Wiestler B, Beisel A, Gieffers C, Hill O, Thiemann M, Mueller W, Sykora J, Kuhn A, Schreglmann N, Letellier E, Zuliani C, Klussmann S, Teodorczyk M, Grone HJ, Ganten TM, Sultmann H, Tuttenberg J, von Deimling A, Regnier-Vigouroux A, Herold-Mende C, Martin-Villalba A. Cancer Cell. 2008;13:235–248. doi: 10.1016/j.ccr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 41.McMichael BK, Meyer SM, Lee BS. J Biol Chem. 2010;285:26641–26651. doi: 10.1074/jbc.M110.119909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kotelevets L, van Hengel J, Bruyneel E, Mareel M, van Roy F, Chastre E. Faseb J. 2005;19:115–117. doi: 10.1096/fj.04-1942fje. [DOI] [PubMed] [Google Scholar]
- 43.Ye X, Ishii I, Kingsbury MA, Chun J. Biochim Biophys Acta. 2002;1585:108–113. doi: 10.1016/s1388-1981(02)00330-x. [DOI] [PubMed] [Google Scholar]
- 44.Oh YS, Jo NW, Choi JW, Kim HS, Seo SW, Kang KO, Hwang JI, Heo K, Kim SH, Kim YH, Kim IH, Kim JH, Banno Y, Ryu SH, Suh PG. Mol Cell Biol. 2004;24:5069–5079. doi: 10.1128/MCB.24.11.5069-5079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C, Dandridge KS, Di A, Marrs KL, Harris EL, Roy K, Jackson JS, Makarova NV, Fujiwara Y, Farrar PL, Nelson DJ, Tigyi GJ, Naren AP. J Exp Med. 2005;202:975–986. doi: 10.1084/jem.20050421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bharti AC, Aggarwal BB. Biochem Pharmacol. 2002;64:883–888. doi: 10.1016/s0006-2952(02)01154-1. [DOI] [PubMed] [Google Scholar]
- 47.Schreck R, Albermann K, Baeuerle PA. Free Radic Res Commun. 1992;17:221–237. doi: 10.3109/10715769209079515. [DOI] [PubMed] [Google Scholar]
- 48.McCarthy JV, Ni J, Dixit VM. J Biol Chem. 1998;273:16968–16975. doi: 10.1074/jbc.273.27.16968. [DOI] [PubMed] [Google Scholar]
- 49.Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, Janeway CA, Medzhitov R, Flavell RA. Nature. 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 50.Jonat C, Rahmsdorf HJ, Park KK, Cato AC, Gebel S, Ponta H, Herrlich P. Cell. 1990;62:1189–1204. doi: 10.1016/0092-8674(90)90395-u. [DOI] [PubMed] [Google Scholar]
- 51.Ray A, Prefontaine KE. Proc Natl Acad Sci U S A. 1994;91:752–756. doi: 10.1073/pnas.91.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diefenbacher ME, Litfin M, Herrlich P, Kassel O. Mol Cell Endocrinol. 2010;320:58–66. doi: 10.1016/j.mce.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 53.de Lange T. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]