Abstract
Recently, functional and potent RNA interference (RNAi) has been reported in peripheral nerve axons transfected with short-interfering RNA (siRNA). In addition, components of RNA-induced silencing complex (RISC) have been identified in axotomized sciatic nerve fibers as well as in regenerating dorsal root ganglia (DRG) neurons in vitro. Based on these observations, and on the fact that siRNA and microRNAs (miRNA) share the same effector enzymes, we hypothesized that the endogenous miRNA biosynthetic pathway would respond to peripheral nerve injury. To answer this question, we investigated changes in the expression of miRNA biosynthetic enzymes following peripheral nerve crush injury in mice. Here we show that several pivotal miRNA biosynthetic enzymes are expressed in an injury-regulated pattern in sciatic nerve in vivo, and in DRG axons in vitro. Moreover, the sciatic nerve lesion induced expression of mRNA-processing bodies (P-bodies), which are the local foci of mRNA degradation in DRG axons. In addition, a group of injury-regulated miRNAs was identified by miRNA microarray and validated by qPCR and in situ hybridization analyses. Taken together, our data support the hypothesis that the peripheral nerve regeneration processes may be regulated by miRNA pathway.
Keywords: miRNA, RISC, P-body, sciatic nerve, axon growth, regeneration, axon varicosities
INTRODUCTION
One of the unique features of the peripheral nervous system (PNS) is its ability to regenerate axons after injury (Yoo et al., 2009). To initiate a regenerative response, the PNS neuron must shift its physiology from synaptic transmission and maintenance of structure to the growth of the axon (Benowitz and Yin, 2007). Shifting to the growth phenotype requires both activation of a growth program via gene transcription as well as activation of local signaling cascades that regulate axon assembly (Snider et al., 2002, Vogelaar et al., 2009). Recent observations suggest that the microRNA (miRNA) pathway may be involved in regulation of these processes (Hengst et al., 2006, Murashov et al., 2007).
The miRNA pathway is an important layer of post-transcriptional gene regulation (Jackson et al., 2010). MiRNAs are initially processed in the nucleus by biosynthetic enzymes, Drosha and DGCR8/Pasha, while in cytoplasm Dicer cleaves them into the mature miRNAs (Bernstein et al., 2001, Lee et al., 2003). MiRNAs then trigger formation of RNA-induced silencing complex (RISC), which is implicated in a sequence-specific translational repression called RNA interference (RNAi) (Bagga et al., 2005, Tan et al., 2009). Several subunits of RISC have been conclusively identified to date: Argonaute2 (Ago2) nuclease (Hammond et al., 2001, Meister and Tuschl, 2004), fragile X mental retardation protein (FMRP) (Ishizuka et al., 2002), p100 (Caudy et al., 2003), TRBP (Chendrimada et al., 2005), PACT (Lee et al., 2006), and RCK/p54 (Chu and Rana, 2006). Recent studies showed that Argonaute proteins interact with the RNA-binding protein GW182 (Sen and Blau, 2005, Eulalio et al., 2009a, Eulalio et al., 2009b, Takimoto et al., 2009). Observations revealed that GW182-containing foci, known as GW bodies (GWBs), coincide with mRNA-processing bodies (P-bodies) where GW182 co-localizes with the de-capping complex (Dcp1, Dcp2), Rap55 and Ro52 (Sen and Blau, 2005, Bhanji et al., 2007, Eulalio et al., 2009a, Eulalio et al., 2009b, Takimoto et al., 2009). Importantly, depleting GW182 suppressed silencing of miRNA targets. Therefore, these observations suggest that in addition to RISC, target silencing by miRNAs is regulated by P-bodies.
Recent studies revealed the differential expression of a number of miRNAs following traumatic injury in CNS, including the brain and spinal cord (Lei et al., 2009). Reduction in the expression of several sensory organ-specific miRNAs was also observed in the injured ipsilateral dorsal root ganglion (DRG) following spinal nerve ligation (SNL) (Aldrich et al., 2009). These evidences suggest that miRNAs are likely to be important mediators of plasticity. Functional RNAi machinery has been reported in axons of PNS neurons (Yoo et al., 2009). Components of RISC were observed in severed sciatic nerve fibers and cultured DRG axons (Hengst et al., 2006, Murashov et al., 2007). Based on these observations and on the fact that siRNA and miRNA share the same effector enzymes, it is logical to presume that the endogenous miRNAs machinery could be involved in the regulation of molecular response to peripheral nerve injury.
In the current study, we asked whether miRNA might be one of the pathways that respond to peripheral nerve crush. Here we show an injury-regulated expression of several pivotal miRNA biosynthetic enzymes in murine sciatic nerve in vivo, and in DRG axons in vitro. In addition, a group of miRNAs, which were expressed in an injury-regulated pattern in regenerating sciatic nerve and DRG was identified by miRNA microarrays, qPCR and in situ hybridization. These data provide further evidence in support of the hypothesis that the peripheral nerve regeneration processes may be regulated by miRNA pathway.
EXPERIMENTAL PROCEDURES
Animals
Experiments were performed on 8-wk-old CD-1 mice, obtained from Charles River laboratories (Wilmington, MA). Animals were housed one per cage under standard laboratory conditions, with a 12h light/dark schedule and unlimited access to food and water. All experimental procedures and the care of the animals were administered according to the guidelines set forth by the Animal Care and Use Committee of East Carolina University, an AAALAC-accredited facility.
Conditioning nerve lesion
Anesthesia was induced using intraperitoneal Ketamine (18/mg/ml)-Xylazine(2mg/ml) mixture (0.05 ml/10g of body wt). The procedure followed a protocol described previously (Islamov et al., 2004). Exposure of the right sciatic nerve was performed with sterile surgical instruments. Approximately 5mm of nerve was exposed from the sciatic notch to the trifurcation of the nerve. The exposed sciatic nerve was crushed in the mid-thigh for 15 sec with a fine hemostat. The wounds were closed with 3M Vetbond tissue adhesive and mice were allowed to recover for 4, 5 or 7 days. For the following experiment, the injured ipsilateral nerves were called injured nerves and the ipsilateral DRGs were called injured DRGs. The contralateral nerves were called naïve nerves and DRGs from contralateral side were called naïve DRGs. Nerves and DRGs from intact animals were called intact nerves and intact DRGs.
Sciatic nerve collection
After specified time periods, animals were euthanized and sciatic nerves were quickly dissected out, snap frozen in liquid nitrogen and stored at −80°C. The naïve nerve was taken from the contralateral side (sciatic nerve from left side). The excised crush sample was taken from the injury side approximately 5 mm in both directions from the point of injury. The intact nerve sample was taken from mice at the matched age without sciatic nerve crush surgery.
Dissociated DRG culture
Mouse L4/5 dorsal root ganglion (DRG) neurons were collected 5 days after a conditioning sciatic nerve crush from both the intact side and injured side. DRGs were dissociated with collagenase and 0.25% trypsin in Dulbecco’s modified Eagle’s medium (DMEM). The dissociated DRGs were plated on Poly-L-Lysine and Laminin (Invitrogen, Carlsbad, CA), coated plates. DRGs were grown in DMEM/F12 containing 10% horse serum, L-glutamine and N2 supplement at 37°C for 18hrs. Cytosin β-D-arabinofuranoside and 5,6-dichlorobenzimidazole riboside (Sigma, Saint Louis, Missouri) were added to the final concentration of 10μM and 80μM respectively in the growth medium to inhibit the growth of glial cells.
Protein lysates
Proteins were extracted from the sciatic nerves collected at specific time points. To obtain adequate proteins for these experiments, at least 5 animals were pooled per time-point. Samples of sciatic nerves were homogenized in ice-cold homogenization buffer, containing protease inhibitor cocktail (Sigma, St. Louis, MO), 1mM PMSF, 20mM Tris, 2mM EGTA, 2mM EDTA, 6mM β-mercaptoethanol and 10% Triton and centrifuged at 10,000g for 10 min at 4°C. The supernatant was transferred to a fresh tube and stored at −20 °C until ready for use.
Immunoblotting
Proteins for Western Blotting analysis were quantified using the BioRad reagent, and 20 μg of solubilized proteins were loaded per lane on sodium dodecyl sulfate gels and separated by SDS-PAGE. The separated proteins were then transferred to immobilon P membranes (Millipore Corporation; Bedford, MA). Membranes were blocked in 5% non-fat milk/PBST for 1 hr at room temperature on a shaker, and then probed with primary antibody against FMRP (Darnell et al., 2005), Ago2 (Ikeda et al., 2006), P-100 (Keenan et al., 2000), Dcp1 (Lykke-Andersen, 2002) or from Abnova; Walnut, CA, Dcp2 (Lykke-Andersen, 2002, Wang et al., 2002), Rap55 (Yang et al., 2006), Ro52 and GWB IC-6 (Pauley et al., 2006) in 5% nonfat milk/PBST at 4°C overnight. For negative control groups, membranes were incubated with pre-immune serum. The membranes were washed three times with TBS, and then incubated with secondary antibody in 5% milk/PBST for two hours at room temperature. After incubation with horseradish peroxidase (HRP) conjugated secondary antibodies, bound antibodies were detected using a chemiluminescence detection system (ECL plus Western blotting detection reagent, Amersham; Arlington Heights, IL, USA). Densitometry was performed using Kodak 1D Image Analysis software. Band values were normalized to alpha-tubulin, to obtain the relative densitometric intensity. One-way ANOVA were performed on injured, naive versus intact relative densitometric intensity for each antibody tested.
List of antibodies
The following antibodies were used for immunodetection procedures: Rabbit polyclonal anti-AGO2- specific antibody (kindly donated by Tom Hobman, University of Alberta, Canada), Guinea pig polyclonal anti-P100 antibody (kindly donated by Tom Keenan, VA Polytechinic Institute and State University, Blacksburg, VA). Mouse monoclonal anti-FMRP antibody was purchased from Developmental Studies Hybridoma Bank(Iowa City, Iowa). Western blot analysis of P-body components in the sciatic nerve utilized rabbit polyclonal primary antibody against Dcp1, Dcp2 from Jens Lykke-Anderson (University of Colorado, Colorado), rabbit polyclonal anti-Dcp2 primary antibody from M. Kilejian (Rutgers University, New Jersey), Mouse Anti-Human Dcp1A Monoclonal Antibody from Abnova (Walnut, CA), human anti-Rap55 antibody (from Donald Bloch, Harvard University, MA) or antibody against GWB IC6 from Marvin Fritzler (University of Calgary, Alberta, Canada), or Rabbit anti-Ro52 polyclonal antibody from Millipore Corporation (Bedford, MA). Mouse monoclonal neuro-specific beta III tubulin antibody (TUJ-1) was obtained from Covance Research Products, Inc. (Denver, PA). The specificities of the antibodies, provided by individual investigators, have been validated by original research groups and the corresponding papers have been published elsewhere. For secondary antibodies, we used HRP conjugated anti-rabbit IgG and HRP conjugated anti-mouse IgG from R&D system (Minneapolis, MN). IRDye 800CW goat anti-Mouse IgG and IRDye, 680LT goat anti-Rabbit IgG secondary antibodies were also used (LI-COR Corporate, Nebraska USA). For fluorescence studies, secondary FITC-, TX Red-, Cy-3, or Cy5-conjugated IgG (Jackson ImmunoResearch Laboratories, Inc; West Grove, PA) or Alexa Fluor 350 (anti-mouse IgG) and Alexa Fluor 594 goat anti-human IgG from Invitrogen (Eugene, Oregon, USA) were applied.
Indirect Immunofluorescence
After culturing lumbar DRGs on coverslips overnight, the DRGs were fixed with 4% paraformaldehyde for 5 min and washed with PBST. After blocking with 10% goat serum for 30 min at room temperature, the samples were incubated with the indicated primary antibodies (1:100 dilution) at 4 °C overnight or for 2 hours at 37 °C. Bound antibodies were detected using FITC-, TX Red-, AMCA- or Alexa Fluor® 594 (Invitrogen) conjugated IgG in 1:100 dilutions. Negative controls included samples processed in parallel with non-immune serum or without primary antibodies. After mounting the slides with anti-fading media (Invitrogen), images were viewed with an Olympus IMT-2 fluorescent microscope and recorded using the Spot digital camera system (Diagnostic Instruments, Sterling Heights, MI) or viewed and captured with Leica DMI6000 B Imaging System powered by MetaMorph. The images we acquired were all single plane fluorescent images. For high magnification images and Z-stacks, images were taken with Olympus FluoView™ FV1000 confocal microscope at UNC Neuroscience Center (Chapel Hill, NC).
MiRNA microarrays
Total RNA for the microarray expression analysis was isolated from sciatic nerves, pooled from ten mice, at 4, and 7 days after the sciatic nerve crush. miRNA expression profiling. Five micrograms of total RNA was labeled with RNA ligase and a Cy3-conjugated dinucleotide, and hybridized to custom oligonucleotide microarrays as described previously (Thomson et al., 2004). Cy3 median intensity values were filtered to remove data points that did not exceed background levels by twofold (He et al., 2005). Values were log2-transformed and median-centred by array. Clustering was performed with Cluster (Stanford University), using values that were median-centred by gene. Dendrograms and expression maps were generated using Treeview (Stanford). Some pooled samples were sent to UNC Lineberger Comprehensive Cancer Center Genomics Core. Following a quality check, they were hybridized to an 8×15K miRNA one color arrays (Agilent, Santa Clara, CA). Both custom and Agilent arrays were done in duplicates and repeated two times. Normalization and analyses were performed with GeneSpring software (Agilent). The GeneSpring analysis software was used for the identification of statistically significant upregulated or downregulated miRNAs at selected time points. The analysis also allowed for identification of a different temporal pattern of expression in the crushed groups compared with the control groups at the chosen time points. For further analyses we selected three miRNAs that were significantly upregulated and one that was significantly downregulated in both custom arrays as well as in Agilent arrays. The selected miRNAs were validated by qRT-PCR, performed on the same total RNA preparations as used for microarrays experiments (Please see supplementary data).
Real-time RT-PCR
Total RNA was isolated from intact and crushed sciatic nerves (n=6). Samples from the experimental and control tissue were run two to three times in triplicates using the NCode™ miRNA Amplification System (Invitrogen) in a BioRad PCR thermocycler. Primers for miRNA include miR124a (5′-TAAGGCACGCGGTGAATGCC-3′), miR221 (5′-AGCTACATTGTCTGCTGGGTTTC-3′), U6 (Ambion mirVana™ qRT-PCR Primer Sets, catalog #30303), miR142-5p (5′-CATAAAGTAGAAAGCACTACTAAAA-3′), miR-199a (5′-ACAGTAGTCTGCACATTGGTTA-3′), miR9 (5′-TCTTTGGTTATCTAGCTGTATGA-3′) and miR21 (5′-TAGCTTATCAGACTGATGTTGA-3′). To ensure equal loading of RNA, relative quantification of gene expression was calculated against reference gene S12. Data were normalized to S12 using the 2−ΔΔCT method.
In situ hybridization
Similar to the immunofluorescence protocol described above, dissociated DRG cultures were postfixed in 4% paraformaldehyde solution in PBS at room temperature for 5 min, washed in PBS twice for 5 min and incubated in 100% methanol + 0.3% hydrogen peroxide solution for 10 min at 4°C. After two washes in PBS for 10 min, cell cultures were prehybridized at 42°C for 1 hour in hybridization buffer, containing 600 mM sodium chloride, 50mM sodium phosphate buffer (PH 7.0), 5.0 mM EDTA, 0.02% Ficoll (Sigma), 0.02% BSA (Sigma), 0.02% polyvinylpyrrolidone (Sigma), 200ng/ml sheared and denatured salmon sperm DNA (Sigma), and 40% formamide (Fisher Scientific, Pittsburgh, PA, USA). Hybridization was performed at 42°C in the same buffer with the addition of dextran sulfate to 7%, tRNA(baker’s yeast) to 0.1 mg/ml, poly-A to 10μg/ml, and LNA antisense or sense oligonucleotide probes against miR-21, miR-142-5p and U6 to a final concentration of 1 ng/μl. Mir-21 FITC –labeled LNA antisense and sense probes were obtained from RiboTask (Odense, Denmark). Mir-142-5p FITC –labeled LNA antisense and scrambled probes, and a U6 LNA probe were obtained from Exiqon (Woburn, MA). After hybridization coverslips were incubated for 1 hour with primary antibodies against FITC, neuronal β-tubulin and GWB. To visualize the signal, FITC-, TX Red-, and AMCA- labeled secondary antibodies (Invitrogen) were applied for 1 hour in 1:100 dilution. Slides were viewed on an Olympus IMT-2 fluorescent microscope.
Statistical analysis
Statistical analysis was performed with one-way ANOVA and Newman-Keuls Multiple Comparison Test using Prism (GraphPad Prism version 3.00 for Windows, GraphPad Software; San Diego, CA, USA). All values are represented as the Mean ± SE.
RESULTS
Levels of miRNA biosynthetic proteins in sciatic nerve are regulated by injury
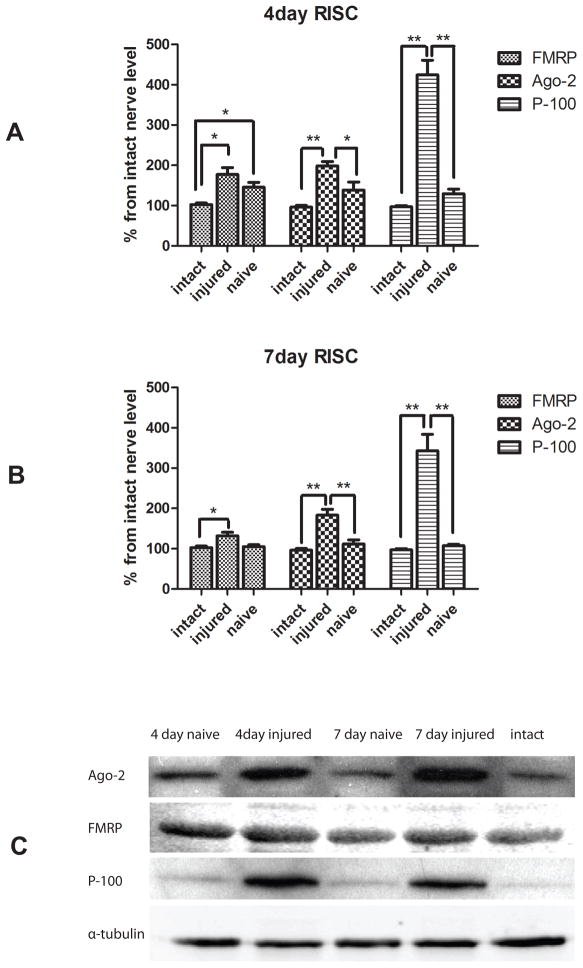
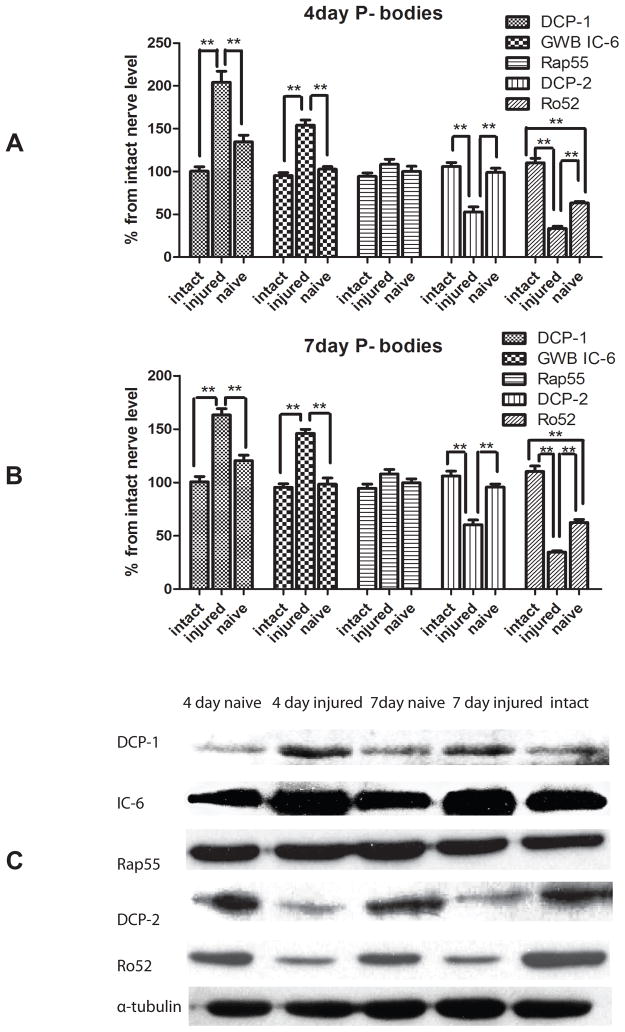
To investigate whether the expression of miRNA biosynthetic proteins was correlated with injury, we first determined the expression of protein components of RISC (FMRP, Ago2 and P-100) and P-body (Dcp1, Dcp2, Rap55, Ro52 and GWB IC-6) at 4 and 7 days post-crush. In a mouse, the process of nerve regeneration has been shown to be active over this period, justifying the time-points for these experiments (Garbay et al., 2000, McMurray et al., 2003). Previous studies in our lab already showed the existence of the components of RISC in peripheral nerve axon in vivo and vitro (Murashov et al., 2007). Here we used immunoblot analyses to determine the injury-regulated expression of biosynthetic components of RISC and P-body. Immunoblot analysis revealed the expression of the components of these multiprotein complexes in response to injury, indicating the potential role of miRNA biosynthetic machinery in peripheral nerve regeneration. Figure 1 indicates that the expressions of the RISC components, including FMRP, Ago-2, and P-100 were induced in response to injury, when compared to intact nerve. The densitometry showed significantly elevated levels of Ago2, FMRP, and P-100 respectively at 4 days (Fig 1A) and 7 days (Fig 1B) after injury. The contralateral naïve side of the nerve had the comparable expression level of RISC components as the intact nerve. The results indicated the induction of RISC in sciatic nerve upon injury. In addition, the injury differentially affected levels of P-body proteins, another miRNA-associated multiprotein complex (Figure 2). We observed upregulation of Dcp1 and GWB IC-6 at 4 days following sciatic nerves injury (Fig 2A) and downregulation of Dcp2 and Ro2 respectively, at 4 day post injury (Fig 2A). In contrast, the expression of Rap55 was not affected. Therefore, the results indicated that injury to sciatic nerve selectively affected the expression of the P-body components.
Figure 1.
Immunoblot analyses for FMRP, Ago2, and p-100 protein in sciatic nerve.
Proteins were extracted from 3 groups of pooled samples (intact sciatic nerves, the contralateral naive nerve and the injured sciatic nerve) at 4 days or 7 days after a conditioning nerve lesion. Immunoblot analyses were conducted with primary antibodies against FMRP, AGO2, and p-100. Using the expression level in intact nerve as a control, relative densitometric intensities were normalized to alpha-tubulin. Densitrometry showed a more abundant expression of RISC component (FMRP, Ago2, p-100) in injured sciatic nerve at both 4 days (A) and 7 days (B) after injury (N=3, * indicate P<0.05, ** indicate P<0.01). Representative immunoblot of FMRP, Ago2, and p-100 protein is shown in C.
Figure 2.
Western Blot analyses of P-body components in the sciatic nerve after injury.
Western Blot analysis of P-body components in the sciatic nerve utilized the primary antibody against Dcp1, Dcp2, Rap55, Ro52, and GWB IC6. Pooled samples (6 nerves per each sample) were extracted from intact sciatic nerves, the contralateral naive nerve and the injured sciatic nerve at 4 days or 7 days after a conditioning nerve lesion. Protein levels were quantified by band densitomery and normalized to alpha-tubulin at 4 days (A) and 7 days (B) after conditional lesion. Immunostaining for Dcp1, the catalytic factor of Dcp2, showed an increase in expression in response to injury, whereas the expression of Dcp2, the decapping enzyme, decreased after injury at both time points. Immunostaining for GWB IC-6 showed much higher GWB IC-6 expression at 4 days and 7 days after injury, while Rap-55 showed no change in expression in response to injury. Ro52 expression level was significantly decreased following injury (N=3, * indicates P<0.05, ** indicates P<0.01). Representative immunoblots for Dcp1, Dcp2, Rap55, Ro52, and GWB IC6 are shown in C.
Components of miRNA machinery are differentially expressed in dissociated DRG neurons after injury
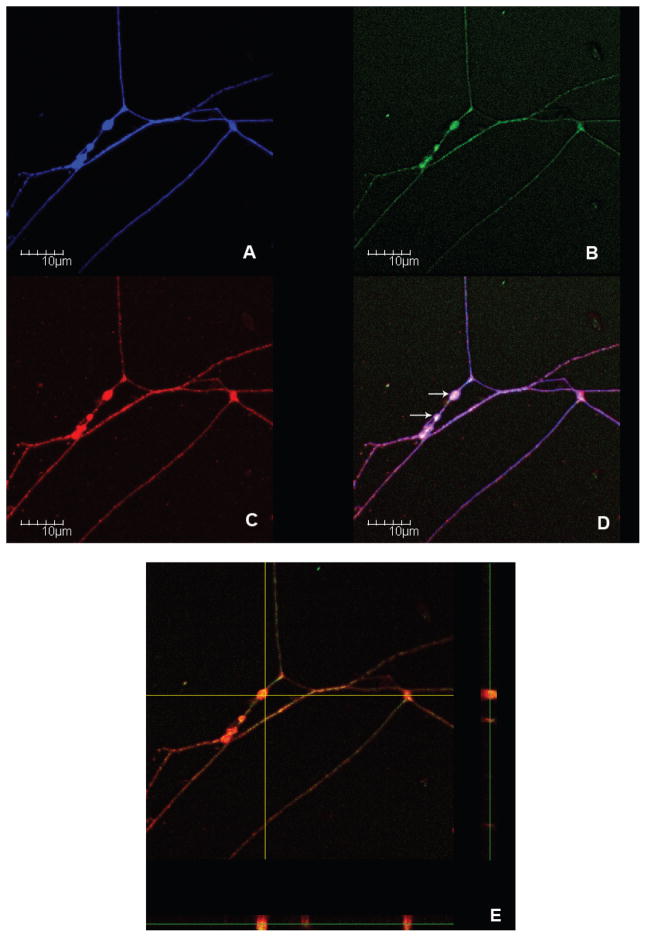
In the previous experiments we showed that miRNA machinery was present and induced in peripheral nerve after injury. In the next experiment we asked whether the source of the differentially expressed miRNA biogenesis enzymes might be of axonal origin. To answer this question, we performed immuofluorescence staining on murine dissociated DRG neurons to visualize the presence of P-body proteins in axons. The DRG neurons plated at a low density were stained with FITC conjugated antibodies against Dcp1 (Figure 3B) and Alexa 594-conjugated antibodies against GWB IC-6 (Figure 3C). The human polyclonal sera against GWB was used previously to detect P-bodies (Cougot et al., 2008). Since the polyclonal antibodies could detect other proteins associated with GW182 protein in GWB (Moser et al., 2007), antibodies against Dcp1, another major component in P-bodies was used to further confirm the localization of P-bodies in axon. AMCA staining against TUJ1, a neuronal marker indicated the location of neuronal soma and axons (Figure 3A). Confocal immunofluorescence showed colocalization of GWB IC-6 with Dcp1 in the varicosities (VR) along axons (Figure 3D). A Z-scanning showed a complete overlap of immuoreactivities against Dcp1 and GWB in axon.
Figure 3.
Colocalization of P-body proteins in the VR of regenerating DRG axons.
Dissociated cultures of mouse DRG neurons collected 5 days after a conditioning lesion were co-labeled with FITC (Green) conjugated antibodies to Dcp1 (B) and Alexia 594 (Red) conjugated antibodies to GWB IC-6 (C). TUJ1 (Blue) staining is used to visualize the axon (A). The merged images revealed co-localizations of investigated proteins in regenerating axons (D). The Z-scanning further confirmed their overlapping expression in the VR (E). White arrows in image D point at P-bodies in the VR.
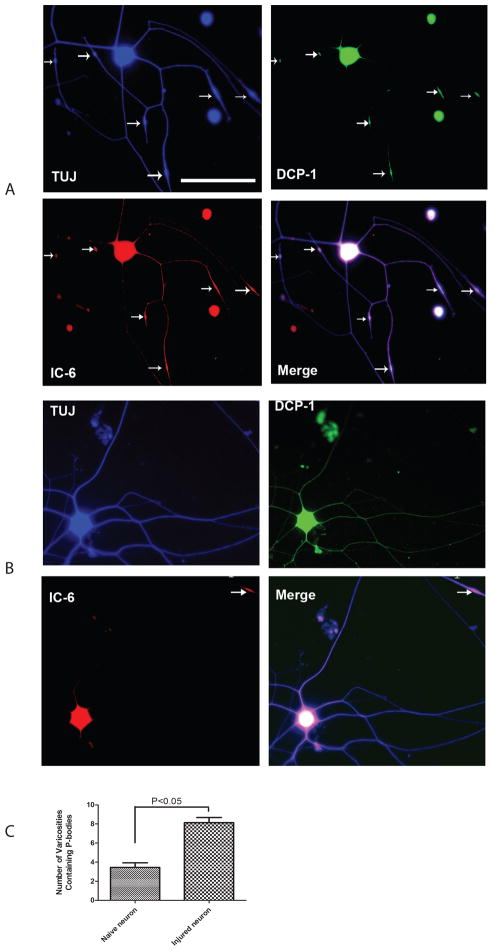
These results clearly demonstrated the colocalization of P-bodies proteins in the VR along axons. By comparing the images of DRG neurons collected from the injured side (Fig. 4A) and the DRG neurons collected from the naïve side (Fig. 4B), we revealed that preconditioning by crush injury not only induces more robust axon regenerative growth but also induces P-bodies containing VR in regenerating axons. Quantifications of the mean number of the VR containing P-bodies per neuron in naïve versus regenerating DRG cultures, revealed that the average number of VR containing P-bodies per neuron increased over ~2.4 fold, from around 3 (n=30) to around 8 (n=30) following injury (Fig. 4C). These findings were consistent with the Western Blot results, which indicated that the expression of miRNA biogenesis enzymes followed an injury-regulated pattern.
Figure 4.
Number of VR containing P-bodies along the axon markedly increases after conditioning lesion.
Low density dissociated DRG cultures were labeled with antibodies to neuronal marker TUJ1 (Blue), Dcp1 (Green) and GWB IC-6 (Red). Merges of the images revealed the localization of the P-bodies in soma and axon. The appearance of P-bodies was limited to the VR along the axon. The fluorescent images in the upper panel show neuronal cultures after sciatic conditioning lesion (A). The images in the lower panel show neurons from naïve side (B). After lesion, the average number of VR containing P-bodies (white arrows) per neuron has significantly increased (C). Scale bar on microphotographs is 100μm.
MiRNAs are expressed in an injury-regulated pattern after conditioning nerve lesion
Since we observed an injury-regulated expression of miRNA biosynthetic enzymes after the sciatic nerve crush, we asked if the pattern of miRNA expression could be also injury-regulated. Total RNA was isolated from pooled crushed and contralateral intact sciatic nerves and subjected to custom (Thomson et al., 2004) and Agilent miRNA expression arrays. Genespring analysis of array data revealed a set of differentially expressed miRNAs. The miRNAs that were following injury-regulated expression pattern in both custom and Agilent arrays at both 4 and 7 days time points were selected for subsequent analyses. Three most highly upregulated miRNAs miR-21, miR-142-5p, and miR-221, as well as brain-specific miR-124a, which was significantly downregulated, were selected for further analyses (see Supplementary data).
qPCR confirms injury-regulated expression of miR-21, miR-142-5p, miR-221, and miR-124a
To verify miRNA array data, we next determined the expression of the selected miRNAs in the same total RNA samples that were used for arrays. Real-Time RT-PCR demonstrated the same pattern of expression of miR-124a, miR-221, miR-142-5p, and miR-21, after sciatic nerve crush (Figure 5). In agreement with the microarray data, the real time RT-PCR demonstrated that miR-124a is significantly downregulated after nerve injury. At the same time, we observed an upregulation of miR-221, miR-142-5p, and miR-21. These data provided further evidence that these miRNAs are injury-regulated.
Figure 5.
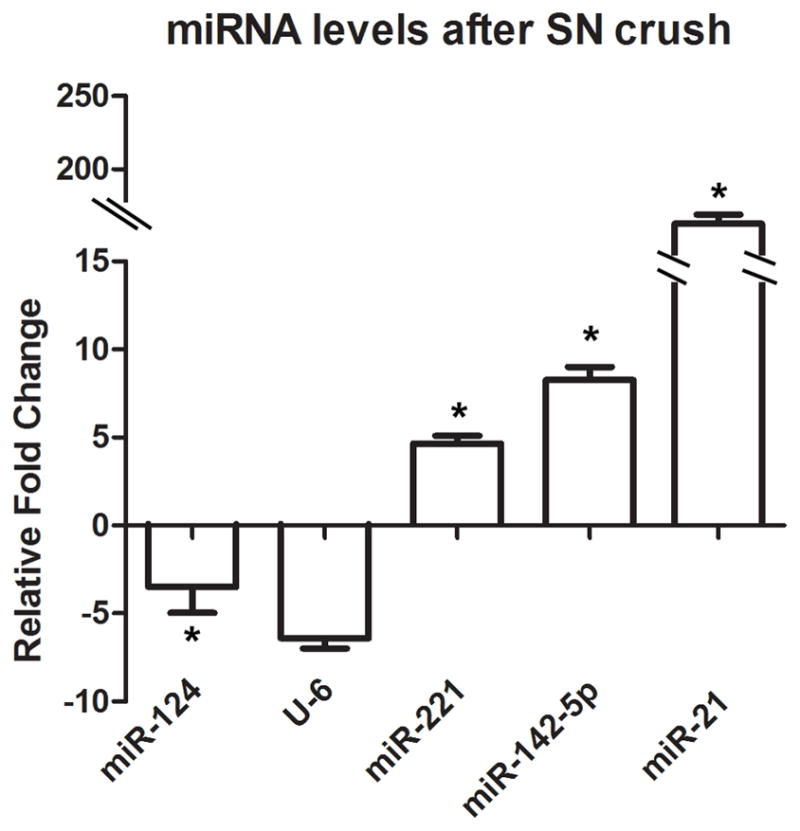
Real-time qPCR for miRNAs. Graph indicates relative change in miRNA level. Total RNA was isolated from intact and crushed sciatic nerves (n=6). Relative quantitation of miRNA level was calculated against the reference gene (S12), and a relative fold change was determined using the 2 ΔΔCT method (n=3, *p<0.05). The levels of miR-221, miR-142-5p and miR-21 showed significant increase in sciatic nerve after crush.
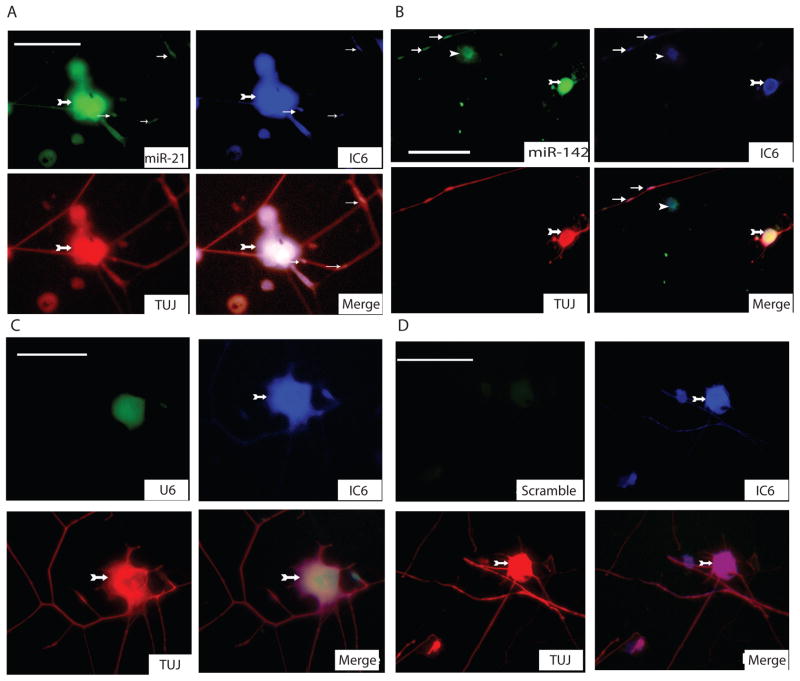
In situ hybridization analysis localized injury-regulated miRNAs miR-21 and miR-142-5p to regenerating DRG axons
In the previous experiments we have shown that several miRNAs are expressed in an injury-regulated pattern in peripheral nerve. The potential sources of miRNAs in crushed nerve samples could include, glial cells, connective tissue, macrophages and axons. In the next experiment we asked whether the identified differentially expressed miRNAs might be present in regenerating DRG neurons. To answer this question, we performed fluorescent in situ hybridization (FISH) on murine dissociated DRG neurons plated at a low density to visualize the presence of miRNAs in cell bodies and axons. For this experiment we selected miR-21 and miR-142-5p, which showed an opposite regulation by injury in the miRNA arrays. After FISH, DRG preparations were immunostained with TUJ1 antibody against neuron-specific β-tubulin. The localization of P-bodies was visualized by immunostaining against GWB IC-6. Analyses of these immunofluorescent images revealed presence of miR-21 and miR-142-5p in DRG cells and their axons (Figure 5). Interestingly, the distribution of miRNAs in the axon was overlapping with P-body immunostaining including axon VR. These results demonstrate an interesting possibility that miRNAs could be induced in cell body upon crush and subsequently translocated into the axon.
DISCUSSION
Our study demonstrated that the levels of several miRNA biosynthetic proteins, as well as miRNAs, were affected by sciatic nerve injury. This observation is supported by previous studies, which demonstrated functional RNAi machinery in peripheral nerve axons (Buckingham et al., 2004, Hengst et al., 2006, Murashov et al., 2007) and one recent observation that has revealed expression of miRNAs in the superior cervical ganglia axons (Natera-Naranjo et al., 2010). While these reports demonstrated that miRNAs and miRNA biosynthetic enzymes are present in peripheral nerve axons in vivo and in vitro, there were no systematic studies addressing physiological role of the miRNA biosynthetic pathway in the PNS. In the current paper, we asked whether miRNA might be one of the pathways that is activated in response of the PNS to peripheral nerve injury. To answer this question, we investigated levels of miRNA biosynthetic proteins and miRNAs after nerve crush, as well as their localization in regenerating cells and axons by FISH and indirect immunofluorescence. Here we showed that several components of RISC and P-bodies, as well as miRNAs were expressed in an injury-regulated pattern in the sciatic nerve and DRG axons after crush injury, supporting a potential physiological role of miRNA pathway in regulation of regenerative nerve growth.
Components of RISC and P-bodies respond to injury of peripheral nerve
In the previous observation we detected expression of several RISC components in murine sciatic nerve (Murashov et al., 2007). In the current report, we observed increased protein levels of Ago2, FMRP, and p100 in response to sciatic nerve crush. The observed injury-regulated expression of these RISC components may indicate that the induction of RISC may be a part of the PNS regenerative response. Interestingly, Ago2, the pivotal RISC protein, was recently detected in P-bodies, as well as, in the stress granules (SGs), which suggested a potential functional relationship between RISC, SGs and P-bodies in physiological response to cellular stresses (Leung et al., 2006, Didiot et al., 2009). P-bodies are an important component of the miRNA machinery as the sites for either degrading or protecting and sequestering mRNA (Parker and Sheth, 2007, Balagopal and Parker, 2009). P-bodies are dynamic complexes which assembly are dependent on, and proportional to, the cytoplasmic pool of translationally repressed messenger ribonucleoproteins (mRNPs) (Teixeira et al., 2005, Franks and Lykke-Andersen, 2008). Although the complete composition of P-bodies is not yet established, they are known to include the decapping enzymes Dcp1 and Dcp2, the activator of decapping Dhh1/RCK/p54, Pat1, Scd6/RAP55, Edc3, the Lsm1-7 complex and the exonuclease Xrn1 (Parker and Sheth, 2007, Balagopal and Parker, 2009). In agreement with previous observation that various cellular stresses initiate the formation of P-bodies (Balagopal and Parker, 2009), we detected the change in the expression level of P-bodies components in sciatic nerve after crush injury using immunoblot analyses.
In particular, we made an interesting observation that sciatic nerve injury led to elevated levels of GWB IC-6 and Dcp1 but decreased expression of Dcp2. The apparent discrepancy may be partially explained by recent report in which, in vitro depletion of the decapping enzyme Dcp2 caused increased P-bodies assembly (Franks and Lykke-Andersen, 2008). While Dcp1 and Dcp2 are both core components of P-bodies, which catalyze removal of the 5′ cap structure from mRNA, they are structurally and functionally different. Dcp1 behaves as a decapping enzyme regulatory subunit, while Dcp2 functions as a catalytic subunit of the decapping enzyme (Sheth and Parker, 2003, She et al., 2008). Therefore, decreased level of Dcp2 may results in accumulation of undigested mRNAs and thus in additional P-bodies assembly. At the same time, another recent observation points out that mammalian cells possess multiple mRNA decapping enzymes and Dcp2 protein may only modestly contribute to bulk mRNA decay in mouse and human tissues (Song et al., 2010).
We also observed an increase in the number and the size of P-bodies after conditioning nerve lesion in the regenerating DRG axons. It is well known that conditioning sciatic nerve crush produces robust regenerative response with concomitant increase in mRNA and protein synthesis (Willis and Twiss, 2006). We presume that the increase in the pool of mRNAs my initiate P-bodies assembly. Indeed, overexpression of a nontranslating mRNAs in yeast (Teixeira et al., 2005) was shown to potentiate P-bodies formation. Moreover, the size and number of P-bodies were proportional to the pool of untranslated mRNAs (Teixeira et al., 2005, Parker and Sheth, 2007). Therefore, when mRNAs are trapped in association with ribosomes, P-bodies decline; conversely, when mRNAs dissociate from ribosomes due to inhibition of translation initiation, P-bodies increase in size and numbers (Teixeira et al., 2005, Parker and Sheth, 2007, Balagopal and Parker, 2009). Consequently, nerve crush may lead to a larger pool of untranslated mRNAs and to increase in the number and size of functional P-bodies (Kedersha et al., 2005, Koritzinsky et al., 2006).
We made another interesting observation that P-bodies were preferentially localized to the axon VR. The number of VR was markedly higher in the regenerating versus naïve DRG axons. While the physiological role of VR is not clear their alleged function has been associated with presynaptic terminals, places of action potential waveform modulation and organelle accumulation and sequestration (Bennett and Muschol, 2009). In addition VR were described as the sites of mRNA concentration and protein synthesis (Lee and Hollenbeck, 2003). Interestingly, it was recently suggested that VR are the sites of clustering of excess growth resources along developing neuritis (Malkinson and Spira, 2010). Our data are complimentary to these observations suggesting that P-body machinery localized to VR can regulate the growth resources by managing mRNAs pool.
Identification of miRNAs specifically expressed in injured sciatic nerve
We used microarrays and RT-PCR to profile miRNA expression changes in the sciatic nerve of mice after injury. The results revealed a group of miRNAs expressed in an injury-regulated pattern. For analysis, we chose three most highly upregulated miRNAs miR-21, miR-142-5p, and miR-221, as well as brain-specific miR-124a, which was significantly downregulated after nerve crush. Interestingly, miRNAs miR-21 and miR-142-5p were reported to be also upregulated following brain injury (Lei et al., 2009). In addition, miR-221 was found to be relatively abundant in distal axons of SCG and had elevated expression level after spinal cord injury (Liu et al., 2009, Natera-Naranjo et al., 2010). The brain-enriched miRNA miR-124 is an important regulator of the temporal progression of adult neurogenesis in mice (Cheng et al., 2009b). Knockdown of endogenous miR-124 maintained subventricular zone stem cells as dividing precursors, whereas ectopic expression led to increased neuron formation. Furthermore, blocking miR-124 function during regeneration led to hyperplasias, followed by a delayed burst of neurogenesis (Cheng et al., 2009a). Although the changes in the miRNA expression profile following injury and development have been reported, the mechanism that regulates miRNA expression has not been yet elucidated. In the current paper we revealed that expression of several miRNA biosynthetic enzymes were injury-regulated. This let us to propose that the change in levels of miRNA biosynthetic proteins may play a role in the injury-regulated expression of miRNAs.
Taken together, our data implicate miRNA biosynthetic pathway in PNS response to injury. Further studies should provide important insight into the role of specific miRNAs in peripheral nerve regeneration. This finding may have significant clinical potential in the treatment of neurodegenerative diseases, specifically those involving peripheral nerves.
Studies show that minimal RISC, basically an Argonaute 2 protein and a single-strand RNA, is sufficient to catalyze target RNA recognition and cleavage (Yang and Yuan, 2009). Ago2 and FMRP are both localized to the stress granules (SGs) where RNA integrated stress response are proposed to occur.
Although no previous study has directly associated P-bodies with peripheral nerve health, identification of patients with auto-immunity to GWBs suggests such a role. Clinical studies indicate that these patients most often suffer from motor and sensory peripheral neuropathy (Bhanji et al., 2007). Although it is not known whether the resulting neuropathy is axonal or glial in its origin, it is logical to presume that autoimmune decrease of P-bodies in axon may negatively impact intra-axonal translation and become detrimental to axon physiology.
CONCLUSIONS
Taken together, this work offers insights into the functional interplay between miRNAs and their biosynthetic enzymes in peripheral nerve injury. In conclusion, we show that injury to peripheral axons induces changes in the content and distribution of some biosynthetic proteins of the miRNA machinery in cultured DRG neurons and their axons. Together with previous findings, these observations suggest that, in addition to RISC, other factors such as P-bodies might underlie the changes in regenerating axons induced by injury. Those factors probably act in a synergistic way thus leading to the promoting regenerative axon growth.
Figure 6.
Colocalization of miRNAs and GWB in neurons and axons.
Fluorescent in-situ hybridization for miR-21 (A), miR-142-5p (B) revealed the distribution of miRNAs in axon. Immunostaining for neuronal β-tubulin served as a neuronal marker and immunostaining for GWB protein marked the localization of P-bodies. U6, a small nuclear RNA was used as a positive control (C). Scrambled probes were used as another negative control. . Green fluorescence shows the distribution of miRNAs and U6. Red fluorescence indicates immunodetection of neuronal β-tubulin protein with TUJ1 antibodies (1:100). Blue signals indicate GWB protein. White arrows indicate fluorescent signals within the axons where miRNA and P-bodies components resided. Arrows with tail indicate neuronal cell bodies, small arrows show P-bodies, arrow heads indicates the non-neuronal cells (Scale bar=75μm).
Table 1.
Regulation of miRNAs at 4 days after nerve crush
Total RNA for the microarray expression analysis was isolated from sciatic nerves, pooled from ten mice, at 4, and 7 days after the sciatic nerve crush. Both custom and Agilent arrays were done in duplicates and repeated two times including dye swap. Normalization and analyses were performed with GeneSpring software (Agilent). Data from custom and Agilent arrays were log2-transformed and median-centred by array. The GeneSpring analysis software was used for the identification of statistically significant upregulated or downregulated miRNAs at selected time points. The analysis also allowed for identification of a different temporal pattern of expression in the crushed groups compared with the control groups at the chosen time. For further analyses we selected three miRNAs that were significantly upregulated and one that was significantly downregulated in both custom arrays as well as in Agilent arrays.
| Custom array | Fold Diff | Regulation | Agilent array | Fold Diff | Regulation | Average of both arrays | Fold Diff | Regulation |
|---|---|---|---|---|---|---|---|---|
| miR-424 | 2.05 | up | miR-29b | 2.14 | up | miR-29b | 2.26 | up |
| miR-223 | 2.22 | up | miR-142-5p | 2.27 | up | miR-142-5p | 2.42 | up |
| miR-29b | 2.38 | up | miR-221 | 2.63 | up | miR-424 | 2.43 | up |
| miR-142-5p | 2.56 | up | miR-223 | 2.70 | up | miR-223 | 2.46 | up |
| miR-221 | 2.77 | up | miR-424 | 2.81 | up | miR-221 | 2.70 | up |
| miR-142-3p | 5.21 | up | miR-142-3p | 4.56 | up | miR-142-3p | 4.88 | up |
| miR-21 | 13.67 | up | miR-21 | 13.04 | up | miR-21 | 13.36 | up |
| miR-133b | 1.23 | down | miR-99a | 2.02 | down | miR-99a | 1.67 | down |
| miR-99a | 1.32 | down | miR-124a | 2.04 | down | miR-30d | 1.77 | down |
| miR-30d | 1.39 | down | miR-125b | 2.08 | down | miR-143 | 1.86 | down |
| miR-143 | 1.60 | down | miR-100 | 2.09 | down | miR-124a | 1.92 | down |
| miR-9* | 1.66 | down | miR-422b | 2.11 | down | miR-9* | 1.94 | down |
| miR-9 | 1.72 | down | miR-143 | 2.12 | down | miR-100 | 1.95 | down |
| miR-124a | 1.79 | down | miR-30d | 2.15 | down | miR-422b | 1.98 | down |
| miR-100 | 1.81 | down | miR-9* | 2.22 | down | miR-125b | 2.03 | down |
| miR-30a-3p | 1.82 | down | miR-138 | 2.40 | down | miR-9 | 2.13 | down |
| miR-422b | 1.84 | down | miR-338 | 2.44 | down | miR-30a-3p | 2.18 | down |
| miR-150 | 1.95 | down | miR-9 | 2.54 | down | miR-338 | 2.24 | down |
| miR-125b | 1.98 | down | miR-30a-3p | 2.55 | down | miR-138 | 2.26 | down |
| miR-338 | 2.04 | down | miR-145 | 2.75 | down | miR-150 | 2.52 | down |
| miR-182 | 2.07 | down | miR-150 | 3.08 | down | miR-145 | 2.64 | down |
| miR-138 | 2.11 | down | miR-183 | 3.23 | down | miR-204 | 2.78 | down |
| miR-204 | 2.32 | down | miR-204 | 3.23 | down | miR-182 | 2.79 | down |
| miR-183 | 2.44 | down | miR-182 | 3.51 | down | miR-183 | 2.83 | down |
| miR-145 | 2.53 | down | miR-133b | 4.47 | down | miR-133b | 2.85 | down |
Table 2.
Regulation of miRNAs at 7 days after nerve crush
Total RNA for the microarray expression analysis was isolated from sciatic nerves, pooled from ten mice, at 4, and 7 days after the sciatic nerve crush. Both custom and Agilent arrays were done in duplicates and repeated two times including dye swap. Normalization and analyses were performed with GeneSpring software (Agilent). Data from custom and Agilent arrays were log2-transformed and median-centred by array. The GeneSpring analysis software was used for the identification of statistically significant upregulated or downregulated miRNAs at selected time points. The analysis also allowed for identification of a different temporal pattern of expression in the crushed groups compared with the control groups at the chosen time. For further analyses we selected three miRNAs that were significantly upregulated and one that was significantly downregulated in both custom arrays as well as in Agilent arrays.
| Custom array | Fold Diff | Regulation | Agilent array | Fold Diff | Regulation | Average of both arrays | Fold Diff | Regulation |
|---|---|---|---|---|---|---|---|---|
| miR-29b | 1.13 | up | miR-29b | 2.17 | up | miR-29b | 1.13 | up |
| miR-223 | 1.49 | up | miR-142-5p | 2.30 | up | miR-223 | 1.49 | up |
| miR-221 | 1.87 | up | miR-221 | 2.72 | up | miR-221 | 1.87 | up |
| miR-142-5p | 1.94 | up | miR-223 | 2.75 | up | miR-142-5p | 1.94 | up |
| miR-424 | 2.11 | up | miR-424 | 2.79 | up | miR-424 | 2.11 | up |
| miR-142-3p | 3.06 | up | miR-142-3p | 4.55 | up | miR-142-3p | 3.06 | up |
| miR-21 | 8.80 | up | miR-21 | 13.75 | up | miR-21 | 8.80 | up |
| miR-30d | 2.01 | down | miR-143 | 1.94 | down | miR-30d | 2.01 | down |
| miR-183 | 2.04 | down | miR-9* | 1.97 | down | miR-183 | 2.04 | down |
| miR-150 | 2.04 | down | miR-125b | 1.98 | down | miR-150 | 2.04 | down |
| miR-99a | 2.05 | down | miR-422b | 1.98 | down | miR-99a | 2.05 | down |
| miR-133b | 2.05 | down | miR-30d | 2.11 | down | miR-133b | 2.05 | down |
| miR-9* | 2.06 | down | miR-124a | 2.18 | down | miR-9* | 2.06 | down |
| miR-422b | 2.07 | down | miR-99a | 2.21 | down | miR-422b | 2.07 | down |
| miR-30a-3p | 2.18 | down | miR-338 | 2.23 | down | miR-30a-3p | 2.18 | down |
| miR-100 | 2.18 | down | miR-138 | 2.28 | down | miR-100 | 2.18 | down |
| miR-182 | 2.25 | down | miR-100 | 2.33 | down | miR-182 | 2.25 | down |
| miR-124a | 2.34 | down | miR-9 | 2.59 | down | miR-124a | 2.34 | down |
| miR-204 | 2.35 | down | miR-30a-3p | 2.75 | down | miR-204 | 2.35 | down |
| miR-138 | 2.36 | down | miR-145 | 2.93 | down | miR-138 | 2.36 | down |
| miR-125b | 2.47 | down | miR-204 | 3.23 | down | miR-125b | 2.47 | down |
| miR-338 | 2.47 | down | miR-182 | 3.28 | down | miR-338 | 2.47 | down |
| miR-9 | 2.50 | down | miR-183 | 3.32 | down | miR-9 | 2.50 | down |
| miR-143 | 2.81 | down | miR-150 | 3.84 | down | miR-143 | 2.81 | down |
| miR-145 | 3.23 | down | miR-133b | 4.26 | down | miR-145 | 3.23 | down |
Highlights.
MiRNAs and miRNA biosynthetic enzymes are injury-regulated in sciatic nerve.
Conditioning nerve lesion increases the number of P-bodies in axon varicosities.
MiRNA pathway may play a regulatory role in the regenerative axon growth.
Acknowledgments
We are grateful to Marvin Fritzler (Univ. of Calgary), Tom Keenan (Virginia State Univ.), Tom Hobman (Univ. of Alberta), Donald Bloch (Harvard Univ.), Jennifer Darnell (Rockefeller Univ.), Jens Lykke-Anderson (Univ. of Colorado), and Mike Kiledjian (Rutgers Univ.) for providing primary antibodies. We thank Vladimir Ghukasyan (Univ. of North Carolina, Chapel Hill) for assisting with confocal microscopy. We are grateful to Robert Carroll (East Carolina Univ.) for help in revising the manuscript.
The work was supported in part by Brody Brothers Endowment Grant MT7779 (AKM), the Wooten Laboratory grant (AKM), and NIEHS grant R15 ES019760-01 (AKM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aldrich BT, Frakes EP, Kasuya J, Hammond DL, Kitamoto T. Changes in expression of sensory organ-specific microRNAs in rat dorsal root ganglia in association with mechanical hypersensitivity induced by spinal nerve ligation. Neuroscience. 2009;164:711–723. doi: 10.1016/j.neuroscience.2009.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CB, Muschol M. Large neurohypophysial varicosities amplify action potentials: results from numerical simulations. Endocrinology. 2009;150:2829–2836. doi: 10.1210/en.2008-1636. [DOI] [PubMed] [Google Scholar]
- Benowitz LI, Yin Y. Combinatorial treatments for promoting axon regeneration in the CNS: strategies for overcoming inhibitory signals and activating neurons’ intrinsic growth state. Dev Neurobiol. 2007;67:1148–1165. doi: 10.1002/dneu.20515. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bhanji RA, Eystathioy T, Chan EK, Bloch DB, Fritzler MJ. Clinical and serological features of patients with autoantibodies to GW/P bodies. Clin Immunol. 2007;125:247–256. doi: 10.1016/j.clim.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham SD, Esmaeili B, Wood M, Sattelle DB. RNA interference: from model organisms towards therapy for neural and neuromuscular disorders. Hum Mol Genet. 2004;13(Spec No 2):R275–288. doi: 10.1093/hmg/ddh224. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB, Silva JM, Myers MM, Hannon GJ, Plasterk RH. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009a;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009b;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Bhattacharyya SN, Tapia-Arancibia L, Bordonne R, Filipowicz W, Bertrand E, Rage F. Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J Neurosci. 2008;28:13793–13804. doi: 10.1523/JNEUROSCI.4155-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiot MC, Subramanian M, Flatter E, Mandel JL, Moine H. Cells lacking the fragile X mental retardation protein (FMRP) have normal RISC activity but exhibit altered stress granule assembly. Mol Biol Cell. 2009;20:428–437. doi: 10.1091/mbc.E08-07-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Helms S, Fritzsch C, Fauser M, Izaurralde E. A C-terminal silencing domain in GW182 is essential for miRNA function. Rna. 2009a;15:1067–1077. doi: 10.1261/rna.1605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Tritschler F, Izaurralde E. The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing. Rna. 2009b;15:1433–1442. doi: 10.1261/rna.1703809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbay B, Heape AM, Sargueil F, Cassagne C. Myelin synthesis in the peripheral nervous system. Prog Neurobiol. 2000;61:267–304. doi: 10.1016/s0301-0082(99)00049-0. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengst U, Cox LJ, Macosko EZ, Jaffrey SR. Functional and selective RNA interference in developing axons and growth cones. J Neurosci. 2006;26:5727–5732. doi: 10.1523/JNEUROSCI.5229-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Satoh M, Pauley KM, Fritzler MJ, Reeves WH, Chan EK. Detection of the argonaute protein Ago2 and microRNAs in the RNA induced silencing complex (RISC) using a monoclonal antibody. J Immunol Methods. 2006;317:38–44. doi: 10.1016/j.jim.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islamov RR, Chintalgattu V, Pak ES, Katwa LC, Murashov AK. Induction of VEGF and its Flt-1 receptor after sciatic nerve crush injury. Neuroreport. 2004;15:2117–2121. doi: 10.1097/00001756-200409150-00024. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan TW, Winter S, Rackwitz HR, Heid HW. Nuclear coactivator protein p100 is present in endoplasmic reticulum and lipid droplets of milk secreting cells. Biochim Biophys Acta. 2000;1523:84–90. doi: 10.1016/s0304-4165(00)00106-9. [DOI] [PubMed] [Google Scholar]
- Koritzinsky M, Magagnin MG, van den Beucken T, Seigneuric R, Savelkouls K, Dostie J, Pyronnet S, Kaufman RJ, Weppler SA, Voncken JW, Lambin P, Koumenis C, Sonenberg N, Wouters BG. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. Embo J. 2006;25:1114–1125. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Hollenbeck PJ. Organization and translation of mRNA in sympathetic axons. J Cell Sci. 2003;116:4467–4478. doi: 10.1242/jcs.00745. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. Embo J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei P, Li Y, Chen X, Yang S, Zhang J. Microarray based analysis of microRNA expression in rat cerebral cortex after traumatic brain injury. Brain Research. 2009;1284:191–201. doi: 10.1016/j.brainres.2009.05.074. [DOI] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci U S A. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N-K, Wang X-F, Lu Q-B, Xu X-M. Altered microRNA expression following traumatic spinal cord injury. Exp Neurol. 2009;219:424–429. doi: 10.1016/j.expneurol.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkinson G, Spira ME. Clustering of excess growth resources within leading growth cones underlies the recurrent “deposition” of varicosities along developing neurites. Exp Neurol. 2010;225:140–153. doi: 10.1016/j.expneurol.2010.06.005. [DOI] [PubMed] [Google Scholar]
- McMurray R, Islamov R, Murashov AK. Raloxifene analog LY117018 enhances the regeneration of sciatic nerve in ovariectomized female mice. Brain Res. 2003;980:140–145. doi: 10.1016/s0006-8993(03)02984-6. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Moser JJ, Eystathioy T, Chan EK, Fritzler MJ. Markers of mRNA stabilization and degradation, and RNAi within astrocytoma GW bodies. J Neurosci Res. 2007;85:3619–3631. doi: 10.1002/jnr.21439. [DOI] [PubMed] [Google Scholar]
- Murashov AK, Chintalgattu V, Islamov RR, Lever TE, Pak ES, Sierpinski PL, Katwa LC, Van Scott MR. RNAi pathway is functional in peripheral nerve axons. FASEB J. 2007;21:656–670. doi: 10.1096/fj.06-6155com. [DOI] [PubMed] [Google Scholar]
- Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA. 2010;16:1516–1529. doi: 10.1261/rna.1833310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Pauley KM, Eystathioy T, Jakymiw A, Hamel JC, Fritzler MJ, Chan EK. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006;7:904–910. doi: 10.1038/sj.embor.7400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- She M, Decker CJ, Svergun DI, Round A, Chen N, Muhlrad D, Parker R, Song H. Structural basis of dcp2 recognition and activation by dcp1. Mol Cell. 2008;29:337–349. doi: 10.1016/j.molcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider WD, Zhou FQ, Zhong J, Markus A. Signaling the pathway to regeneration. Neuron. 2002;35:13–16. doi: 10.1016/s0896-6273(02)00762-6. [DOI] [PubMed] [Google Scholar]
- Song MG, Li Y, Kiledjian M. Multiple mRNA decapping enzymes in mammalian cells. Mol Cell. 2010;40:423–432. doi: 10.1016/j.molcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto K, Wakiyama M, Yokoyama S. Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. Rna. 2009;15:1078–1089. doi: 10.1261/rna.1363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Zhang B, Wu T, Skogerbo G, Zhu X, Guo X, He S, Chen R. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol Biol. 2009;10:12. doi: 10.1186/1471-2199-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. Rna. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- Vogelaar CF, Gervasi NM, Gumy LF, Story DJ, Raha-Chowdhury R, Leung KM, Holt CE, Fawcett JW. Axonal mRNAs: characterisation and role in the growth and regeneration of dorsal root ganglion axons and growth cones. Mol Cell Neurosci. 2009;42:102–115. doi: 10.1016/j.mcn.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci U S A. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DE, Twiss JL. The evolving roles of axonally synthesized proteins in regeneration. Curr Opin Neurobiol. 2006;16:111–118. doi: 10.1016/j.conb.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Yang J, Yuan YA. A structural perspective of the protein-RNA interactions involved in virus-induced RNA silencing and its suppression. Biochim Biophys Acta. 2009;1789:642–652. doi: 10.1016/j.bbagrm.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Yang WH, Yu JH, Gulick T, Bloch KD, Bloch DB. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA. 2006;12:547–554. doi: 10.1261/rna.2302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S, van Niekerk EA, Merianda TT, Twiss JL. Dynamics of axonal mRNA transport and implications for peripheral nerve regeneration. Exp Neurol. 2009 doi: 10.1016/j.expneurol.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]