Abstract
There is strong evidence that deregulation of prolactin (PRL) signaling contributes to pathogenesis and chemoresistance of breast cancer. Therefore, understanding cross-talk between distinct signal transduction pathways triggered by activation of the prolactin receptor (PRL-R), is essential for elucidating the pathogenesis of metastatic breast cancer.
In this study, we applied a sequential inhibitory analysis of various signaling intermediates to examine the hierarchy of protein interactions within the PRL signaling network and to evaluate the relative contributions of multiple signaling branches downstream of PRL-R to the activation of the extracellular signal-regulated kinases ERK1 and ERK2 in T47D and MCF-7 human breast cancer cells.
Quantitative measurements of the phosphorylation/activation patterns of proteins showed that PRL simultaneously activated Src family kinases (SFKs) and the JAK/STAT, phosphoinositide-3 (PI3)-kinase/Akt and MAPK signaling pathways. The specific blockade or siRNA-mediated suppression of SFK/FAK, JAK2/STAT5, PI3-kinase/PDK1/Akt, Rac/PAK or Ras regulatory circuits revealed that (1) the PI3-kinase/Akt pathway is required for activation of the MAPK/ERK signaling cascade upon PRL stimulation; (2) PI3-kinase-mediated activation of the c-Raf-MEK1/2-ERK1/2 cascade occurs independent of signaling dowstream of STATs, Akt and PKC, but requires JAK2, SFKs and FAK activities; (3) activated PRL-R mainly utilizes the PI3-kinase-dependent Rac/PAK pathway rather than the canonical Shc/Grb2/SOS/Ras route to initiate and sustain ERK1/2 signaling. By interconnecting diverse signaling pathways PLR may enhance proliferation, survival, migration and invasiveness of breast cancer cells.
Keywords: Prolactin signaling, ERK activation, PI3-kinase, Rac/PAK, cross-talk, breast cancer
INTRODUCTION
Prolactin (PRL)1, a hormone secreted by the pituitary gland and to a lesser extent by other tissues, is involved in many diverse physiological processes, including reproduction and lactation, growth and development, metabolism, brain functioning, immunomodulation and osmoregulation [1–2]. PRL acts as a growth, differentiating and survival factor in normal human mammary epithelial cells [3]. The levels of serum PRL and its receptor expression are increased in human breast cancer tissues [4–8]. PRL promotes neoplastic transformation by increasing cell proliferation in pre-invasive lesions, potentiates the transition to invasive carcinoma and is implicated in breast tumor resistance to chemotherapy [9–10].
PRL binding initiates conformational changes in the intracellular domains of dimerized class I cytokine family prolactin receptors (PRL-R) which leads to autophosphorylation and activation of their associated Janus family kinases (JAKs), followed by phosphorylation of PRL-R [11–12] and stimulation of signal transducers and activators of transcription (STAT), phosphoinositide 3 (PI3)-kinase/Akt, Ras/mitogen activated protein kinase (MAPK) and other signaling pathways that control mitogenic, apoptotic, motogenic and cell differentiation responses [3, 13]. Aberrant activation of the three-tiered MAPK signaling cascade comprised of c-Raf, MEK1/2 (mitogen-activated protein kinase kinase 1/2) and ERK1/2 (extracellular signal-regulated kinase 1/2) is common in many types of human cancers. Thus, the routes that positively regulate ERK1/2 activity toward its numerous cytosolic and nuclear effectors represent an attractive target for the development of anticancer drugs [14].
Studying the regulatory connections in the PRL-R signaling network is essential for understanding the pathogenesis of metastatic breast cancer. Yet, the features of intra- and inter-pathway interactions (cross-talk) that lead to the emergent properties of the integrated cellular response are poorly understood. Therefore, with the goal of mapping the PRL-R signaling network architecture from receptor to ERK1/2, we examined the activation patterns of ERK1/2 in response to PRL and upon perturbations at different levels of network hierarchy in human breast cancer cell lines, derived from patients with invasive/infiltrative ductal carcinoma. Here, we unravel a pathway whereby the propagation of signals originating from PRL-R and leading to ERK1/2 activation via c-Raf, is largely controlled by a PI3-kinase-dependent, but Akt and STAT-independent, Rac/PAK (p21-activated kinase) route.
MATERIALS AND METHODS
Ligands, inhibitors, antibodies and chemicals
Prolactin was obtained from Peprotech Inc. (Rocky Hill, NJ). The stock solutions of inhibitors were prepared as recommended by the manufacturers. Lists of specific inhibitors and antibodies used in this study and their commercial sources are shown in Supplemental Table 1S and Table 2S, respectively. All other common chemicals, solvents and reagents were of highest grade available from various commercial sources.
Cell lines and culture conditions
T47D (ATCC No. HTB-133) cells were cultured in a complete RPMI-1640 media with L-glutamine and 25 mM HEPES (Mediatech Inc., Manassas, VA), supplemented with 10% fetal bovine serum (FBS) (Gemini Bio-Products, West Sacramento, CA), 20 μg/ml bovine insulin (Sigma-Aldrich, St Louis, MO) and penicillin–streptomycin solution (100 μg/ml each) (Mediatech Inc.). MCF7 (ATCC No. HTB-22) cells were grown in a complete DMEM/F-12 media (Mediatech Inc.) containing 10% FBS and 1% penicillin–streptomycin solution. All cells were cultivated in a humidified 5% CO2 incubator at 37 C. Cells were grown for 4–5 days and after reaching confluency were harvested by exposure to 0.25% Trypsin–EDTA solution (GIBCO) and then passed into new T-75 tissue culture flasks (Denville Scientific, Metuchen, NJ). Starvation media did not contain FBS and insulin.
Cell stimulation, protein extraction and immunoprecipitation
Cells were plated in 60×15cm (for total cell lysates) or in 150×20 cm (for protein immunoprecipitation) tissue culture dishes (Denville Scientific, Metuchen, NJ) in appropriate complete cell culture media and grown until they reached ~80% confluency. Cells were starved overnight in respective starvation media, (when applicable) preincubated with inhibitors or solvent alone, left unstimulated or were stimulated with a ligand for different periods of time at 37°C. Inhibitors and a ligand were diluted to final concentrations in starvation media. 10 sec just before the end of stimulation, the media was removed by vacuum suction and cells were scraped either in ice-cold lysis buffer (150 mM NaCl, 50 mM HEPES (pH 7.4), 1 mM EGTA, 1% Triton-X100, 10% glycerol diluted in dH2O) or lysed in immunoprecipitation buffer (150 mM NaCl, 25 mM HEPES (pH 7.4), 2.5 mM EGTA, 1% Triton-X100, 1% Igepal CA-630, 5% glycerol diluted in dH2O) for subsequent protein pull-down assay. Total cell lysates were vigorously vortexed and subjected to centrifugation at 10,000×g for 10 min at 4°C to remove detergent-insoluble material. Equal amounts of solubilized proteins in supernatant were dissolved in 4× NuPAGE lithium-dodecyl sulphate (LDS) sample buffer supplemented with NuPAGE sample reducing agent (50 mM dithiothreitol, DTT) (Invitrogen, Carlsbad, CA) in a ratio of 130:50:20 and heated for 5 min at 75°C. Proteins of interest were pulled down by gently mixing cell supernatants for 4 hrs at 4°C with a 50:50 mixture of recombinant protein A and protein G Sepharose 4B bead slurry (Zymed Laboratories, Invitrogen), which was firstly pre-incubated with 5 μg of primary antibody against a specific protein for 2 hrs at RT. Alternatively, tyrosine-phosphorylated proteins were collected from supernatants with 50 μl of monoclonal anti-phosphotyrosine-agarose beads (pY-20) (Sigma-Aldrich) for 4 hrs at 4°C. Thereafter, the beads were washed twice in ice-cold immunoprecipitate washing buffer (150 mM NaCl, 20 mM HEPES (pH 7.4), 0.1% Triton-X100, 10% glycerol diluted in dH2O) and twice in ice-cold PBS. The immunoprecipitated proteins were eluted in 1× NuPAGE LDS sample buffer (25% 4× NuPAGE LDS sample buffer, 75% PBS) and heated for 5 min at 75°C.
Subcellular fractionation
Cells were grown in 100×20mm tissue culture dishes to confluence, starved overnight, stimulated as described above in the presence or absence of small molecule inhibitors, and incubated with 1.2 ml of ice-cold permeabilization buffer (150 mM NaCl, 2.5 mM EGTA, 50 mM HEPES, 10% glycerol, 150 μg/ml digitonin (Sigma-Aldrich) diluted in dH2O) for 10 min. Afterwards, cells were gently collected into Eppendorf tubes and spun down for 2 min in an ultracentrifuge at max speed (10,000×g) at 4°C. Supernatant (cytosolic extract) was transferred into separate tube, and cell pellet, representing a crude membrane fraction, was resuspended in 150 μL of ice-cold lysis buffer. Extracts were centrifuged at 10,000×g for 10 min at 4°C to remove debris and the supernatants were resuspended in Laemmli buffer as indicated above. Cytosolic and particulate fractions were then assayed for protein translocation and activation.
Ras and Rac1 activation assays
Active (GTP-bound) Ras and Rac1 from 500 μg total cell lysates were captured with 30 μl of Raf-1 Ras binding domains (RBD) or PAK1 p21-binding domains (PBD), respectively, bound to glutathione-agarose beads (Millipore) for 3 h at 4°C. In vitro GTPγS (non-hydrolyzable GTP analog from Millipore) protein loading (final concentration 100 μM) was performed according to manufacturer’s recommendations. Protein complexes were collected by brief centrifugation and washed three times with ice-cold immunoprecipitation buffer supplemented with 10 mM MgCl2. Ras-GTP or Rac1-GTP proteins were released from agarose beads with NuPAGE LDS sample buffer and heated for 5 min at 75°C.
Transient cell transfection with siRNA
Half an hour or less before transfection MCF-7 cells were trypsinized and resuspended in antibiotic-free complete media. 1.2×106 cells per sample were aliquoted into Eppendorf tubes and centrifuged at 90×g for 10 min at RT. Supernatant was removed and the cell pellet was resuspended in 100 μl of Ingenio Electroporation solution (Mirus Bio, Madison, WI) containing siRNA. The responses of cells transfected with 100 nM of validated RAC1, K-RAS, PAK1, PAK2, PAK3, PAK4, PAK6 and PAK7 siRNA or their combinations were compared to those transfected with AllStars non-targeting negative control siRNA (NT siRNA) (all from Qiagen, Valencia, CA). siRNA sequences are shown in Supplemental Table 3S. Cell suspensions containing siRNA were electroporated using the P-020 program on Amaxa’s Nucleofector II device (Lonza Cologne AZ, Basel, Switzerland). Immediately after electroporation, 0.5 ml of the pre-equilibrated antibiotic-free complete media was added to the cuvette and the cell suspension was gently transferred into 6-well plates (final volume 2.0 ml media per well). Cells were allowed to attach for 6 hours before the addition of penicillin-streptomycin solution. Total proteins were isolated from cells harvested 72 hours post-transfection.
Real-time qPCR
To estimate the level of suppression of target mRNAs in response to siRNA transfection, total RNA was isolated using RNeasy Plus kit (Qiagen) according to the manufacturer’s protocol from total cell lysates prepared 72 hours post-transfection. RNA (0.5–2 μg) was reverse-transcribed with SuperScript III First-strand reverse transcriptase system using oligo-dT primers (Invitrogen) to generate cDNA. Real-time qPCR was performed with an ABI Prism 7000 and Power SYBR Green PCR Master mix (Applied Biosystems) to detect amplified products. Each 25-μl PCR reaction mix contained 1 μl of cDNA, 12.5 μl of Master mix and 10 pmol of each primer (all from Applied Biosystems). Sequences of primers are shown in Supplemental Table 4S. Relative mRNA abundance was normalized to the internal standard GAPDH by the ΔΔCT method, as described by the manufacturer (Invitrogen).
Gel electrophoresis, immunoblotting and data evaluation
Protein separation by gel electrophoresis and Multistrip Western blotting procedures were performed as described previously [15]. Briefly, samples were subjected to LDS-PAGE under reducing conditions. Resolved proteins were transferred onto nitrocellulose membranes, which were afterwards blocked in 3% bovine serum albumin (BSA) solution for 1 h at RT, kept with indicated primary antibodies overnight at RT, extensively washed with TBS-T washing buffer (10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5% (w/v) Triton X-100), and incubated with appropriate horseradish peroxidase-conjugated horse anti-mouse (Cell Signaling Technology, Danvers, MA) or goat anti-rabbit (Pierce Biotechnology/Thermo Fisher Scientific, Rockford, IL) secondary antibodies at dilutions 1:10,000 and 1:50,000, respectively) for 1 h at RT before final wash with TBS-T washing buffer and subsequent detection of protein bands by enhanced chemiluminescence system using SuperSignal West Dura Extended Duration Substrate (Pierce Biotechnology). Bands were visualized and their signal net intensities were quantified via computer-assisted densitometry analysis by KODAK Image Station 440CF (Kodak Scientific Imaging Systems, New Haven, CT). The signal intensities of phosphorylated protein were normalized by signal intensities of the total (phosphorylated and non-phosphorylated) protein at each time point and were expressed as fold changes over basal levels (in unstimulated cells). Kinetic curves were plotted based on fold changes in SigmaPlot v.10 (Systat Software, Inc., San Jose, CA). All experiments were performed in triplicates. Representative blots and/or their quantitative values were shown.
Wound healing assay
Cells grown to confluence in 6-well plates were scratched using a pipette tip to create a wound, washed with serum-free medium to remove loosened cells, and then cultured in serum-free medium that contained PRL with or without small molecule inhibitors, with medium being replaced every 24 hours. Cells were photographed at 0 and 72 hours after wounding with an inverted light microscope at 4× magnification. Experiments were performed in triplicate. Area of each wound surface was quantitated using Adobe Photoshop software (Mountain View, CA). The mean percentage wound closure was calculated using the equation (S2−S1)/S2*100, where S2 is cell-free scratch area at 0 h after wounding, S1 – cell-free scratch area at 72 hours after wounding.
Cell growth/viability assay
Cells were plated in quadruplicates into 24-well plates at a density of 20,000 cells/ml. After 24 hours, complete culture medium was changed into fresh low serum-containing medium (1% FBS) supplemented with 10 nM PRL with or without inhibitors. To evaluate cell growth and viability 72 hours after inhibitor treatment, the AlamarBlue assay (Invitrogen, Carlsbad, CA) was performed as described previously [16]. Results are expressed as percentage of control and presented as the mean ± SD obtained from three independent experiments.
RESULTS
Prolactin concomitantly activates c-Src, JAK/STAT, PI3K/Akt and MAPK signaling cascades
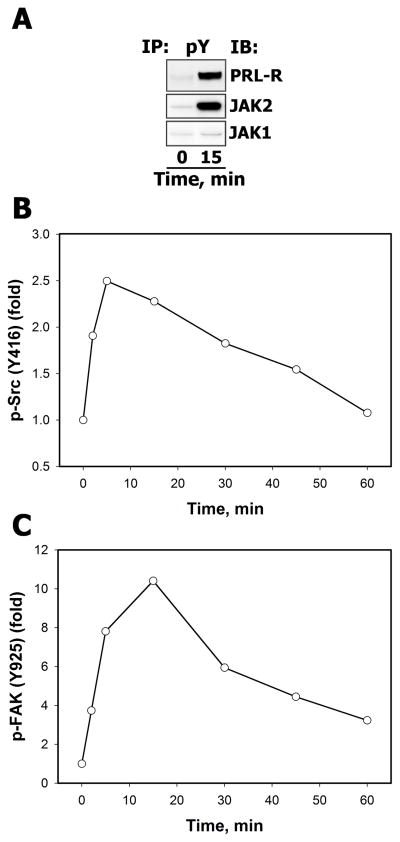
The ability of recombinant human PRL to stimulate its cognate receptor and activate Janus family kinases (JAKs) was examined by probing the immunoprecipitates of tyrosine-phosphorylated proteins from lysates of non-stimulated and PRL-treated T47D cells with specific anti-PRL-R, anti-JAK2 or anti-JAK1 antibodies. The results show that PRL induced a strong tyrosine phosphorylation of PRL-R and JAK2, but not JAK1, compared to non-stimulated cells (Fig. 1A).
Figure 1.
A. PRL induced activation of PRL-R and JAK family tyrosine kinases Serum-starved T47D cells were either left unstimulated or were stimulated with 10 nM PRL for 15 min. Tyrosine phosphorylated proteins were immunoprecipitated (IP) from total cell lysates (TCL) using the monoclonal agarose-conjugated anti-phosphotyrosine antibody (Ab), pY-20. Resolved proteins were transferred onto nitrocellulose membrane, which was immunoblotted (IB) with Abs against the proteins, indicated on the right. B–C. PRL activates Src family tyrosine kinases and their target FAK. Total and phosphorylated forms of Src family protein kinases (B) and FAK (C) of unstimulated and PRL-stimulated (10 nM, for the indicated time intervals) T47D cells were detected by IB of the TCL with anti-c-Src, anti-phospho-Src (Tyr416), anti-FAK and anti-phospho-FAK (Tyr925) Abs, respectively. The signal intensities of phosphorylated protein were normalized by signal intensities of the total (phosphorylated and non-phosphorylated) protein at each time point and then expressed as fold changes over basal levels (in unstimulated cells). Densitometric quantitation of representative blots is shown (n=3).
Because PRL-R and JAK2 colocalize with cytosolic src avian sarcoma viral oncogen homolog (c-Src) in lipid-rich fractions of the plasma membrane [17], we assessed whether c-Src was activated in response to PRL in breast cancer cells by measuring the phosphorylation state of c-Src at Tyr416, located in the activation loop of the kinase domain, which is required for maximum c-Src enzyme activity [18]. Western blotting analysis using the phosphospecific Src Tyr416 antibody showed that c-Src phosphorylation increased nearly 2-fold above basal level after 2 min PRL treatment, reached a peak at 5 min (2.5-fold) and returned to the basal level by 60 min (Fig. 1B). As further evidence for increased c-Src activity, we also followed the phosphorylation kinetics of its effector focal adhesion kinase (FAK) on Tyr925 (Fig. 1C), a major target site for c-Src [19].
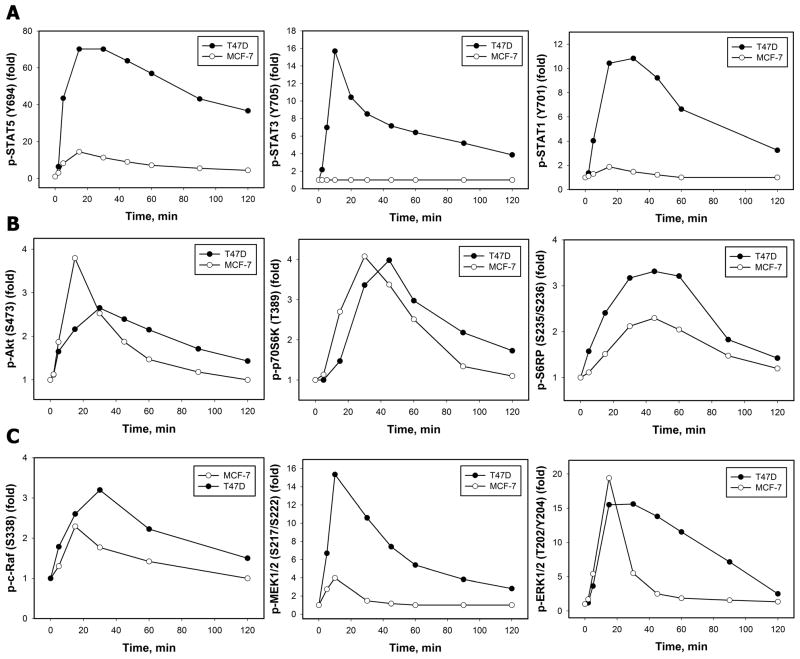
The potency of PRL to transduce the signals through its receptor to multiple branches of intracellular signaling pathways was then verified by monitoring the activation patterns of the STATs, PI3-kinase/Akt and MAPK signaling cascades (Fig. 2). Our results demonstrate that stimulation of T47D cells with PRL promoted an increase in the phosphorylation of STAT5 at Tyr694 (Fig. 2A, left panel), STAT3 at Tyr705 (Fig. 2A, middle panel) and STAT1 at Tyr701, as revealed by site-specific antibodies that recognized the phosphorylated state of respective residues. Phosphorylation of these sites on STATs is obligatory for their homo- and hetero-dimerization, nuclear translocation and binding to specific DNA elements in the promoters of signal-responsive genes [20]. Consistent with other observations [21], no detectable levels of phospho-STAT3 were detected in MCF-7 cells, which also had less pronounced phosphorylation of STAT5 and STAT1 proteins compared to T47D cells (Fig. 2A). Phosphorylation levels of the serine/threonine kinase Akt on Ser473 were assessed as readout of PI3-kinase activity in response to PRL (Fig. 2B, left panel). Simultaneously, PRL treatment induced phosphorylation and activation of p70 S6 kinase (p70S6K) (Fig. 2B, middle panel) and its effector ribosomal protein S6 (S6RP) (Fig. 2B, right panel), which lie downstream of 3-Phosphoinositide-dependent kinase 1 (PDK1) and Akt and which are key enzymes in the regulation of protein synthesis and the G(1)/S transition of the cell cycle [22–23]. One of the explanations for the dissimilar levels of response of these signaling pathways may be the difference in endogenous PRL-R levels between in MCF-7 and T47D cells.
Figure 2. Time-courses of PRL-induced activation of STAT (A), PI3-kinase (B) and MAPK (C) signaling pathways in breast cancer cells.
Both total and phosphorylated forms of STAT5 (Tyr694) (A, left panel), STAT3 (Tyr705) (A, middle panel), STAT1 (Tyr701) (A, right panel), Akt (Ser473) (B, left panel), p70S6K (Thr389) (B, middle panel), S6RP (Ser235/Ser236) (B, right panel), c-Raf (Ser338) (C, left panel), MEK1/2 (Ser217) (C, middle panel) and ERK1/2 (Thr202/Tyr204) (C, right panel) proteins were detected by IB with Abs against respective antigens in TCL of unstimulated and PRL-stimulated (10 nM, for the indicated time intervals) T47D (black circles) or MCF-7 (white circles) cells. The ratio of phospho-protein:total protein at each time point was expressed as fold changes over basal levels. Quantitation of representative blots is shown (n=3).
PRL caused an apparent increase in phosphorylation levels of c-Raf (Fig. 2C, left panel), MEK1/2 (Fig. 2C, middle panel), ERK1/2 (Fig. 2C, right panel) and its major effector p90 ribosomal S6 kinase (p90RSK) (Supplemental Fig. 1S), which is known to phosphorylate a broad array of substrates in different cellular locations, regulating immediate early gene response, translation, cell-cycle progression, cell proliferation, survival and motility [24]. A much more transient and less robust activation of the MAPK cascade proteins occurred in MCF-7 cells compared to T47D cells.
Decrease in activation of STAT5, Akt and ERK1/2 upon inhibition of Src family kinases is partially mediated by FAK
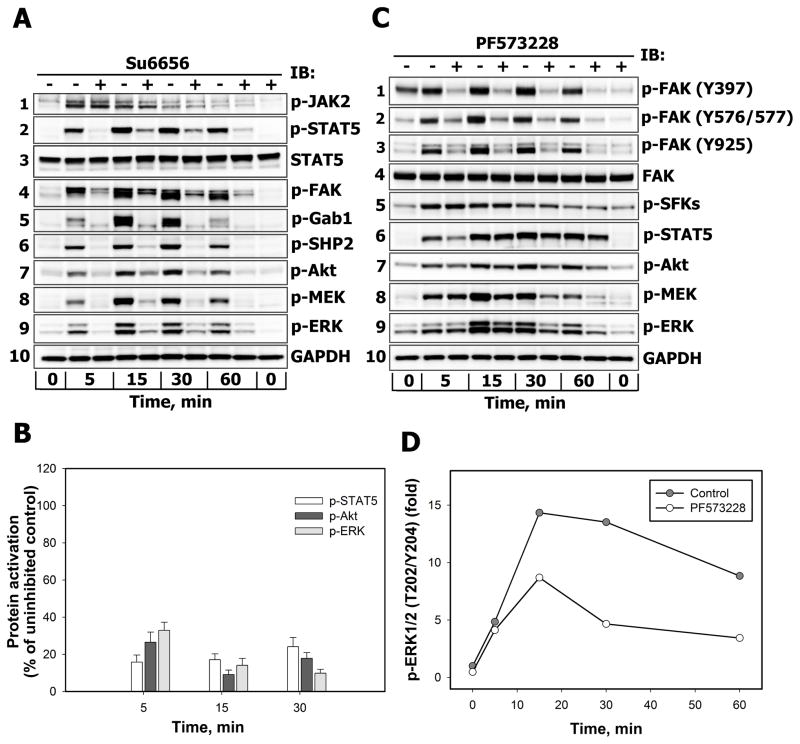
Src family kinases (SFKs) have been shown to play a critical role in many cytokine receptor pathways [25–26]. To examine the role of SFKs in PRL signaling network, we examined the activation of JAK/STAT, PI3-kinase/Akt and MAPK signaling pathways in T47D (Fig. 3A) and MCF-7 (Fig. 3B) breast cancer cells following PRL stimulation in the presence or absence of Su6656, a selective inhibitor of SFKs, including c-Src, Yes, Lyn and Fyn [27]. This treatment potently suppressed PRL-induced activation of SFK as shown in Supplemental Fig. 2S. Although inhibition of SFKs did not change the autophosphorylation status of JAK2 on Tyr1007/Tyr1008 residues (Fig. 3A, blot 1), which lie within the kinase domain and regulate kinase activity [28], the phosphorylation of STAT5 on Tyr694 (Fig. 3A, blot 2) and focal adhesion kinase (FAK) on Tyr925 (Fig. 3A, blot 4) were significantly attenuated. This observation suggests that SFKs lie upstream of these proteins, but may be downstream of JAK2. Once phosphorylated on Tyr925, FAK is predicted to recruit growth factor receptor-bound protein 2 (Grb2), an adaptor protein known to be involved in Ras/MAPK signaling [29]. In the canonical Ras/MAPK signaling pathway, Grb2 binds phosphotyrosine motifs via the Src homology 2 (SH2) domain, while two flanking Src homology 3 (SH3) domains bind Son of Sevenless (SOS), the guanine nucleotide exchange factor (GEF) for small GTPase Ras which acts upstream of the Raf/MEK/ERK cascade [30].
Figure 3.
A–B. The effects of SFK inhibition on the activation of downstream effectors of PRL-R Serum-starved T47D (A) or MCF-7 (B) cells were either left untreated (−) or were treated (+) with Su6656 (10 μM, 30 min) before stimulation with 10 nM PRL for the indicated time intervals. Phosphorylated forms of JAK2 (Tyr Tyr1007/1008), STAT5 (Tyr694), FAK (Tyr925), Gab1 (Tyr627), SHP2 (Tyr542), Akt (Ser473), MEK1/2 (Ser217/Ser221) and ERK1/2 (Thr202/Tyr204) proteins were detected by IB of the TCL with Abs against respective antigens. GAPDH levels were detected with anti-GAPDH Abs and used as protein loading control. Representative blots are shown (n=3). B. Phosphorylated STAT5 (white bars), Akt (dark grey bars) and ERK1/2 (light grey bars) in TCL of Su6656-treated cells were quantified by densitometry and are expressed as percent of control of the corresponding phospho-protein measured in TCL of PRL-stimulated cells incubated without Su6656. Data represent the mean ± SD of three independent experiments. C–D. The effects of FAK inhibition on the activation of downstream effectors of PRL-R. Serum-starved T47D cells were either left untreated (−) or were treated (+) with PF573228 (0.5 μM, 2 h) before stimulation with 10 nM PRL for the indicated time intervals. Phosphorylated forms of FAK (Tyr397), FAK (Tyr576/577), FAK (Tyr925), SFKs (Tyr416), STAT5 (Tyr694), Akt (Ser473), MEK1/2 (Ser217/Ser221) and ERK1/2 (Thr202/Tyr204) proteins were detected by IB of the TCL with Abs against respective antigens. GAPDH levels were used as protein loading control. Representative blots are shown (n=3). D. The ratio of phospho-ERK1/2:total ERK1/2 at each time point was expressed as fold changes over basal levels.
Su6656 treatment abrogated the phosphorylation of Grb2-associated binder 1 (Gab1) on Tyr627 residue (Fig. 3A, blot 5), which is required for binding of the protein tyrosine phosphatase SHP2, with its subsequent phosphorylation on Grb2-binding sites (Tyr542 and Tyr580) by upstream kinases and an increase in phosphatase activity [31–33]. SHP2 can positively regulate STAT5 signaling [34–35] and activate Ras through multiple mechanisms [31]. Our data show that SFKs are responsible for SHP2 phosphorylation in PRL signaling (Fig. 3A, blot 6). At the same time Su6656 treatment dramatically suppressed the PRL-induced activation of Akt (Fig. 3A, blot 7), MEK (Fig. 3A, blot 8) and ERK1/2 (Fig. 3A, blot 9). Glyceraldehyde-3-Phosphate Dehyrogenase (GAPDH) protein levels were used as a control for equal protein loading (Fig. 3A, blot 10). Similar inhibitory effects of Su6656 treatment on PRL-induced phosphorylation of STAT5 (Fig. 3B, white bars), Akt (Fig. 3B, dark grey bars) and ERK1/2 (Fig. 3B, light grey bars) were obtained in PRL-stimulated MCF-7 cells.
Based on these results, indicating that SFKs are required for PRL-mediated ERK1/2 activation in breast cancer cells, we further determined the quantitative contribution of immediate SFK substrate FAK to major signaling pathways by using the specific FAK inhibitor PF573228 [36].
Growth factors facilitate autophosphorylation of FAK at Tyr397, which is a critical residue for the activation and function of FAK, and serves as a docking site for SFKs and p85 regulatory subunit of PI3-kinase [37–38]. Recruitment of SFKs results in the phosphorylation of Tyr407, Tyr576 and Tyr577 in the catalytic domain, and Tyr871 and Tyr925 in the carboxy-terminal region of FAK [39–40]. PRL-induced phosphorylation of FAK at Tyr397, Tyr576, Tyr577 and Tyr925 residues was suppressed by treating T47D cells with PF573228 (Fig. 3C, blots 1–3) without affecting total levels of FAK (Fig. 3C, blot 4) and GAPDH (Fig. 3C, blot 10). PF573228 treatment did not interfere with the activation of SFKs (Fig. 3C, blot 5), but slightly reduced tyrosine phosphorylation of STAT5 (Fig. 3C, blot 6) as well as attenuated Akt (Fig. 3C, blot 7) and MEK/ERK responses (Fig. 3C, blots 8–9 and Fig. 3D), suggesting that FAK only partially accounts for the ERK1/2 responses downstream of SFKs through PI3-kinase/Akt-dependent or –independent mechanisms.
Prolactin-induced ERK activation depends on JAK2 activity, but is uncoupled from STAT signaling
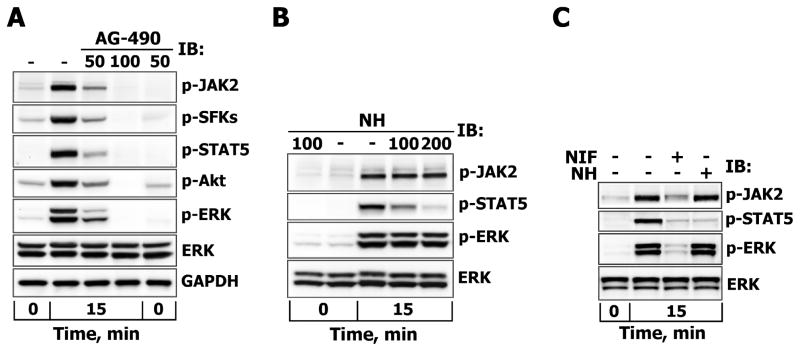
To examine the involvement of the JAK/STAT signaling pathway in the SFK/FAK-dependent activation of ERK1/2, T47D cells were pretreated with AG-490, an inhibitor of JAK2/JAK3 or with cell-permeable nonpeptidic nicotinoyl hydrazone (NH) compound, which prevents STAT5 and, to a lesser extent, STAT1/3 phosphorylation and dimerization by selectively targeting their Src homology 2 (SH2) domains [41]. AG-490 treatment abrogated PRL-induced phosphorylation of JAK2, SFKs, STAT5, Akt and ERK1/2 in a dose-dependent manner ((Fig. 4A), indicating that JAK2 acts upstream of these proteins. By contrast, the inhibition of STAT5 did not reduce the activation levels of JAK2 and did not block PRL-induced phosphorylation of ERK1/2 (Fig. 4B). Similar results with AG-490 and NH were obtained in MCF-7 cells (Fig. 4C and data not shown). In addition, MCF-7 cells were pretreated with nifuroxazide (NIF), a cell-permeable nitrofuran-based agent that suppresses the activation of cellular STAT1/3/5 transcription activity by inhibiting autophosphorylation of JAK2 and Tyk2, another member of the JAK family, but not those of JAK1 and c-Src [42]. As expected, NIF treatment decreased JAK2 and STAT5 tyrosine phosphorylation and greatly reduced ERK1/2 activation (>90%) in PRL-stimulated MCF-7 cells, whereas total ERK1/2 protein levels remained unaffected (Fig. 4C). Of note, T47D appeared to be much more resistant to NIF treatment. These data indicate that JAK2-dependent activation of proteins other than STATs mediate the PRL-induced activation of ERK1/2 in breast cancer cells.
Figure 4. The effects of JAK2 and STAT5 inhibition on PRL-induced activation of ERK1/2.
Serum-starved T47D cells were either left untreated (−) or were treated (+) with AG-490 (50 or 100 μM) for 3 hours (A) or with STAT5 inhibitor (NH, 100 or 200 μM) for 1 hour (B), while MCF-7 cells were treated with NH (200 μM) or nifuroxazide (NIF, 100 μM) for 1 hour (C) before stimulation with 10 nM PRL for 15 min. Phosphorylated forms of JAK2 (Tyr1007/1008), SFKs (Tyr416), Akt (Ser473), STAT5 (Tyr694) and ERK1/2 (Thr202/Tyr204) proteins were detected by IB of the TCL with Abs against respective antigens. Total ERK1/2 protein levels were detected with anti-ERK1/2 Abs and were used as protein loading control. Representative blots are shown.
PI3-kinase-mediated ERK activation via c-Raf occurs regardless of downstream Akt signaling
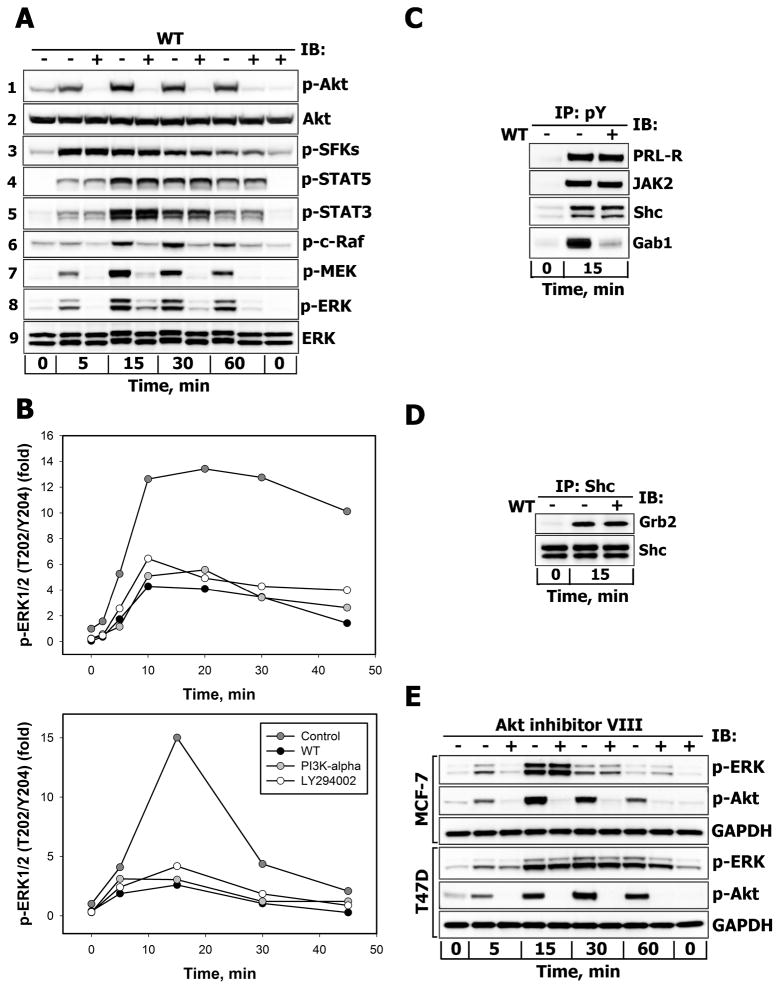
We next explored the possibility that SFK/FAK-dependent ERK1/2 responses could be modulated by the PI3-kinase/Akt signaling pathway, which, as shown above, is strongly suppressed by SFKs inhibition (Fig. 3A, blot 7) and partially depends on FAK activity (Fig. 3C, blot 7). For this purpose, T47D cells were pretreated with wortmannin (WT), a specific covalent inhibitor of class I, II and III PI3-kinases, and stimulated with PRL for different time intervals. The complete inhibition of inducible Akt phosphorylation at Ser473 (Fig. 5A, blot 1) in the presence of WT upon PRL stimulation confirmed that the 200 nM WT dose effectively inhibited the production of phosphoinositol-triphosphate PI(3,4,5)P3 by PI3-kinase and activation of the PI3-kinase/Akt pathway. PI3-kinase inhibition nearly completely prevented early and late signal propagation throughout the whole MAPK cascade, starting with c-Raf on its activating Ser338 residue (Fig. 5A, blot 6) to MEK (Fig. 5A, blot 7) and to ERK1/2 (Fig. 5A, blot 8). This effect was not due to inhibitor-induced changes in the expression levels of Akt (Fig. 5A, blot 2) or ERK1/2 (Fig. 5A, blot 9). PI3-kinase inhibition did not reduce the phosphorylation of SFKs at Tyr416 (Fig. 5A, blot 3), indicating that SFKs act upstream of PI3-kinase and are not responsible for WT-induced changes in ERK1/2 activation. Of note, the PRL-induced increases in STAT5 and STAT3 tyrosine phosphorylation levels were not inhibited by WT (Fig. 5A, blots 4–5), in agreement with the observation from the inhibition studies shown in Fig. 4 that STATs do not participate in MAPK activation.
Figure 5.
A. Effects of PI3-kinase inhibition on the activation of downstream effectors of PRL-R Serum-starved T47D cells were either left untreated (−) or were treated (+) with wortmannin (WT, 200 nM, 30 min) before stimulation with 10 nM PRL for the indicated time intervals. Phosphorylated forms of Akt (Ser473), SFKs (Tyr416), STAT5 (Tyr694), STAT3 (Tyr705), c-Raf (Ser338), MEK1/2 (Ser217/Ser221) and ERK1/2 (Thr202/Tyr204) proteins were detected by IB of the TCL with Abs against respective antigens. Total Akt and ERK1/2 protein levels were detected with anti-Akt1 or anti-ERK1/2 Abs, respectively, and served as protein loading controls. Representative blots are shown (n=3). B. Effects of structurally different inhibitors of PI3-kinase on PRL-induced activation of ERK1/2. Serum-starved T47D (upper panel) and MCF-7 (lower panel) cells were either left untreated (control, dark grey circles) or were preincubated with WT (200 nM, black circles), PI3K-α inhibitor 2 (2 μM, grey circles) or LY 294002 (25 μM, white circles) for 30 min before stimulation with 10 nM PRL for the indicated time intervals. Both total and phosphorylated forms of ERK1/2 were detected by IB of the TCL with anti-ERK1/2 or anti-phospho-ERK1/2 (Thr202/Tyr204) Abs, respectively. The ratio of phospho-ERK1/2:total ERK1/2 at each time point is expressed as fold changes over basal levels. Quantitation of represenatative blots is shown (n=3). C. Effects of PI3-kinase inhibition on tyrosine phosphorylation levels of PRL-R, JAK2, Shc and Gab proteins. Tyrosine phosphorylated proteins were immunoprecipitated (IP) from TCL of unstimulated or PRL-stimulated (10 nM, 15 min) and WT-treated (+) (200 nM, 30 min) or untreated (−) T47D cells and probed for proteins indicated on the right. D. Effects of PI3-kinase inhibition on Shc association with Grb2. Shc proteins were immunoprecipitated (IP) from TCL of unstimulated or PRL-stimulated (10 nM, 15 min) and WT-treated (+) (200 nM, 30 min) or untreated (−) T47D cells and probed with anti-Grb2 or anti-Shc Abs. E. Effects of Akt inhibition on PRL-induced activation of ERK1/2. Serum-starved MCF-7 or T47D cells were either left untreated (−) or were treated (+) with Akt1/2/3 inhibitor (Akt-VIII, 10 μM, 30 min) before stimulation with 10 nM PRL for the indicated time intervals. Phosphorylated forms of Akt (Ser473) and ERK1/2 (Thr202/Tyr204) proteins were detected by IB of the TCL with Abs against respective antigens. GAPDH levels served as protein loading control. Representative blots are shown (n=3).
In order to obtain further evidence for the involvement of class I PI3-kinase in ERK1/2 activation in PRL signaling, we used a selective inhibitor for the α-isoform of PI3-kinase, PI3K-α inhibitor 2. As a result of this treatment, peak ERK1/2 phosphorylation was decreased by 60% in T47D cells and by 80% in MCF-7 cells. This level of inhibition was similar to that obtained upon treatment with WT (70% and 83%) or LY294002 (63% and 72%) in T47D cells (Fig. 5B, upper panel) and MCF-7 (Fig. 5B, lower panel) cells, respectively.
Importantly, cell treatment with WT did not change overall tyrosine phosphorylation levels of PRL-R, JAK2 and p52/p46 Shc adaptor proteins, which are presumed to bind the Grb2-SOS complex, which couples Shc to the Ras activated MAPK pathway (Fig. 5C). Consistently, PI3-kinase inhibition failed to reduce the amounts of Grb2 that inducibly associated with Shc upon PRL stimulation, as shown by immunoblotting of Shc precipitates with anti-Grb2 antibodies (Fig. 5D), indicating that suppression of Shc-Grb2 complex formation was not responsible for the inhibition of ERK1/2 activation. By contrast, suppressing PI(3,4,5)P3 formation by PI3-kinase inhibition significantly reduced the membrane recruitment (Supplemental Fig. 3S) and tyrosine phosphorylation of pleckstrin homology (PH) domain-containing Gab proteins (Fig. 5C), which could potentially affect SHP2 activation. However, neither tyrosine phosphorylation of SHP2 nor its recruitment to the plasma membrane were significantly altered by WT (Supplemental Fig. 3S), implying that the functioning of SHP2 may depend on proteins that lack PH-domain(s) and therefore are independent of PI3-kinase. Thus, neither of these well-established mechanisms of activation of the MAPK cascade could account for the sensitivity of PRL-induced ERK activation to PI3-kinase inhibitors.
Next, in order to evaluate the contribution of Akt, an immediate effector of PI3-kinase, and its downstream targets to ERK1/2 activation, the cells were pretreated with an isozyme-selective Akt1/2/3 inhibitor (Akt-VIII), which does not interfere with the PI3-kinase activity per se. As shown in Fig. 5E, Akt inhibition had no significant effect on ERK1/2 phosphorylation in T47D and MCF-7 cells upon PRL-treatment. This observation demonstrates that the proteins that are critical for ERK1/2 activation either operate downstream of PI3-kinase, but upstream of Akt (e.g. PDK1), or belong to a distinct PI3-kinase-dependent signaling branch such as Rac/Cdc42/PAK. Therefore, next we examined the contribution of group I PAK kinases (PAK1/2/3) and their upstream effectors to ERK1/2 activation.
Prevalence of Rac/PAK pathway in prolactin-induced ERK activation
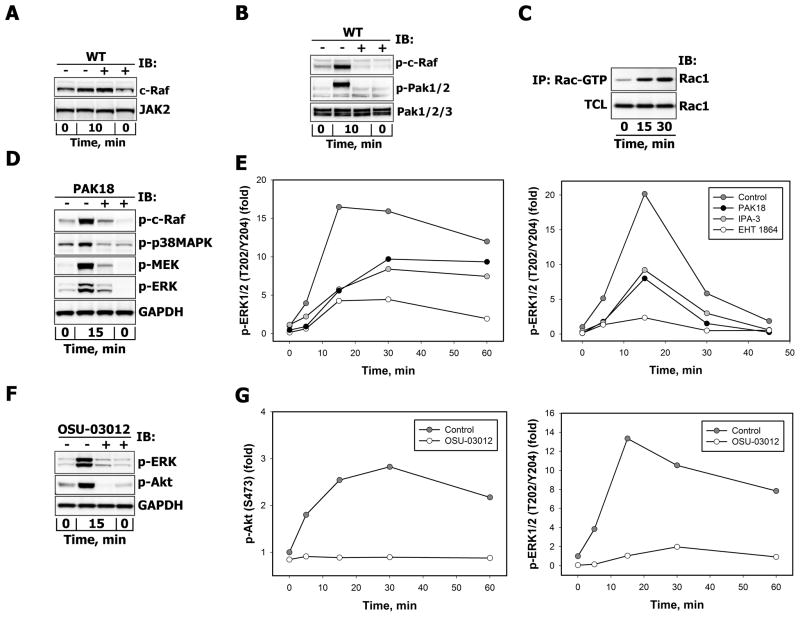
Although inhibition of PI3-kinase did not prevent c-Raf recruitment to the plasma membrane (Fig. 6A), it considerably reduced PRL-induced c-Raf phosphorylation at Ser338 (Fig. 6B and Fig. 5A, blot 6), which correlated with a decreased phosphorylation of serine/threonine kinases PAK1/2 on activating Thr423/Thr402 residues (Fig. 6B), supporting the notion that Ser338 is a target site for PAK1 [43].
Figure 6.
A. Effects of PI3-kinase inhibition on PRL-induced recruitment of c-Raf Serum-starved T47D cells were either left untreated (−) or were treated (+) with WT (200 nM, 30min) before stimulation with 10 nM PRL for 10 min. c-Raf proteins were detected by IB of the particulate/membrane fraction, which was isolated as described in “Materials and Methods”. Simultaneous detection of JAK2 in the same samples served as loading control. B. Effects of PI3K inhibition on PRL-induced c-Raf and PAK activation. Serum-starved T47D cells were either left untreated (−) or were treated (+) with wortmannin (WT, 200 nM, 30 min) before stimulation with PRL (10 nM, 10 min). Phosphorylated c-Raf (Ser338), PAK1/2 (Thr423/Thr402) and total PAK1/2/3 protein levels were detected by IB with Abs against respective antigens. C. PRL stimulation of Rac1 activation. After MCF-7 cell stimulation with 10 nM PRL, Rac-GTP levels were measured at indicated time points by affinity precipitation with p21-Rac1 binding domain of human PAK1 bound to glutathione agarose beads as described in “Materials and Methods” and detected with anti-Rac1Abs, which were also used to determine total Rac1 protein levels in corresponding TCL. D. Effects of PAK-18 on PRL-induced activation of MAPK cascades. Serum-starved T47D cells were either left untreated (−) or were treated (+) with PAK18 peptide (10 μM, 60 min) before stimulation with 10 nM PRL for 15 min. Phosphorylated forms of c-Raf (Ser338), p38MAPK (Thr180/Tyr182), MEK1/2 (Ser217/221) and ERK1/2 (Thr202/Tyr204) proteins were detected by IB of the TCL with Abs against respective antigens. GAPDH levels served as protein loading control. Representative blots are shown (n=3). E. Effects of Rac/PAK inhibition on PRL-induced activation of ERK1/2. Serum-starved T47D (left panel) or MCF-7 (right panel) cells were either left untreated (control, dark grey circles) or were treated (+) with EHT 1864 (10 μM, 60 min, white circles), IPA-3 (10 μM, 30 min, grey circles) or PAK18 peptide (10 μM, 60 min, black circles) before stimulation with 10 nM PRL for the indicated time intervals. Both total and phosphorylated forms of ERK1/2 (Thr202/Tyr204) were detected by IB of the TCL with Abs against respective antigens. The ratio of phospho-ERK1/2:total ERK1/2 at each time point was expressed as fold changes over basal levels. Quantitation of representative blots is shown (n=3). F–G. Effects of concurrent inhibition of PDK1 and PAK on PRL-induced activation of Akt and ERK1/2. Serum-starved MCF-7 (F) or T47D (G) cells were either left untreated (−) or were treated (+) with OSU-03012 (25 μM, 30 min) before stimulation with 10 nM PRL for indicated periods of time. Phosphorylated forms of Akt (Ser473) and ERK1/2 (Thr202/Tyr204) proteins were detected by IB of the TCL with Abs against respective antigens. GAPDH levels served as protein loading control. Representative blots are shown (n=3). (F). The ratio of phospho-protein:total protein at each time point was expressed as fold changes over basal levels (G).
The multi-step activation of PAK involves its interaction with PAK-interacting exchange factor (PIX), which recruits PAK to the small GTPases Rac and Cdc42, resulting in relief from autoinhibition, autophosphorylation and/or phosphorylation by exogenous kinases. In addition, a GTPase-independent PAK activation mechanisms also exist [44–45].
Pull-down experiments using the p21-binding domain (PBD) of PAK to selectively isolate the GTP-bound form of Rac1 showed that PRL was able to induce activation of Rac1 in breast cancer cells (Fig. 6C). Next, T47D and MCF-7 cells were stimulated with PRL for various periods of time in the presence or absence of PAK18, which is composed of the cell permeant TAT peptide sequence and an 18-mer proline-rich PIX-interacting motif of PAK that disrupts PIX-PAK interaction and thereby reduces PAK activation by Rac1 and Cdc42 [43]. Alternatively, cells were pretreated with the allosteric PAK inhibitor IPA-3, which promotes an inactive conformation of PAK1, but does not inhibit the enzymatic activity of preactivated PAKs [46–47].
Along with its well-known downstream target p38 MAPK [48], PAK inhibition significantly reduced the activation of c-Raf, MEK and ERK1/2 following 15 min of PRL stimulation in T47D cells (Fig. 6D). More detailed measurements of the temporal phosphorylation response of ERK1/2 revealed that treatment with PAK18 and IPA-3 decreased peak ERK1/2 activation by 66% and 65% in T47D cells (Fig. 6E, left panel) and by 60% and 54% in MCF-7 cells (Fig. 6E, right panel), respectively. Next, we used EHT 1864, a small molecule inhibitor of Rac family small GTPases (Rac1, Rac1b, Rac2 and Rac3), which prevents Rac interaction with all effectors, including PAKs [49]. In comparison with untreated cells, EHT 1864 treatment reduced maximal ERK1/2 activation by 74% in T47D (Fig. 6E, left panel) and by 88% in MCF-7 (Fig. 6E, right panel) cells.
PRL enhances the motility of T47D, MCF-7, and MDA-231 breast cancer cell lines and potentiates EGF-induced migration [50]. In our study, the inhibitory effect of EHT 1864 on cell migration was examined using a wound-healing assay. Rac inhibition significantly reduced PRL-induced motility of MCF-7 cells (Supplemental Fig. 4S). In addition, Rac/PAK inhibition by IPA-3 and EHT 1864 significantly reduced PRL-mediated T47D breast cancer cell growth in vitro (Supplemental Fig. 5S).
Apart from its activation by small GTPases Rac and Cdc42, PAKs can also bind to Grb2 [51] and be recruited to activated receptors on the plasma membrane, where PDK1 can phosphorylate PAK1 at Thr423, a residue that is critical for PAK1 activation, by disrupting the folded autoinhibitory conformation [52–54]. Therefore, to ensure a complete suppression of PAK1 activation, we used OSU-03012, a novel Celecoxib derivative, which simultaneously inhibits PDK1 activity, thus blocking the PDK1-mediated PAK1 activation route, as well as inhibiting PAK activity via competitive inhibition of ATP binding [55]. OSU-03012 treatment nearly abolished PRL-induced Akt and ERK1/2 responses in MCF-7 cells (Fig. 6F) and T47D cells (Fig. 6G), as well as in more invasive SK-BR-3 breast cancer cells (unpublished observations). This effect was not mediated by PDK1-dependent activation of protein kinases C (PKC), since cell treatment with bisindolylmaleimide I, a potent and selective inhibitor of multiple PKC isozymes, did not reduce ERK1/2 activation in response to PRL (data not shown).
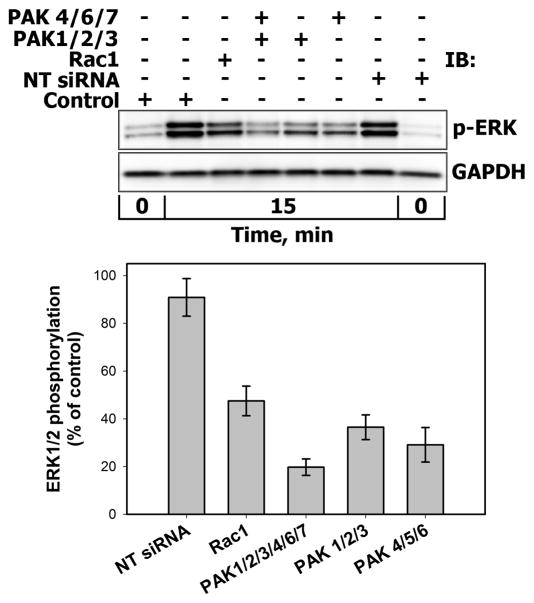
The involvement of the Rac/PAK pathway in PRL-induced activation of ERK1/2 were verified by siRNA-mediated suppression of Rac1, PAK1/2/3, PAK4/6/7 and all PAK family members. The remaining relative mRNA or protein levels were measured 72 hours post-transfection by Western blotting or quantitative PCR analysis, respectively (Supplemental Fig. 6S) as described in “Materials and Methods” section. In comparison with untransfected control or non-targeting (NT) negative control siRNA-treated cells, ERK1/2 phosphorylation peak decreased by approximately 50% and 80% upon suppression of Rac1 and PAK1/2/3/4/6/7, respectively (Fig. 7). Inability to block ERK1/2 activation by 100% by Rac1 siRNA may indicate that ERK1/2 can be also activated by the other isoforms and members of the Rac family, such as Rac1b, Rac2, Rac3 and/or Cdc42.
Figure 7. Effects of siRNA-mediated suppression of Rac1 and PAK family members on PRL-induced activation of ERK1/2.
MCF-7 cells were either left untrasfected (Control) or were transiently transfected in dublicates with non-targeting (NT) negative control siRNA, siRNA duplexes specific for Rac1, group I PAKs (PAK1/2/3), group II PAKs (PAK4/6/7) or group I/II PAKs for 72 hours. Cells were then stimulated with 10 nM PRL for 15 min. Phosphorylated forms of ERK1/2 (Thr202/Tyr204) were detected by IB of the TCL. GAPDH levels served as protein loading control. Representative blots are shown in the upper panel. The lower panel shows a graph with the calculated averages ±SD of remaining ERK1/2 phosphorylation (normalized to GAPDH) from two independent experiments.
Ras plays a minor role in prolactin-induced ERK activation
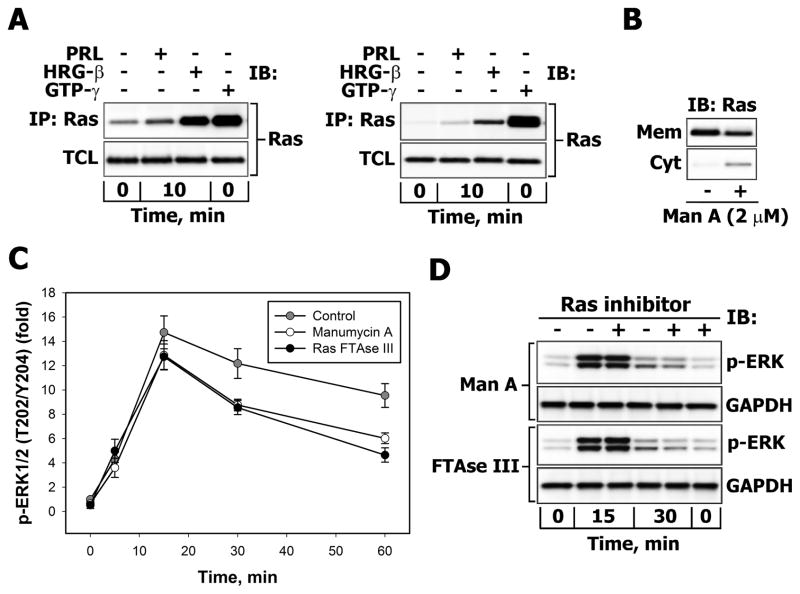
Our data show that PRL-stimulated T47D and MCF-7 cells display very low activation of the small GTPase Ras over a basal level compared to the potent Ras-inducer heregulin-β (HRG-β) (Fig. 8A). Moreover, PRL-activated Ras corresponds to only a small fraction of the total pool of Ras-GTP (Fig. 8A). Next, to estimate the relative contribution of the parallel route of PRL-induced activation of ERK1/2, involving Ras-GTP, T47D and MCF-7 breast cancer cells were pretreated with farnesyltransferase inhibitors (manumycin A or Ras FTase III), which interfere with the post-translational processing of Ras and its proper targeting to the plasma membrane thus blocking the Ras-mediated signaling pathways. The amounts of Ras present in the insoluble (membrane) and soluble (cytosolic) subcellular fractions were evaluated by Western blotting. Under basal conditions, Ras was absent from the soluble fraction. Treatment with 2 μM manumycin A for 7 hours decreased Ras levels in the membrane fraction by approximately 25% and simultaneously increased Ras protein levels in cytosol (Fig. 8B). However, manumycin A treatment had no effect on the initial rate of increase in ERK1/2 phosphorylation and only a moderately suppressed it at time points of 30 minutes or longer in either T47D (Fig. 8C) or MCF-7 (Fig. 8D) cells. Similar effects were obtained with another farnesyltransferase inhibitor Ras FTase III (Fig. 8C–D) and siRNA against K-RAS, which downregulated the K-Ras protein levels by 70%, but failed to block the phosphorylation of ERK1/2 and Akt (Supplemental Fig. 7S). These results could mean that the inhibition of Ras signaling by drugs or siRNA may not have sufficed to block ERK1/2 activation. However, in conjunction with the observation that PRL only leads to a modest activation of Ras, we suggest that the Rac/PAK signaling pathway is the predominant route of PRL-induced ERK1/2 activation.
Figure 8.
A. Comparison of Ras activation in PRL and HRG-β-treated breast cancer cells Serum-starved T47D (left panel) or MCF-7 (right panel) were either left unstimulated or were stimulated with 10 nM PRL or HRG-β for 10 min. GTP-gamma-S loading of Ras in non-stimulated cells was used as a positive control for Ras activation. Affinity precipitation of activated Ras was carried out as described in “Materials and Methods”. Ras was detected by IB of the TCL and corresponding immunoprecipitates (IP) with monoclonal anti-Ras Abs. B. Effects of the farnesyl transferase inhibitor manumycin A on membrane accumulation of Ras. T47D cells were either left untreated (−) or were treated with manumycin A (2 μM) for 7 hours in serum-free medium. Ras was detected by IB of cytosolic (Cyt) and particulate/membrane (Mem) fractions with monoclonal anti-Ras Abs. C–D. Effects of farnesyl transferase inhibitors on PRL-induced ERK1/2 activation. C. Serum-starved T47D cells were either left untreated (control, grey circles) or were treated with RasFTase III (2 μM, black circles) or manumycin A (2 μM, white circles) for 7 hours before stimulation with 10 nM PRL for indicated time intervals. Both total and phosphorylated forms of ERK1/2 (Thr202/Tyr204) were detected by IB with Abs against respective antigens. The ratio of phospho-ERK1/2:total ERK1/2 at each time point was expressed as fold changes over basal levels. Quantitation of representative blots is shown (n=3). D. Phosphorylated ERK1/2 and total GAPDH proteins were detected by IB the TCL of similarly treated MCF-7 cells. Representative blots are shown (n=3).
DISCUSSION
In the present study, we examined the architecture of the PRL-R signaling network in breast cancer cells. We shown that PRL concurrently activates distinct signaling pathways, including the JAK/STAT, PI3-kinase/Akt and MAPK cascades, both in T47D and MCF-7 breast cancer cells, although to a different extent. Differences in the activation patterns of the key intermediates in the two cell lines are qualitatively minor and may be attributed to the different amounts of PRL-R expressed in each (T47D - 25,800 sites/cell and MCF-7 - 8,310 sites/cell) [4, 56], as well as to varying expression levels, constitutive activation status, deregulation mechanisms or degree of engagement of specific signaling intermediates between these two cell lines. Similar to other studies [57–58], we detected only a weak increase in Ras-GTP levels in response to PRL treatment, despite the fact that PRL induced Shc phosphorylation on Grb2-binding sites (Supplemental Fig. 8S). Possible reasons for the low Ras activation could involve transient, weak and/or delayed complex formation between Shc, Grb2 and SOS [57–58], as well as a much less efficient recruitment of these proteins to the plasma membrane compared to HRG-β, which is a potent inducer of Ras (see Fig. 8A), Rac and ERK1/2 activation in breast cancer cells [59–60] (Supplemental Fig. 9S).
It has been reported that c-Src mediates PRL-dependent proliferation of T47D and MCF-7 breast cancer cells via the activation of FAK/ERK1/2 and PI3K/Akt signaling pathways (35). We confirmed the positive roles of SFKs and FAK in regulating ERK1/2 responses, and provided additional evidence that, in fact, SFK/FAK-mediated activation of PI3-kinase, but not its effector Akt or STAT5, is a critical determinant of PRL-stimulated activation of the MAPK cascade. We found that PI3-kinase positively regulates ERK1/2 phosphorylation at the level of c-Raf. Inhibition of PI3-kinase, Rac and PAK activities or Rac1 and PAK1/2/3 and PAK4/6/7 protein levels markedly reduced ERK1/2 phosphorylation, supporting the previously reported roles for various PAK family members in activation of MAPK cascade in other signaling networks [61–63].
Moreover, simultaneous inhibition of PDK1 and PAKs abrogated the ERK1/2 responses to PRL in T47D, MCF-7 and SK-BR-3 breast cancer cell lines, thereby generalizing our observations that activated PRL-R largely utilizes the PI3-kinase-dependent Rac/PAK pathway rather than the canonical Shc/Grb2/SOS/Ras route to initiate, augment and sustain ERK1/2 signaling.
This conclusion is further supported by the minimal effect of Ras inactivation by the use of farnesyl transferase inhibitors or K-RAS siRNA. However, we cannot exclude that Ras inhibition was incomplete or that the contribution of K-RAS to ERK and Akt activation may be readily compensated by other Ras isoforms (e.g. H-Ras, N-Ras). Moreover, using higher concentrations of the farnesyl transferase inhibitors to eliminate all functional Ras from the plasma membrane caused significant Akt dephosphorylation, followed by ERK1/2 deactivation (Supplemental Fig. 10S) and cell detachment and death, possibly due to deregulation of anti-apoptotic pathways as a consequence of Ras inhibition or other effects of defarnesylation [64]. Therefore, these approaches could not be used to quantify more accurately the contributions of Ras-dependent and Ras-independent inputs into ERK1/2.
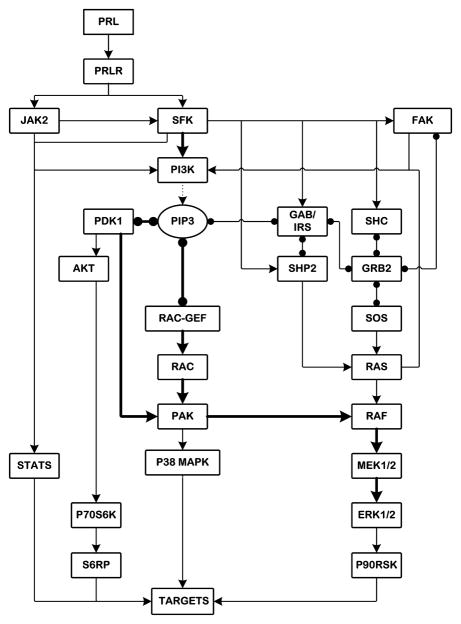
A general picture emerges from our results, in conjunction with prior studies, in which the signaling from PRL-R/JAKs/SFKs diverges into four major pathways: STATs, PI3-kinase/Akt, Rac/PAK and Shc/Grb2-SOS/Ras (Fig. 9). The signal to ERK1/2 is predominantly routed through the PI3-kinase/PDK1-dependent Rac/PAK/c-Raf/MEK route. In parallel, the Rac/PAK pathway also feeds into the stress response MAPK cascades, such as p38MAPK. By activating these pathways, PLR can perform important functions in controlling cell cycle entry, apoptosis, cell shape, polarity, adhesion as well as migration. Given that more than 50% of human breast cancers display overexpression and hyperactivation of PAK1/2 and/or PI3-kinase, which correlate with increased invasiveness and survival of breast cancer cells [65–67], our findings provide a more detailed roadmap by which these pathways are integrated, which is likely to be relevant for therapeutic interventions that target these pathways and therefore may have clinical significance.
Figure 9. Suggested architecture of PRL-R signaling network with potential routes of MAPK activation in breast cancer cells.
Solid lines with arrows show the activation or phosphorylation of proteins. Dotted lines indicate the formation of phosphatidylinositol-trisphosphate (PIP3) by PI3-kinase. Solid lines with circular ends represent protein-protein and protein-lipid interactions. Thick solid lines show prevailing routes of ERK1/2 activation.
Clearly, further studies are required to accurately quantify the contributions of these different signaling routes leading to ERK1/2 activation and related downstream cascades in tumor and non-malignant cells and to assess their impact on physiological outcomes related to tumorigenesis and metastatic potential. Such studies will form the basis for a more complete computational analysis of the integrated PRL-R signaling network in which the roles of protein phosphatases and multiple feedback loops can be quantified.
CONCLUSION
In conclusion, our systems-level analysis of PRL signaling network demonstrates the interplay between the PI3-kinase and MAPK signaling cascade, which, to the best of our knowledge, has never been studied in the context of PRL signaling. Our data reveal that the signal from the activated PRL receptor to ERK1/2 predominantly uses the PI3-kinase-dependent Rac/PAK/c-Raf/MEK pathway rather than the canonical Shc/Grb2/SOS/Ras route. In turn, the PI3-kinase-dependent ERK1/2 activation is controlled by JAK2, Src family kinases and FAK, whereas STATs, Akt and PKC do not regulate PRL-induced ERK1/2 responses. At the same time, Rac/PAK inhibiton or silencing by siRNA significantly suppresses PRL-mediated breast cancer cell growth and motility. Thus our study highlights the rationale for targeting Rac/PAK signaling pathway alone or in combination with PI3-kinase and/or Src-directed therapies in breast cancer.
Supplementary Material
Acknowledgments
This work was supported by the NIH Grant GM059570.
Footnotes
ABBREVIATIONS: PRL- Prolactin, PRL-R - prolactin receptor, JAKs - Janus family kinases, MEK1/2 - Mitogen-activated protein kinase kinase 1/2, ERK1/2 - extracellular signal-regulated kinase 1/2, MAPK - Mitogen activated protein kinase, STAT- signal transducers and activators of transcription, PI3K - phosphoinositide 3-kinase, PAK - p21-activated kinase, PDK1 - 3-Phosphoinositide-dependent kinase 1, p70S6K – p70 ribosomal protein S6 kinase, S6RP – S6 Ribosomal protein, SFKs - Src family kinases, FAK – focal adhesion kinase, Grb2 - growth factor receptor-bound protein 2, SH3 - Src homology 3, SH2 - Src homology 2, SOS - Son of Sevenless, GEF - guanine nucleotide exchange factor, GAPDH – Glyceraldehyde-3-Phosphate Dehyrogenase, Gab1 - Grb2-associated binder 1, WT - wortmannin, PH - pleckstrin homology, PIX - PAK-interacting exchange factor, PBD - p21-binding domain, Shc – SH2 and collagen domain protein C1.
AUTHORS’ CONTRIBUTIONS
Listed by author order on the manuscript: EA designed and carried out the experiments, analyzed the data, prepared tables and figures, drafted and participated in writing of the manuscript. SA carried out the experiments. WK, BNK and JBH interpreted the results, conceived some experiments and edited the manuscript. AK designed and conceived of the study, was responsible for the acquisition of all data, interpreted the results, drafted and wrote the manuscript. All authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Edita Aksamitiene, Email: edita.aksamitiene@jefferson.edu.
Sirisha Achanta, Email: sirisha.achanta@jefferson.edu.
Walter Kolch, Email: walter.kolch@ucd.ie.
Boris N. Kholodenko, Email: boris.kholodenko@ucd.ie.
Jan B. Hoek, Email: jan.hoek@jefferson.edu.
Anatoly Kiyatkin, Email: anatoly.kiyatkin@jefferson.edu.
References
- 1.Binart N, Ormandy CJ, Kelly PA. Adv Exp Med Biol. 2000;480:85–92. doi: 10.1007/0-306-46832-8_10. [DOI] [PubMed] [Google Scholar]
- 2.Fresno Vara JA, Caceres MA, Silva A, Martin-Perez J. Mol Biol Cell. 2001;12(7):2171–2183. doi: 10.1091/mbc.12.7.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clevenger CV, Furth PA, Hankinson SE, Schuler LA. Endocr Rev. 2003;24(1):1–27. doi: 10.1210/er.2001-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peirce SK, Chen WY. J Endocrinol. 2001;171(1):R1–4. doi: 10.1677/joe.0.171r001. [DOI] [PubMed] [Google Scholar]
- 5.Bernichtein S, Touraine P, Goffin V. J Endocrinol. 2010;206(1):1–11. doi: 10.1677/JOE-10-0069. [DOI] [PubMed] [Google Scholar]
- 6.Tworoger SS, Eliassen AH, Sluss P, Hankinson SE. J Clin Oncol. 2007;25(12):1482–1488. doi: 10.1200/JCO.2006.07.6356. [DOI] [PubMed] [Google Scholar]
- 7.Gill S, Peston D, Vonderhaar BK, Shousha S. J Clin Pathol. 2001;54(12):956–960. doi: 10.1136/jcp.54.12.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swaminathan G, Varghese B, Fuchs SY. J Mammary Gland Biol Neoplasia. 2008;13(1):81–91. doi: 10.1007/s10911-008-9068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oakes SR, Robertson FG, Kench JG, Gardiner-Garden M, Wand MP, Green JE, Ormandy CJ. Oncogene. 2007;26(4):543–553. doi: 10.1038/sj.onc.1209838. [DOI] [PubMed] [Google Scholar]
- 10.LaPensee EW, Ben-Jonathan N. Endocr Relat Cancer. 2010;17(2):R91–107. doi: 10.1677/ERC-09-0253. [DOI] [PubMed] [Google Scholar]
- 11.Clevenger CV, Gadd SL, Zheng J. Trends Endocrinol Metab. 2009;20(5):223–229. doi: 10.1016/j.tem.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Rane SG, Reddy EP. Oncogene. 2000;19(49):5662–5679. doi: 10.1038/sj.onc.1203925. [DOI] [PubMed] [Google Scholar]
- 13.Lee RC, Walters JA, Reyland ME, Anderson SM. J Biol Chem. 1999;274(15):10024–10034. doi: 10.1074/jbc.274.15.10024. [DOI] [PubMed] [Google Scholar]
- 14.Roberts PJ, Der CJ. Oncogene. 2007;26(22):3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 15.Kiyatkin A, Aksamitiene E. Methods Mol Biol. 2009;536:149–161. doi: 10.1007/978-1-59745-542-8_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aksamitiene E, Kholodenko BN, Kolch W, Hoek JB, Kiyatkin A. Cell Signal. 2010;22(9):1369–1378. doi: 10.1016/j.cellsig.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piazza TM, Lu JC, Carver KC, Schuler LA. Mol Endocrinol. 2009;23(2):202–212. doi: 10.1210/me.2008-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rucci N, Susa M, Teti A. Anticancer Agents Med Chem. 2008;8(3):342–349. doi: 10.2174/187152008783961905. [DOI] [PubMed] [Google Scholar]
- 19.Schlaepfer DD, Hunter T. Mol Cell Biol. 1996;16(10):5623–5633. doi: 10.1128/mcb.16.10.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heim MH. J Recept Signal Transduct Res. 1999;19(1–4):75–120. doi: 10.3109/10799899909036638. [DOI] [PubMed] [Google Scholar]
- 21.Schaber JD, Fang H, Xu J, Grimley PM, Rui H. Cancer Res. 1998;58(9):1914–1919. [PubMed] [Google Scholar]
- 22.Brazil DP, Hemmings BA. Trends Biochem Sci. 2001;26(11):657–664. doi: 10.1016/s0968-0004(01)01958-2. [DOI] [PubMed] [Google Scholar]
- 23.Meyuhas O. Int Rev Cell Mol Biol. 2008;268:1–37. doi: 10.1016/S1937-6448(08)00801-0. [DOI] [PubMed] [Google Scholar]
- 24.Anjum R, Blenis J. Nat Rev Mol Cell Biol. 2008;9(10):747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 25.Thomas SM, Brugge JS. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 26.Reddy EP, Korapati A, Chaturvedi P, Rane S. Oncogene. 2000;19(21):2532–2547. doi: 10.1038/sj.onc.1203594. [DOI] [PubMed] [Google Scholar]
- 27.Blake RA, Broome MA, Liu X, Wu J, Gishizky M, Sun L, Courtneidge SA. Mol Cell Biol. 2000;20(23):9018–9027. doi: 10.1128/mcb.20.23.9018-9027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng J, Witthuhn BA, Matsuda T, Kohlhuber F, Kerr IM, Ihle JN. Mol Cell Biol. 1997;17(5):2497–2501. doi: 10.1128/mcb.17.5.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gabarra-Niecko V, Keely PJ, Schaller MD. Biochem J. 2002;365(Pt 3):591–603. doi: 10.1042/BJ20020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Downward J. FEBS Lett. 1994;338(2):113–117. doi: 10.1016/0014-5793(94)80346-3. [DOI] [PubMed] [Google Scholar]
- 31.Dance M, Montagner A, Salles JP, Yart A, Raynal P. Cell Signal. 2008;20(3):453–459. doi: 10.1016/j.cellsig.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Lu W, Shen K, Cole PA. Biochemistry. 2003;42(18):5461–5468. doi: 10.1021/bi0340144. [DOI] [PubMed] [Google Scholar]
- 33.Lu W, Gong D, Bar-Sagi D, Cole PA. Mol Cell. 2001;8(4):759–769. doi: 10.1016/s1097-2765(01)00369-0. [DOI] [PubMed] [Google Scholar]
- 34.Ali S, Chen Z, Lebrun JJ, Vogel W, Kharitonenkov A, Kelly PA, Ullrich A. EMBO J. 1996;15(1):135–142. [PMC free article] [PubMed] [Google Scholar]
- 35.Chughtai N, Schimchowitsch S, Lebrun JJ, Ali S. J Biol Chem. 2002;277(34):31107–31114. doi: 10.1074/jbc.M200156200. [DOI] [PubMed] [Google Scholar]
- 36.Slack-Davis JK, Martin KH, Tilghman RW, Iwanicki M, Ung EJ, Autry C, Luzzio MJ, Cooper B, Kath JC, Roberts WG, Parsons JT. J Biol Chem. 2007;282(20):14845–14852. doi: 10.1074/jbc.M606695200. [DOI] [PubMed] [Google Scholar]
- 37.Eide BL, Turck CW, Escobedo JA. Mol Cell Biol. 1995;15(5):2819–2827. doi: 10.1128/mcb.15.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen HC, Appeddu PA, Isoda H, Guan JL. J Biol Chem. 1996;271(42):26329–26334. doi: 10.1074/jbc.271.42.26329. [DOI] [PubMed] [Google Scholar]
- 39.Calalb MB, Polte TR, Hanks SK. Mol Cell Biol. 1995;15(2):954–963. doi: 10.1128/mcb.15.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calalb MB, Zhang X, Polte TR, Hanks SK. Biochem Biophys Res Commun. 1996;228(3):662–668. doi: 10.1006/bbrc.1996.1714. [DOI] [PubMed] [Google Scholar]
- 41.Muller J, Sperl B, Reindl W, Kiessling A, Berg T. Chembiochem. 2008;9(5):723–727. doi: 10.1002/cbic.200700701. [DOI] [PubMed] [Google Scholar]
- 42.Nelson EA, Walker SR, Kepich A, Gashin LB, Hideshima T, Ikeda H, Chauhan D, Anderson KC, Frank DA. Blood. 2008;112(13):5095–5102. doi: 10.1182/blood-2007-12-129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maruta H, He H, Nheu T. Methods Mol Biol. 2002;189:75–85. doi: 10.1385/1-59259-281-3:075. [DOI] [PubMed] [Google Scholar]
- 44.Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, Tan I, Leung T, Lim L. Mol Cell. 1998;1(2):183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 45.Bokoch GM. Annu Rev Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 46.Deacon SW, Beeser A, Fukui JA, Rennefahrt UE, Myers C, Chernoff J, Peterson JR. Chem Biol. 2008;15(4):322–331. doi: 10.1016/j.chembiol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viaud J, Peterson JR. Mol Cancer Ther. 2009;8(9):2559–2565. doi: 10.1158/1535-7163.MCT-09-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang S, Han J, Sells MA, Chernoff J, Knaus UG, Ulevitch RJ, Bokoch GM. J Biol Chem. 1995;270(41):23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]
- 49.Shutes A, Onesto C, Picard V, Leblond B, Schweighoffer F, Der CJ. J Biol Chem. 2007;282(49):35666–35678. doi: 10.1074/jbc.M703571200. [DOI] [PubMed] [Google Scholar]
- 50.Maus MV, Reilly SC, Clevenger CV. Endocrinology. 1999;140(11):5447–5450. doi: 10.1210/endo.140.11.7245. [DOI] [PubMed] [Google Scholar]
- 51.Puto LA, Pestonjamasp K, King CC, Bokoch GM. J Biol Chem. 2003;278(11):9388–9393. doi: 10.1074/jbc.M208414200. [DOI] [PubMed] [Google Scholar]
- 52.King CC, Gardiner EM, Zenke FT, Bohl BP, Newton AC, Hemmings BA, Bokoch GM. J Biol Chem. 2000;275(52):41201–41209. doi: 10.1074/jbc.M006553200. [DOI] [PubMed] [Google Scholar]
- 53.Toker A, Newton AC. Cell. 2000;103(2):185–188. doi: 10.1016/s0092-8674(00)00110-0. [DOI] [PubMed] [Google Scholar]
- 54.Fryer BH, Field J. Cancer Lett. 2005;229(1):13–23. doi: 10.1016/j.canlet.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 55.Porchia LM, Guerra M, Wang YC, Zhang Y, Espinosa AV, Shinohara M, Kulp SK, Kirschner LS, Saji M, Chen CS, Ringel MD. Mol Pharmacol. 2007;72(5):1124–1131. doi: 10.1124/mol.107.037556. [DOI] [PubMed] [Google Scholar]
- 56.Shiu RP. Cancer Res. 1979;39(11):4381–4386. [PubMed] [Google Scholar]
- 57.Erwin RA, Kirken RA, Malabarba MG, Farrar WL, Rui H. Endocrinology. 1995;136(8):3512–3518. doi: 10.1210/endo.136.8.7628388. [DOI] [PubMed] [Google Scholar]
- 58.Das R, Vonderhaar BK. Oncogene. 1996;13(6):1139–1145. [PubMed] [Google Scholar]
- 59.Yang C, Liu Y, Lemmon MA, Kazanietz MG. Mol Cell Biol. 2006;26(3):831–842. doi: 10.1128/MCB.26.3.831-842.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thottassery JV, Sun Y, Westbrook L, Rentz SS, Manuvakhova M, Qu Z, Samuel S, Upshaw R, Cunningham A, Kern FG. Cancer Res. 2004;64(13):4637–4647. doi: 10.1158/0008-5472.CAN-03-2645. [DOI] [PubMed] [Google Scholar]
- 61.Wu X, Carr HS, Dan I, Ruvolo PP, Frost JA. J Cell Biochem. 2008;105(1):167–175. doi: 10.1002/jcb.21809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cammarano MS, Nekrasova T, Noel B, Minden A. Mol Cell Biol. 2005;25(21):9532–9542. doi: 10.1128/MCB.25.21.9532-9542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beeser A, Jaffer ZM, Hofmann C, Chernoff J. J Biol Chem. 2005;280(44):36609–36615. doi: 10.1074/jbc.M502306200. [DOI] [PubMed] [Google Scholar]
- 64.Cox AD, Der CJ. Oncogene. 2003;22(56):8999–9006. doi: 10.1038/sj.onc.1207111. [DOI] [PubMed] [Google Scholar]
- 65.Balasenthil S, Sahin AA, Barnes CJ, Wang RA, Pestell RG, Vadlamudi RK, Kumar R. J Biol Chem. 2004;279(2):1422–1428. doi: 10.1074/jbc.M309937200. [DOI] [PubMed] [Google Scholar]
- 66.Dummler B, Ohshiro K, Kumar R, Field J. Cancer Metastasis Rev. 2009;28(1–2):51–63. doi: 10.1007/s10555-008-9168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mira JP, Benard V, Groffen J, Sanders LC, Knaus UG. Proc Natl Acad Sci U S A. 2000;97(1):185–189. doi: 10.1073/pnas.97.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.