Abstract
Members of the Bacteroidales order are among the most abundant gram-negative bacteria of the human colonic microbiota. These species decorate their cell surface glycoproteins with fucosylated glycans, which are believed to play important roles in host intestinal colonization. Currently, there is no method for the enrichment of these glycoproteins for their identification. Here, we describe a chemical approach directed toward labeling and detecting fucosylated glycoproteins from cultured Bacteroidales species, namely Bacteroides fragilis and Parabacteroides distasonis. We treated these bacteria with an alkyne-bearing fucose analog, which is metabolically integrated into the bacterial surface fucosylated glycoproteins. The alkyne-tagged glycoproteins can then react with azide-bearing biophysical probes via bioorthogonal click chemistry for detection or glycoproteomic analysis.
Human symbionts of the Bacteroidales order are among the predominantly represented genera in the mammalian gut.1 Bacteroidales species impart numerous beneficial effects to their hosts: they participate in nutrient processing—hydrolyzing dietary polysaccharides and converting them into short chain fatty acids that can be utilized by the host.2, 3 They are also involved in promoting the development of gut-associated lymphoid tissues4 and regulation of intestinal angiogenesis.5 New evidence suggests that some Bacteroidales species also play an important role in the maintenance of a healthy body weight.6, 7
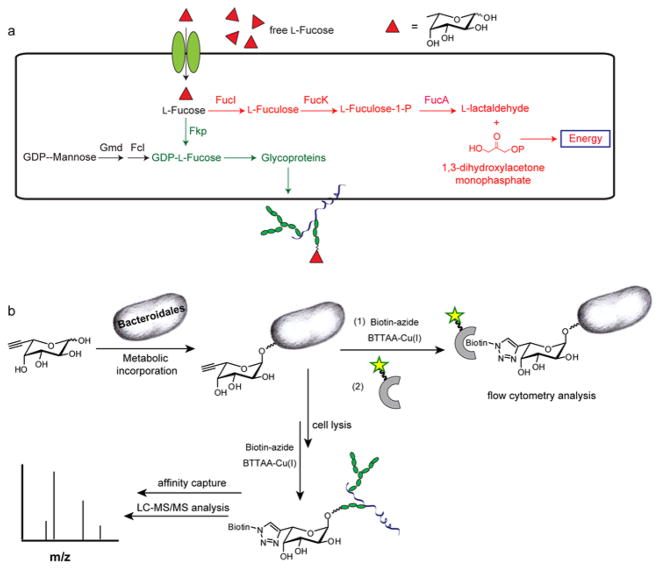
L-Fucose, an abundant component of many host intestinal cell-surface glycoconjugates,8 has risen as a key mediator of interactions between Bacteroidales and their mammalian hosts.9 Discovered by Gordon and coworkers, fucose fulfills a principal nutritional requirement of Bacteroides thetaiotaomicron, a major member of the human distal small intestinal microflora, serving as a host-derived energy source.10 When its intracellular fucose level is low, B. thetaiotaomicron sends a signal to the host, upregulating the production of α1,2-linked fucosides in the host intestinal enterocytes. Fucosidases secreted by the bacteria then cleave L-Fucose from these glycoconjugates, and the cleaved fucose is imported into the bacteria to serve as a carbon source (Fig. 1a). In addition, recent studies have shown that Bacteroides fragilis and Parabacteroides distasonis use a mammalian-like pathway to decorate surface capsular polysaccharides and glycoproteins with fucosylated glycan, a process that is essential for their persistence within the competitive ecosystem of the gut.11, 12 Despite the importance of fucose in host-symbiont interplay, fundamental questions regarding the distribution and biological functions of these fucosylated glycoproteins in the Bacteroidales remain unanswered. For example, how many proteins are fucosylated? How does the fucosylation change in the proteins of the commensal microbes in response to the nutrient changes in their hosts? What are the functional roles of these fucosylated glycoproteins in intestinal colonization and in mediating a symbiotic relationship with the host? Here, we report a chemical reporter strategy for identifying and characterizing the fucosylated glycoproteins in these abundant intestinal symbionts with the ultimate goal of answering these questions.
Figure 1. Detection of fucosylated glycoproteins in Bacteroidales species.
(a) Fucose metabolic pathways in Bacteroidales species. Red: fucose utilization system; Black: GDP-fucose de novo biosynthetic pathway; Green: GDP-fucose salvage pathway. (b) A chemical strategy for the detection and enrichment of fucosylated glycoproteins in Bacteroidales.
Studies by Comstock and coworkers showed that the B. fragilis has both a “host-like” de novo biosynthetic pathway and a salvage pathway for the production of GDP-L-fucose, the universal fucosyl donor. In the salvage pathway, a bifunctional enzyme, Fkp, conserved in all Bacteroidales species converts fucose to Fuc-1-P and thence to GDP-fucose (Fig. 1a).11 Further, we discovered that recombinant Fkp from B. fragilis has relaxed specificity toward fucose analogues bearing unnatural substituents at the C-5 position, both terminal alkyne- and azide-bearing substrates (FucAl and FucAz) can be processed to produce the corresponding GDP-fucose analogs.13 Based on these precedents, we designed the following strategy to label fucosylated glycoproteins in Bacteroidales species. First, the alkyne-bearing unnatural substrate, FucAl,14 is introduced into the culture media, allowing for the metabolic incorporation into the surface glycoconjugates of the microbes (Fig. 1b). In a second step, the tagged sugar on the bacterial surface or lysates is chemically reacted with an exogenously delivered affinity probe bearing the azide, which allows for the enrichment and identification of the labeled glycoproteins via the bioorthogonal Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAc).15–18
To evaluate if FucAl could be metabolized by the target cells, we cultured P. distasonis anaerobically in a defined medium in the presence or absence of 200 μM FucAl supplements. After 24 hours, we harvested the bacteria and performed a ligation reaction with biotin-azide via the biocompatible CuAAC mediated by BTTAA-Cu(I).19 To detect the biotinylated glycoconjugates on the bacterial surface, we stained the labeled P. distasonis with streptavidin-Alexa Fluor 488 and analyzed the surface-associated Alexa Fluor 488 fluorescence by flow cytometry. P. distasonis treated with FucAl showed a marked increase in fluorescence that indicated the conjugation of biotin moieties on the bacterial surface, whereas untreated cells showed only a background level of fluorescence after exposure to the CuAAC reagents (Fig. 2). The background fluorescence was similar to that observed with bacteria that were not exposed to any reagents, which confirmed the specificity of the CuAAC and the integration of the alkyne tag into the bacterial surface.
Figure 2. Detection of fucosylated glycoconjugates on the cell surface of Bacteroidales species by flow cytometry analysis.
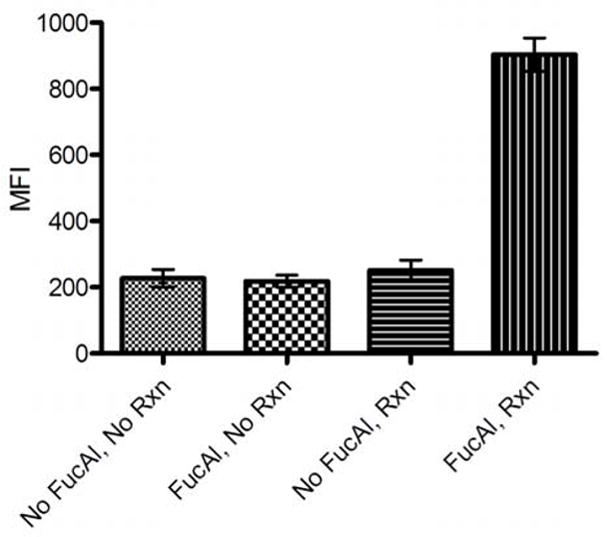
P. distasonis were cultured in the presence or absence of FucAl (200 μM) overnight before reacting with biotin-azide (100 μM) in the presence of the BTTAA-Cu(I) catalyst ([Cu] = 60 μM, [BTTAA]:[Cu]=6:1) for 10 min and probing with streptavidin-Alexa Fluor 488. Errors were based on triplicate runs in one experiment.
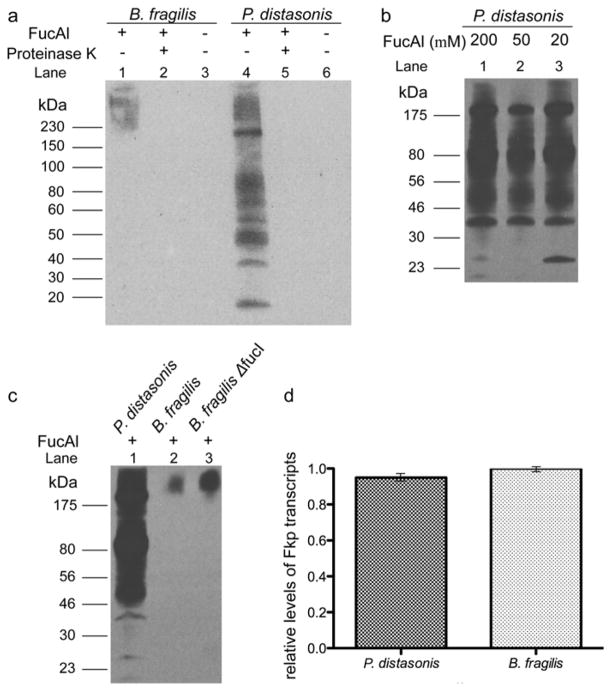
To verify if FucAl was incorporated into P. distasonis glycoproteins, we lysed the bacteria and reacted the bacterial lysates with biotin-azide catalyzed by the BTTAA-Cu(I) complex and analyzed the labeled glycoproteins by SDS-PAGE and anti-biotin Western blot. Significant glycoprotein labeling in lysates from P. distasonis treated with FucAl was observed, whereas no signals were detectable in lysates from untreated bacteria. The signal was abolished upon proteinase K treatment, which confirmed that the unnatural sugar was incorporated into the cellular glycoproteins (Fig. 3a). Multiple labeled glycoproteins were detected with molecular weights ranging from 20–220 kD. Noteworthy, we found that 20 μM of FucAl was sufficient to provide a robust signal in this assay (Fig. 3b). Similar results were obtained from B. fragilis cultured with FucAl, though significantly weaker labeling was observed—only proteins with molecular weights ranging from 175–220 kD were detected in the anti-biotin Western blot (Fig. 3a). Importantly, no signals were detectable for the Δfkp mutant, demonstrating that FucAl was incorporated through the salvage pathway (see Supporting Information Fig. S1).
Figure 3. Analysis of fucosylated glycoproteins and the fucose metabolic pathways in Bacteroidales species.
(a) FucAl is incorporated into Bacteroidales glycoproteins. B. fragilis (lane 1–3) and P. distasonis (lane 4–6) were cultured in the presence (lane 1, 2, 4, 5) or absence (lane 3, 6) of FucAl (200 μM), lysed, treated with (lane 2, 5) or without (lane 1, 3, 4, 6) protease K (100 μg/mL), reacted with biotin-azide via CuAAC and then probed with an α-biotin antibody. (b) P. distasonis is efficiently labeled using low concentrations of FucAl. P. distasonis was cultured in the presence of 200 μM (lane 1), 50 μM (lane 2) and 20 μM (lane 3) FucAl, lysed, reacted with biotin-azide via CuAAC and then probed with an α-biotin antibody. (c) Loss of the B. fragilis fucose isomerase pathway results in greater FucAl incorporation. P. distasonis, B. fragilis and B. fragilis ΔfucI were cultured in the presence of FucAl (200 μM), lysed, reacted with biotin-azide via CuAAC and then probed with an α-biotin antibody. Equal protein loading was confirmed by Coomassie blue staining (data not shown). (d) FKP expression level is not a major factor in comparative FucAl incorporation. Histogram of the relative ratio of FKP mRNA transcript levels between P. distasonis and B. fragilis. Errors were based on triplicate runs in one experiment.
The discrepancy of FucAl incorporation in P. distasonis and B. fragilis may be due to several factors, including i) different fucose metabolic pathways in these two species; ii) different expression levels of relevant enzymes or iii) different substrate specificities of the key fucose salvage pathway enzyme Fkp in these Bacteroidales species.
P. distasonis and B. fragilis share a common fucose acquisition system.20 However, P. distasonis lacks the fucose utilization system that breaks down fucose to generate L-lactaldehyde and dihydroxyacetone phosphate (Fig. 1a). Therefore, exogenous fucose, when imported into the cytosol, is all shunted to be incorporated into glycoconjugates. In order to evaluate if deleting the fucose utilization pathway could result in higher FucAl incorporation in B. fragilis, we generated a ΔfucI mutant by deleting the gene encoding L-fucose isomerase (FucI), the first enzyme in the fucose utilization pathway (Fig. 1a). The ΔfucI mutant grew normally in defined medium supplemented with dihydroxyacetone. We cultured B. fragilis and the ΔfucI mutant in defined medium supplemented with 200 μM FucAl overnight. We then reacted the treated lysates with biotin-azide via CuAAC, and analyzed the FucAl incorporation by anti-biotin Western blot. As quantified by ImageJ, the ΔfucI mutant showed a 3-fold stronger signal in the Western blot, indicating that incorporation of the unnatural sugar into B. fragilis cellular glycoproteins can be boosted by blocking the fucose utilization system (Fig. 3c).
Next, we analyzed the relative expression of fkp in both Bacteroidales species by using RT-PCR to detect the corresponding mRNA transcripts. As shown in Fig 3d, no apparent differences were found when both species were cultured overnight in the defined medium, indicating that FKP expression level is not a major factor responsible for the observed differences of FucAl incorporation.
In summary, we demonstrate that FucAl, an alkynyl analog of fucose, is metabolized by Bacteroidales and incorporated into the cell surface glycoproteins. The alkyne-tagged glycoproteins can then be specifically detected with azido probes via CuAAC. The specificity and sensitivity of this chemical strategy will allow for enrichment of fucosylated glycoproteins for proteomic analysis, which is currently underway in our laboratory.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (GM080585 to P.W.) and startup funds from Albert Einstein College of Medicine (to P.W.). C.B. is supported by the NIH training grant T32 GM007491. We thank Prof. Laura Comstock for providing B. fragilis Δfkp and Δgmd mutants, and Prof. Michael Malamy for providing B. fragilis 638R and pADB242A.
Footnotes
Dedicated to Prof. Carolyn Bertozzi on the occasion of her receiving Tetrahedron Young Investigator Award
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moore WE, Holdeman LV. Appl Microbiol. 1974;27:961. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Science. 2005;307:1955. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 3.Xu J, Gordon JI. Proc Natl Acad Sci U S A. 2003;100:10452. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhee KJ, Sethupathi P, Driks A, Lanning DK, Knight KL. J Immunol. 2004;172:1118. doi: 10.4049/jimmunol.172.2.1118. [DOI] [PubMed] [Google Scholar]
- 5.Stappenbeck TS, Hooper LV, Gordon JI. Proc Natl Acad Sci U S A. 2002;99:15451. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Nature. 2006;444:1022. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. Nature. 2006;444:1027. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 8.Ma B, Simala-Grant JL, Taylor DE. Glycobiology. 2006;16:158R. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- 9.Comstock LE. Cell Host Microbe. 2009;5:522. doi: 10.1016/j.chom.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. Proc Natl Acad Sci U S A. 1999;96:9833. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coyne MJ, Reinap B, Lee MM, Comstock LE. Science. 2005;307:1778. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 12.Fletcher CM, Coyne MJ, Bentley DL, Villa OF, Comstock LE. Proc Natl Acad Sci U S A. 2007;104:2413. doi: 10.1073/pnas.0608797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, Hu T, Frantom PA, Zheng T, Gerwe B, Del Amo DS, Garret S, Seidel RD, 3rd, Wu P. Proc Natl Acad Sci U S A. 2009;106:16096. doi: 10.1073/pnas.0908248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawa M, Hsu TL, Itoh T, Sugiyama M, Hanson SR, Vogt PK, Wong CH. Proc Natl Acad Sci U S A. 2006;103:12371. doi: 10.1073/pnas.0605418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem Int Ed. 2002;41:2596. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Tornoe CW, Christensen C, Meldal M. J Org Chem. 2002;67:3057. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 17.Soriano Del Amo D, Wang W, Jiang H, Besanceney C, Yan AC, Levy M, Liu Y, Marlow FL, Wu P. J Am Chem Soc. 2010;132:16893. doi: 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sletten EM, Bertozzi CR. Angew Chem Int Ed. 2009;48:6974. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Besanceney-Webler C, Jiang H, Zheng T, Feng L, Soriano del Amo D, Wang W, Klivansky LM, Marlow FL, Liu Y, Wu P. Angew Chem Int Ed. doi: 10.1002/anie.201101817. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, Cordum H, Van Brunt A, Kim K, Fulton RS, Fulton LA, Clifton SW, Wilson RK, Knight RD, Gordon JI. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.