Abstract
Metabolic oligosaccharide engineering is powerful approach to altering the structure of cellular sialosides. This method relies on culturing cells with N-acetylmannosamine (ManNAc) analogs that are metabolized to their sialic acid counterparts and added to glycoproteins and glycolipids. Here we employed two cell lines that are deficient in ManNAc biosynthesis and examined their relative abilities to metabolize a panel of ManNAc analogs to sialosides. In addition to measuring global sialoside production, we also examined biosynthesis of the sialic acid-containing glycolipid, GM3. We discovered that the two cell lines differ in their ability to discriminate among the variant forms of ManNAc. Further, our data suggest that modified forms of sialic acid may be preferentially incorporated into certain sialosides and excluded from others. Taken together, our results demonstrate that global analysis of sialoside production can obscure sialoside-specific differences. These findings have implications for downstream applications of metabolic oligosaccharide engineering, including imaging and proteomics.
Keywords: ganglioside, sialic acid, diazirine, sialyltransferase, metabolic engineering
Glycoconjugates are displayed on the surface of mammalian cells where they play critical roles in both normal and pathological recognition events. Efforts to better delineate glycoconjugates' functions are hindered by the fact that the glycan components are not primary gene products, limiting the utility of site-directed mutagenesis to alter glycan structures in predictable ways. Metabolic oligosaccharide engineering1,2 has provided an important alternative approach to studying glycoconjugate function. By introducing small structural modifications to monosaccharides, and then adding these analogs to cultured cells, it is possible to alter the structure of cellular glycoconjugates in a predictable and controlled way. Metabolic oligosaccharide engineering relies on the abilities of mammalian cells to internalize analogs, metabolize them, and incorporate the unnatural metabolites into glycan structures in place of natural sugars. Early work demonstrated that sialic acid analogs in which the N-acyl chain was lengthened with additional methylene units could be readily tolerated and incorporated into cell surface glycan structures in cells and living animals.3,4 Since then, metabolic oligosaccharide engineering has expanded in scope and now enables the introduction of a variety of functional groups with specialized reactivity (e. g. ketone,5 azide,6,7 alkyne,8 diazirine9) into various glycoconjugates, but most successfully into sialic acid-containing glycoconjugates, also known as sialosides.1 Metabolic oligosaccharide engineering has found widespread utility, including the exploration of host-virus interactions,10 the regulation of neuronal cell differentiation,11 and the visualization of developmental changes in glycosylation.12
While metabolic oligosaccharide engineering has proved to be a highly effective method to alter sialoside structure, the majority of studies measure the success of the engineering at a global level and do not distinguish among the types of sialosides that are modified by this technique. In humans, production of sialosides is controlled by twenty different sialyltransferases.13 Sialyltransferases transfer sialic acid from CMP-sialic acid, the nucleotide sugar donor, to acceptor glycoconjugates. Sialyltransferases are classified into families that catalyze the formation of the α2-3-, α2-6-, or α2-8-linked sialic acid and each sialyltransferase has a unique acceptor specificity. Because of this diversity at the sialyltransfer step, it is possible that metabolically incorporated sialic acid variants are not equally well incorporated into all types of sialosides. Therefore, we decided investigate the linkage-specific and acceptor-specific incorporation of sialic acid analogs.
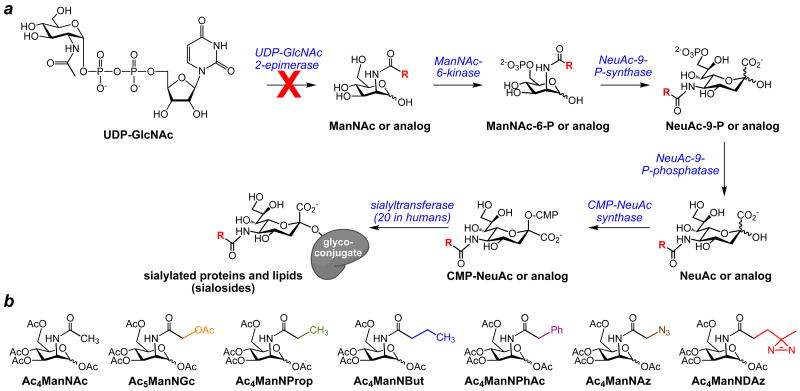
For these experiments, we employed analogs of N-acetylmannosamine (ManNAc), the biosynthetic precursor of sialic acid. In mammalian cells, ManNAc is metabolized to N-acetylneuraminic acid (NeuAc), a common form of sialic acid, in three enzymatic steps (Scheme 1a). Similarly, N-acyl modified ManNAc analogs can be metabolized to their sialic acid counterparts.1 Sialic acids and sialic acid analogs are activated to CMP-sialic acids, then transported into the Golgi, where the sialic acids are transferred to glycoconjugates destined for secretion or cell surface display. Based on previous reports of successful metabolic oligosaccharide engineering, we prepared a small panel of ManNAc analogs (Scheme 1b). ManNAz, a widely-used azide-containing analog, is known to be metabolized into SiaNAz, which has been incorporated into many cell surface glycoconjugates,6 including gangliosides of Jurkat cells.14 ManNProp and ManNPhAc have also been successfully metabolized to SiaNProp and SiaNPhAc, respectively, and incorporated into cell surface gangliosides in several cancer lines. These modified gangliosides have been utilized as immunogenic targets for antibodies.15-18 ManNBut is incorporated into glycoconjugates as SiaNBut, but acts as a chain terminator in polysialylation.19 ManNGc, which can be metabolized into the naturally occurring sialic acid NeuGc, has been used to metabolically produce hydroxylated cell surface gangliosides in neuronal cells to investigate myelin-axon interactions.20 Finally, ManNDAz is metabolized to SiaDAz, which contains a photoactivable crosslinking group that enables covalent crosslinking of sialosides to their binding partners.9,21 In the experiments described here, all ManNAc analogs were peracetylated to enable their facile transport into the cell.22 Protecting groups are assumed to be removed by intracellular nonspecific esterases.
Scheme 1. Metabolism of protected ManNAc analogs to sialosides.
(a) BJAB K20 and CHO Lec3 cells lack UDP-GlcNAc 2-epimerase activity, rendering them unable to biosynthesize ManNAc. When these cells are cultured in serum free conditions, they fail to produce sialosides. The UDP-GlcNAc 2-epimerase-deficient cells maintain the ability to transform ManNAc to sialosides and can also metabolize exogenously supplied ManNAc analogs to their sialoside counterparts. (b) N-acyl-modified ManNAc analogs were prepared and added to the media of cultured cells. All analogs were peracetylated to facilitate their entry into cells. All acetyl groups, including the one on the N-glycolyl side chain of Ac5ManNGc, are believed to be removed by intracellular esterases.
To specifically track the metabolism of exogenously introduced ManNAc analogs, we employed two cell lines, human BJAB K2023,24 and hamster CHO Lec3,25 that are impaired in sialic acid biosynthesis due to lack of UDP-GlcNAc 2-epimerase activity (Scheme 1a). When these epimerase-deficient cell lines are cultured in serum-free conditions, where they cannot scavenge sialic acid, they are unable to synthesize sialosides. When the media is supplemented with Ac4ManNAc, the cells regain the ability to generate sialylated glycoconjugates. Similarly, when the cells are cultured with unnatural ManNAc analogs, any sialoside production is interpreted to reflect successful metabolism of these unnatural analogs to sialosides. As a control, we also examined sialoside production in similar cell lines, BJAB K88 and wt CHO, that maintain UDP-GlcNAc 2-epimerase activity.
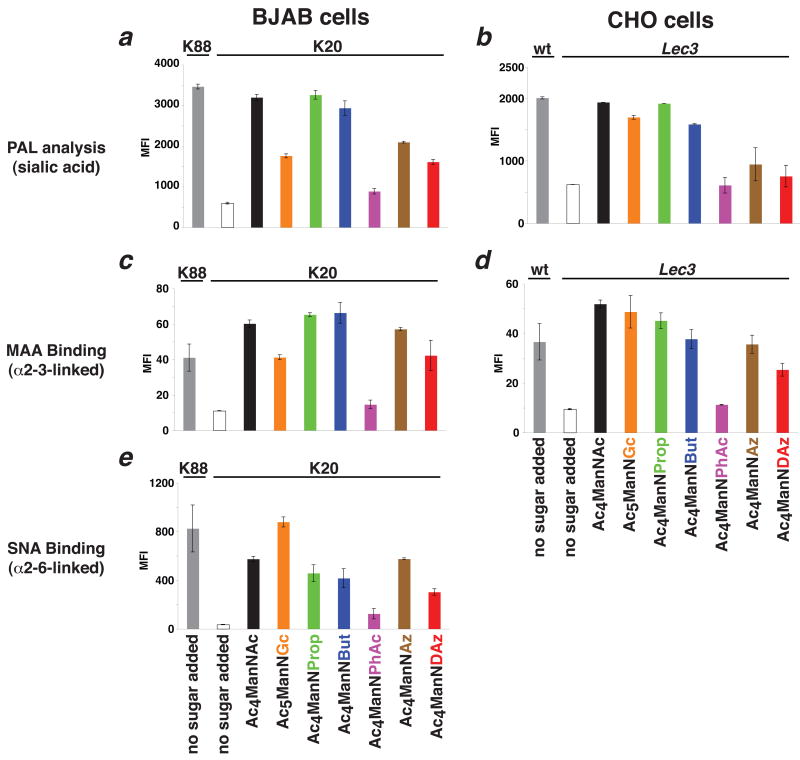
BJAB K20 and CHO Lec3 cells were cultured in serum-free conditions with each of the protected ManNAc analogs. First, we examined total cell surface sialoside levels using the recently reported periodate oxidation and aniline-catalyzed oxime ligation (PAL).26 Consistent with previous reports, we found that Ac4ManNAc, Ac4ManNProp, and Ac4ManNBut were robustly metabolized to sialosides, both in human BJAB K20 cells and hamster CHO Lec3 cells (Figure 1a and b). Ac4ManNAz and Ac4ManNDAz were also metabolized to sialosides in both cell lines, albeit at lower efficiency. Our data do not clearly demonstrate that Ac4ManNPhAc is metabolized to sialosides, but this may reflect the sensitivity limits of the PAL assay since SiaNPhAc-containing glycoconjugates have been detected using antibodies by others.16-18 We noted one significant difference between human BJAB K20 cells and hamster CHO Lec3 cells cultured with Ac5ManNGc: Lec3 cells produce high levels of NeuGc-containing glycoconjugates, while these levels are lower in K20 cells.
Figure 1. Cell surface display of modified sialic acids.
Cell surface sialylation of engineered BJAB K20 and CHO Lec3 cells was analyzed by periodate oxidation and aniline-catalyzed oxime ligation (PAL; measures total cell surface sialylation) and by binding of fluorescently-labeled lectins (MAA recognizes α2-3-linked sialic acid; SNA recognizes α2-6-linked sialic acid). Fluorescence of individual cells, reporting on cell surface sialylation, was measured by flow cytometry. The mean fluorescence intensity (MFI) of at least 10,000 live cells is shown. BJAB K88 and wt CHO cell lines possessing UDP-GlcNAc 2-epimerase activity were used as positive controls. Experiments were performed in biological triplicate; duplication of the entire experiment yielded similar results. (a) PAL analysis of BJAB cells. (b) PAL analysis of CHO cells. (c) MAA lectin analysis of BJAB cells. (d) MAA lectin analysis of CHO cells. (e) SNA lectin analysis of BJAB cells. SNA binding was not assayed in CHO cells because they lack production of α2-6-linked sialosides.
Next, we used lectin binding to examine whether the incorporation of variant sialic acids was linkage-dependent (Figure 1c-e). α2-3-linked sialosides were detected with the Maackia amurensis lectin (MAA) and α2-6-linked sialosides were detected with the Sambucus nigra lectin (SNA). In human BJAB K20 cells, we detected both α2-3- and α2-6-linked sialosides. Generally, the linkage-specific incorporation trends were similar to the global incorporation trend observed by PAL. However, while a relatively low level of MAA binding was observed in BJAB K20 cells cultured with Ac5ManNGc, the level of SNA binding was high and exceeded the level of SNA binding observed for BJAB K20 cells cultured with Ac4ManNAc. In CHO Lec3 cells, we examined only α2-3-linked sialoside production, since these cells do not produce detectable levels of α2-6-linked sialosides.27 In CHO Lec3 cells, the trends we observed for MAA binding for different ManNAc analogs were similar to the trends for global sialoside production detected by PAL. However, we once again noticed a difference between BJAB K20 and CHO Lec3 cells: our MAA binding data suggested that CHO Lec3 cells produce high levels of NeuGc-containing α2-3-linked sialosides, while these levels are lower in BJAB K20 cells.
We considered the possibility that the observed differences SNA and MAA binding that we observed for BJAB K20 cells cultured with Ac5ManNGc reflect an altered lectin affinity for NeuGc-containing sialosides, but feel that this is unlikely. First, previous reports demonstrated that SNA and MAA lectins do not discriminate between NeuAc and NeuGc.28 Second, CHO Lec3 cells cultured with Ac5ManNGc produce high levels of MAA binding, suggesting that MAA is capable of binding NeuGc-containing glycoconjugates with high affinity. Therefore, we favor the interpretation that BJAB K20 cells produce NeuGc-containing α2-3-linked sialosides less efficiently that CHO Lec3 cells do.
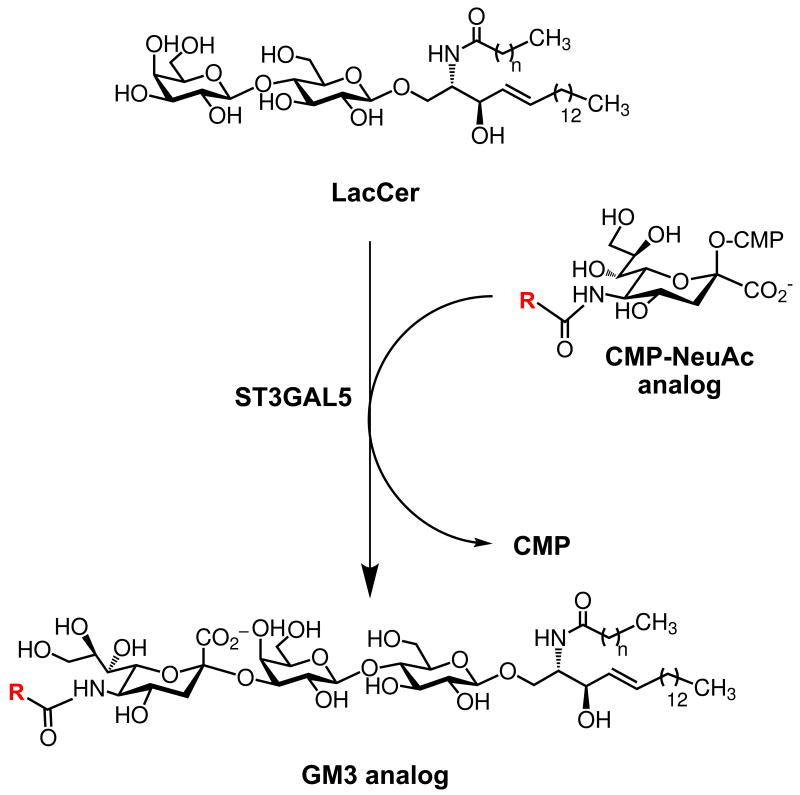
To further investigate the differences in α2-3-linked sialoside production in these two cell lines, we chose to examine production of a single α2-3-linked sialoside produced by a single sialyltransferase. GM3 is a sialylated glycolipid, also known as a ganglioside, produced when the sialyltransferase ST3GAL5 adds sialic acid in an α2-3-linkage to lactose ceramide (LacCer) (Scheme 2).29 Both BJAB K20 and CHO Lec3 cells express ST3GAL5 and, when provided with sialic acid, are capable of producing GM3. Moreover, GM3 is not elaborated to more complex gangliosides at significant levels in either of these cell lines, simplifying the analysis of unnatural sialic acid incorporation levels. Again, we cultured BJAB K20 and CHO Lec3 cells in serum-free conditions with each of the protected ManNAc analogs. Metabolically-labeled cells were harvested and the gangliosides were extracted using established methods.30-32 Gangliosides were resolved by high performance thin layer chromatography (HPTLC) and visualized by resorcinol staining, which specifically detects sialic acid-containing molecules.
Scheme 2. Biosynthesis of GM3 gangliosides.
GM3 is produced by the action of a single sialyltransferase, ST3GAL5, that transfers sialic acid from a CMP-sialic acid to LacCer.
BJAB K20 cells readily produced GM3 analogs containing a variety of modifications to the N-acyl side chain of sialic acid (Figure 2a). Ac4ManNAc, Ac4ManNProp, Ac4ManNBut, and Ac4ManNAz were efficiently metabolized to the corresponding GM3 analogs. Ac4ManNDAz was also metabolized successfully, albeit less efficiently. However, when BJAB K20 cells were cultured with Ac5ManNGc, production of NeuGc-containing GM3 was significantly lower than production of NeuAc-containing GM3. This result is consistent with the MAA binding experiments that suggest that NeuGc is not efficiently incorporated into the α2-3-linkage in this cell line. Only one analog (Ac4ManNPhAc) was not metabolized to a GM3 analog at a level detectable by resorcinol staining. MALDI-TOF-MS analysis (supplementary data) of extracted gangliosides from BJAB K20 cells cultured with Ac4ManNAc, Ac5ManNGc, Ac4ManNAz, and Ac4ManNDAz confirmed the identity of metabolically engineered GM3 molecules.
Figure 2. HPTLC analysis of gangliosides produced by BJAB and CHO cells cultured with protected ManNAc analogs.
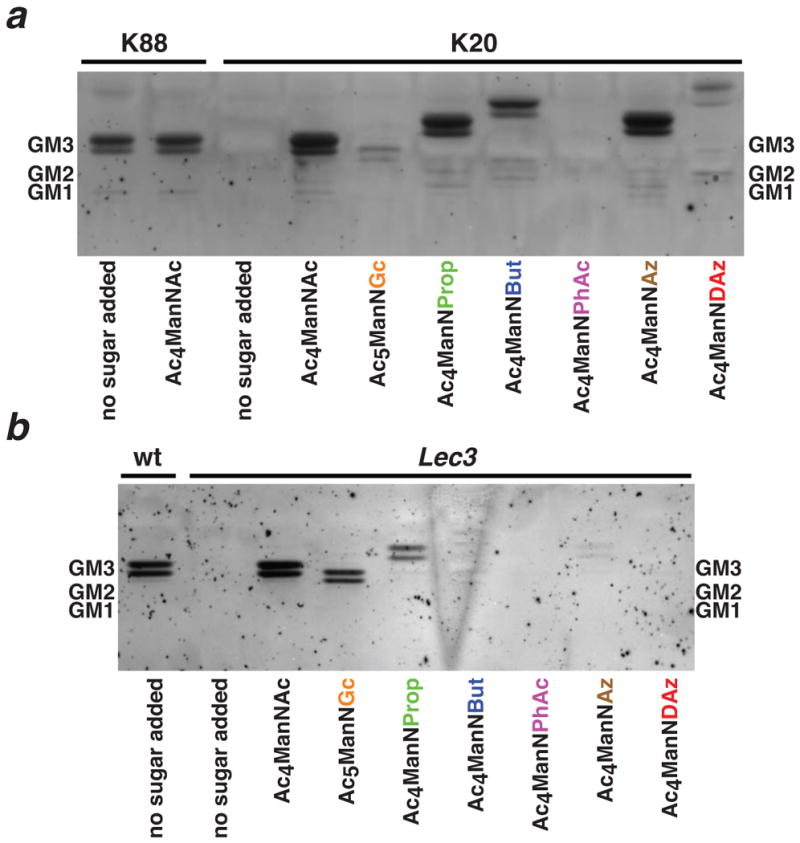
BJAB K20 and CHO Lec3 cells were cultured with protected ManNAc analogs. BJAB K88 and wt CHO cell lines containing UDP-GlcNAc 2-epimerase activity were used as positive controls. Gangliosides were extracted, resolved by HPTLC, and visualized by resorcinol staining. Mobilities of standard gangliosides are indicated on either side of the HPTLC images. Gangliosides appear as doublets, reflecting two different fatty acid lengths that are incorporated into LacCer in these cell lines. (a) BJAB cells generate GM3 and small quantities of GM1a gangliosides. (b) CHO cells produce only GM3 gangliosides. Gangliosides incorporating modified sialic acids have altered mobilities. Incorporation of the modified sialic acids was confirmed by mass spectrometry (supplementary data).
We observed a different pattern of GM3 analog production in CHO Lec3 cells (Figure 2b). CHO Lec3 cells readily metabolized Ac4ManNAc, Ac5ManNGc, and Ac4ManNProp to the corresponding GM3 analogs. Increasing the side chain length (Ac4ManNBut and Ac4ManNAz) led to diminished levels of incorporation and larger N-acyl side chains, found on Ac4ManNPhAc and Ac4ManNDAz, were not tolerated. MALDI-TOF-MS analysis of extracted gangliosides from CHO Lec3 cells cultured with Ac4ManNAc, Ac5ManNGc, and Ac4ManNAz confirmed the identity of metabolically engineered GM3 molecules (supplementary data). In comparison to BJAB K20 cells, CHO Lec3 cells appear to be more efficient at incorporating NeuGc into GM3 and less efficient at incorporating SiaNBut and SiaNAz.
The experiments presented here allowed us to compare the global versus specific sialoside production in two mammalian cell lines. By using cell lines deficient in sialic acid biosynthesis, we specifically tracked production of modified sialosides. Our data reveal sialoside-specific incorporation patterns that depend on the form of sialic acid. For example, BJAB K20 cells seem to discriminate against incorporating NeuGc into α2-3-linked sialosides, including GM3. The low levels of NeuGc incorporation into α2-3-linked sialosides are unlikely to be explained solely by lower levels of CMP-NeuGc production, since the BJAB K20 cells cultured with Ac5ManNGc still show robust levels of SNA binding. Similarly, CHO Lec3 cells produce relatively high levels of cell surface SiaNBut, but are ineffective at producing SiaNBut-containing GM3. These differences between global and specific sialoside production may reflect differences in the activity or expression level of one or more of the enzymes involved in sialoside metabolism. Perhaps certain sialyltransferases discriminate among different CMP-sialic acid analogs, resulting in variant sialic acids being more readily incorporated into some sialosides, but excluded from others. Alternatively, discrimination may occur at the level of sialoside catabolism, with sialidases exhibiting substrate specificity for different forms of sialic acid. In addition, differing rates at which different sialosides are synthesized and degraded may affect our observations.
Our findings also highlight key differences in the utility of different cell lines for performing metabolic oligosaccharide engineering experiments, particularly those aimed at producing modified gangliosides. The BJAB K20 cell line demonstrates the substrate flexibility to introduce larger modifications (such as the N-butanoyl substituent and the diazirine) into gangliosides, but shows attenuated ability to introduce a naturally occurring sialic acid variant (NeuGc). Conversely, the CHO Lec3 cell line exhibits restrictions in substrate scope but, unlike the BJAB cell line, is able to utilize Ac5ManNGc as a precursor for NeuGc-GM3 production. We do not know the molecular basis of the specificity differences between the BJAB K20 and CHO Lec3; these cell lines differ in both their tissue and species of origin. However, we are intrigued by these findings in light of the fact that BJAB K20 cells are derived from humans, an organism that produces NeuAc but not NeuGc, while CHO Lec3 cells are derived from hamsters, an organism that produces both NeuAc and NeuGc.33
Supplementary Material
Acknowledgments
We thank Michael Pawlita (German Cancer Research Center) and James Paulson (The Scripps Research Institute) for sharing BJAB K20 and BJAB K88 cells, Mark Lehrman (UT Southwestern Medical Center) for sharing wt CHO cells, and Pamela Stanley (Albert Einstein College of Medicine) for sharing CHO Lec3 cells. We thank Yan Li (UT Southwestern Medical Center Protein Chemistry Technology Center) for mass spectrometry analysis of sugars, Parastoo Azadi and Roberto Sonon (University of Georgia Complex Carbohydrate Research Center) for mass spectrometry of ganglioside samples, and Angela Mobley (UT Southwestern Medical Center Flow Cytometry Core Facility) for help with flow cytometry. We acknowledge financial support from the NIH (GM090271), the March of Dimes (5-FY06-913), and UT Southwestern Medical Center. This research was supported in part by the National Institutes of Health (NIH/NCRR)-funded grant entitled ‘Integrated Technology Resource for Biomedical Glycomics’ (P41RR018502) to the Complex Carbohydrate Research Center (Athens, GA). J.J.K. is an Alfred P. Sloan Research Fellow.
Footnotes
Supplementary data: Supplementary data associated with this article can be found, in the online version, at doi:XXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Du J, Meledeo MA, Wang Z, Khanna HS, Paruchuri VDP, Yarema KJ. Glycobiology. 2009;19:1382. doi: 10.1093/glycob/cwp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dube DH, Bertozzi CR. Curr Opin Chem Biol. 2003;7:616. doi: 10.1016/j.cbpa.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Kayser H, Zeitler R, Kannicht C, Grunow D, Nuck R, Reutter W. J Biol Chem. 1992;267:16934. [PubMed] [Google Scholar]
- 4.Keppler OT, Stehling P, Herrmann M, Kayser H, Grunow D, Reutter W, Pawlita M. J Biol Chem. 1995;270:1308. doi: 10.1074/jbc.270.3.1308. [DOI] [PubMed] [Google Scholar]
- 5.Mahal LK, Yarema KJ, Bertozzi CR. Science. 1997;276:1125. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 6.Saxon E, Bertozzi CR. Science. 2000;287:2007. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 7.Han S, Collins BE, Bengtson P, Paulson JC. Nat Chem Biol. 2005;1:93. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- 8.Hsu TL, Hanson SR, Kishikawa K, Wang SK, Sawa M, Wong CH. Proc Natl Acad Sci U S A. 2007;104:2614. doi: 10.1073/pnas.0611307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka Y, Kohler JJ. J Am Chem Soc. 2008;130:3278. doi: 10.1021/ja7109772. [DOI] [PubMed] [Google Scholar]
- 10.Herrmann M, von der Lieth CW, Stehling P, Reutter W, Pawlita M. J Virol. 1997;71:5922. doi: 10.1128/jvi.71.8.5922-5931.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt C, Stehling P, Schnitzer J, Reutter W, Horstkorte R. J Biol Chem. 1998;273:19146. doi: 10.1074/jbc.273.30.19146. [DOI] [PubMed] [Google Scholar]
- 12.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. Science. 2008;320:664. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi MA, Samyn-Petit B, Julien S, Delannoy P. Biochimie. 2001;83:727. doi: 10.1016/s0300-9084(01)01301-3. [DOI] [PubMed] [Google Scholar]
- 14.Bussink AP, van Swieten PF, Ghauharali K, Scheij S, van Eijk M, Wennekes T, van der Marel GA, Boot RG, Aerts JM, Overkleeft HS. J Lipid Res. 2007;48:1417. doi: 10.1194/jlr.C700006-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Zou W, Borrelli S, Gilbert M, Liu T, Pon RA, Jennings HJ. J Biol Chem. 2004;279:25390. doi: 10.1074/jbc.M402787200. [DOI] [PubMed] [Google Scholar]
- 16.Pan Y, Chefalo P, Nagy N, Harding C, Guo Z. J Med Chem. 2005;48:875. doi: 10.1021/jm0494422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chefalo P, Pan Y, Nagy N, Guo Z, Harding CV. Biochemistry. 2006;45:3733. doi: 10.1021/bi052161r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Zhang J, Guo Z. Bioorg Med Chem. 2007;15:7561. doi: 10.1016/j.bmc.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahal LK, Charter NW, Angata K, Fukuda M, Koshland DE, Jr, Bertozzi CR. Science. 2001;294:380. doi: 10.1126/science.1062192. [DOI] [PubMed] [Google Scholar]
- 20.Collins BE, Fralich TJ, Itonori S, Ichikawa Y, Schnaar RL. Glycobiology. 2000;10:11. doi: 10.1093/glycob/10.1.11. [DOI] [PubMed] [Google Scholar]
- 21.Bond MR, Whitman CM, Kohler JJ. Mol Biosys. 2010;6:1796. doi: 10.1039/c0mb00069h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones MB, Teng H, Rhee JK, Lahar N, Baskaran G, Yarema KJ. Biotechnol Bioeng. 2004;85:394. doi: 10.1002/bit.10901. [DOI] [PubMed] [Google Scholar]
- 23.Keppler OT, Peter ME, Hinderlich S, Moldenhauer G, Stehling P, Schmitz I, Schwartz-Albiez R, Reutter W, Pawlita M. Glycobiology. 1999;9:557. doi: 10.1093/glycob/9.6.557. [DOI] [PubMed] [Google Scholar]
- 24.Keppler OT, Hinderlich S, Langner J, Schwartz-Albiez R, Reutter W, Pawlita M. Science. 1999;284:1372. doi: 10.1126/science.284.5418.1372. [DOI] [PubMed] [Google Scholar]
- 25.Hong Y, Stanley P. J Biol Chem. 2003;278:53045. doi: 10.1074/jbc.M309967200. [DOI] [PubMed] [Google Scholar]
- 26.Zeng Y, Ramya TN, Dirksen A, Dawson PE, Paulson JC. Nat Methods. 2009;6:207. doi: 10.1038/nmeth.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee EU, Roth J, Paulson JC. J Biol Chem. 1989;264:13848. [PubMed] [Google Scholar]
- 28.Brinkman-Van der Linden ECM, Sonnenburg JL, Varki A. Anal Biochem. 2002;303:98. doi: 10.1006/abio.2001.5539. [DOI] [PubMed] [Google Scholar]
- 29.Ishii A, Ohta M, Watanabe Y, Matsuda K, Ishiyama K, Sakoe K, Nakamura M, Inokuchi J, Sanai Y, Saito M. J Biol Chem. 1998;273:31652. doi: 10.1074/jbc.273.48.31652. [DOI] [PubMed] [Google Scholar]
- 30.Ladisch S, Gillard B. Anal Biochem. 1985;146:220. doi: 10.1016/0003-2697(85)90419-1. [DOI] [PubMed] [Google Scholar]
- 31.Schnaar RL. Methods Enz. 1994;230:348. doi: 10.1016/0076-6879(94)30024-0. [DOI] [PubMed] [Google Scholar]
- 32.Nimrichter L, Burdick MM, Aoki K, Laroy W, Fierro MA, Hudson SA, Von Seggern CE, Cotter RJ, Bochner BS, Tiemeyer M, Konstantopoulos K, Schnaar RL. Blood. 2008;112:3744. doi: 10.1182/blood-2008-04-149641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varki A. Proc Natl Acad Sci USA. 2010;107:8939. doi: 10.1073/pnas.0914634107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.