Abstract
The use of bacteriophages, instead of antibodies, in the ELISA-based detection of bacterial strains was tested. This procedure appeared to be efficient, and specific strains of Salmonella enterica and Escherichia coli could be detected. The sensitivity of the assay was about 105 bacterial cells/well (106/ml), which is comparable with or outperforms other ELISA tests detecting intact bacterial cells without an enrichment step. The specificity of the assay depends on the kind of bacteriophage used. We conclude that the use of bacteriophages in the detection and identification of bacteria by an ELISA-based method can be an alternative to the use of specific antibodies. The advantages of the use of bacteriophages are their environmental abundance (and, thus, a possibility to isolate various phages with different specificities) and the availability of methods for obtaining large amounts of phage lysates, which are simple, rapid, cheap, and easy.
Introduction
Since a large number of infectious diseases are caused by pathogenic bacterial strains, it is obvious that such strains should be specifically detected during diagnostic procedures. Different approaches are used to obtain satisfactory results. The platform used for detection often depends on the kind of molecular target in the pathogen that has been chosen. When nucleic acids are used, a precise detection of bacteria is possible, but the main disadvantage of this method is the requirement for the isolation and purification of target molecules prior to their detection. This may be a time-consuming step, which makes the whole assay relatively long and expensive [1].
Another approach is to recognize antigens on the surface of detected bacteria by using specific antibodies. This approach does not need any time-consuming initial preparation of the sample; however, antibodies are quite costly and cumbersome in preparation. Their performance, especially affinity and avidity, strongly affects the assay sensitivity. Moreover, an additional drawback is their limited shelf life, which limits the viability of ready-to-use antibody-based sensors [2].
Recently, it was demonstrated that antibodies can be replaced with bacteriophages in assays devoted to detecting bacteria [3, 4]. These viruses offer many advantages over standard antibodies in detection procedures, namely, phage proteins engaged in host recognition are extremely stable, phages are easy to isolate and propagate, and they can be stored for a relatively long time. These features make bacteriophages a cheap and fully functional tool that can replace primary antibodies in procedures of immunological detection of bacteria [5, 6].
Bacteriophages have already been widely used for the identification of bacteria in phage typing tests [7–10]. In these assays, one can employ the relatively narrow host range of some phages, which is limited to one or several strains. When multiple phages with partially overlapping host ranges are used, the ability of the tested bacterial strain to support the growth of some of phages from the collection can be considered as a kind of a fingerprint, allowing for easy and inexpensive strain identification. However, classical phage typing is useful to distinguish bacterial strains, but not to detect them in a clinical or environmental sample.
In this work, we aimed to develop an assay for the detection of bacteria using unmodified bacteriophages, which can replace primary antibodies in standard ELISA tests. The main idea of this study was to construct a detection system for the quick identification of Salmonella and Escherichia coli strains with the use of cheap and simple procedures for preparing the reagents and to perform the assay.
Materials and methods
Bacteria and bacteriophages
The bacterial strains and bacteriophages used in this work are listed in Table 1.
Table 1.
Phages and bacterial strains
| Strain | Source/reference |
|---|---|
| Phage T4 CGSC#12143 | E. coli Genetic Stock Center, Yale, USA |
| Phage P1vir | Collection of Dept. Molecular Biology UGa |
| Phage λ | [11] |
| Phage O1 | Collection of Salmonella Microorganismsb |
| Escherichia coli O157:H7 86-24 ∆stx2 | Gail Christie, Department of Microbiology & Immunology, Virginia Commonwealth University |
| Escherichia coli MG1655 CGSC#6300 | E. coli Genetic Stock Center, Yale, USA |
| Escherichia coli B KOS 1466 | Collection of Salmonella Microorganismsb |
| Escherichia coli C1a | [12] |
| Salmonella enterica ser. London KOS 76 | Collection of Salmonella Microorganismsb |
| Salmonella enterica ser. Panama KOS 73 | Collection of Salmonella Microorganismsb |
| Salmonella enterica ser. Heidelberg KOS 16 | Collection of Salmonella Microorganismsb |
| Salmonella enterica ser. Anatum KOS 78 | Collection of Salmonella Microorganismsb |
| Salmonella enterica ser. Tennessee KOS 142 | Collection of Salmonella Microorganismsb |
| Salmonella enterica ser. Reading KOS 19 | Collection of Salmonella Microorganismsb |
| Salmonella enterica ser. Enteritidis KOS 1663 | Collection of Salmonella Microorganismsb |
aUG, University of Gdańsk, Poland
bCollection of National Salmonella Centre at Medical University of Gdańsk, Poland
Antibodies
Anti-E. coli and Salmonella spp. rabbit antibodies were purchased from USBiological (USA). HM serum was obtained from Immunolab (Poland). Anti-O1 serum was prepared in the following way: four rabbits were immunized using a preparation of O1 phage in PBS (1011 pfu/ml). The procedure for immunization was as follows: 0.2 ml injected subcutaneously initially, 0.3 ml injected subcutaneously after one week, and 0.5 ml injected intravenously after another week. At one week after the last immunization, the antibody level was tested and the serum was used for subsequent experiments.
Phage preparation
Phage lysate (2 l) was centrifuged (3,000g, 20 min) and the supernatant was concentrated using 10% polyethylene glycol of molecular weight 8,000 Da (PEG 8000) and 2 M NaCl. After overnight incubation, the phage precipitate was centrifuged (8,000g for 20 min) and then suspended in 10 ml of TM (10 mM Tris-HCl, pH 7.2, 10 mM MgSO4) buffer. The remaining PEG 8000 was extracted by addition of 2 ml of chloroform and vortexing. This step was repeated three times. The resulting phage suspension was purified on a cesium chloride gradient by ultracentrifugation (32,000 rpm for 2 h using a 50 Ti Beckman rotor), and then dialyzed against TM buffer and titrated [13].
Plate coating with phage
Wells of 96-well ELISA plates were coated using 0.1 ml of appropriate phage lysate and incubated overnight at 4°C. The wells were washed three times with PBS buffer (pH 7.4) and blocked for 3 h at 37°C with 5% BSA solution in PBS.
ELISA procedure
Following triple washing of the wells with PBS containing 0.05% Tween 20, bacterial cells in PBS were added. The plates were then incubated for 1 h at room temperature, after which the wells were washed three times with PBS containing 0.05% Tween20. HM serum (Immunolab, Poland), suspended in 5% BSA, was added (1:1,000 v/v) and incubated for 1 h at room temperature. Following triple washing with the PBS buffer containing 0.05% Tween20, secondary biotinylated antibodies were added (1:2,000) and the plates were incubated for another hour at room temperature. Following triple washing with PBS containing 0.05% Tween20, a solution of ExtrAvidin–alkaline phosphatase conjugate (Sigma, USA) (1:500 dilution) was added, and the plates were incubated for 1 h. The wells were washed four times with PBS containing 0.05% Tween20, and 50 μl of Alkaline Phosphatase Yellow (pNPP) Liquid Substrate (Sigma, USA) were added to each well. After yellow color appeared, the reaction was stopped with 15 μl of 3 M NaOH, and absorbance was measured at a 405-nm wavelength.
Results
We aimed to develop an assay for the detection of Salmonella and E. coli strains by using an ELISA-based method, in which bacteriophages are employed as recognition agents. For testing the ability of bacteriophages to immobilize bacterial cells specifically on the ELISA plate, we assessed the performance of phages O1, lambda, P1, and T4 to detect cells of Salmonella enterica ser. Enteritidis KOS 1663, which were inactivated by formaldehyde treatment, according to Paton et al. [14].
The O1 phage is known to be highly specific to salmonellae [15], while phage lambda is known to infect a relatively narrow range of bacteria, including Escherichia coli and Shigella, but not Salmonella [16]. The phage T4 host range is restricted mostly to E. coli and Shigella species [17], and the P1 host range is restricted mostly to Enterobacteriaceae [18]. We expected phage lambda and T4 to be unable to bind Salmonella cells used in this experiment, while the other phages should be able to capture and immobilize the bacterial cells in ELISA plate wells, and, thus, allow for subsequent detection.
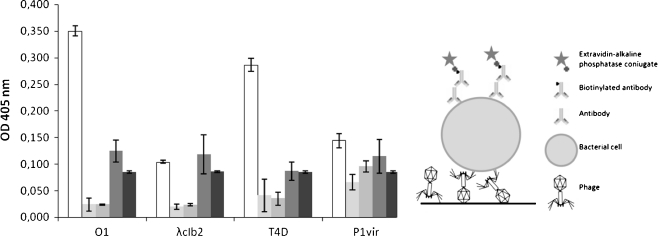
Detection was carried out as follows. Equal amounts of phages (108/well) were adsorbed to the polystyrene surface of ELISA plate wells. The next steps involved the blocking of remaining binding sites by using skimmed milk and subsequent addition of formaldehyde-fixed Salmonella enterica ser. Enteritidis KOS 1663 cells (107/well). Then, primary HM serum was used, and, subsequently, biotinylated anti-rabbit secondary antibodies were added. After incubation with ExtrAvidin–alkaline phosphatase conjugate, the signal was measured. To assess the specificity of the assay, several controls were used, which basically lacked one part of signal-generating complexes: biotynylated secondary antibodies, primary HM serum, phage, or bacteria. When any of these factors were lacking, the signal remained weak, in contrast to the wells in which all components of the assay were present (Fig. 1). Contrary to our expectations based on a literature survey [17], we found that T4 phage binds efficiently to Salmonella enterica ser. Enteritidis cells (Fig. 1). However, it may indicate that some phages can adsorb to bacterial cells without subsequent ability to inject its genetic material or to perform productive infection.
Fig. 1.
Use of phages O1, λ, T4, and P1 as capture agents for Salmonella enterica ser. Enteritidis. In the left panel, the white bars represent signals from wells where complete assay was performed, the pale gray bars represent signals from wells where biotinylated anti-rabbit antibodies were omitted, the gray bars represent signals from wells where HM serum was omitted, the dark gray bars represent signals from wells where no bacteria were added, and the very dark gray bars represent signals from wells where no phage was added. The presented values are average results from four experiments, with error bars representing the standard deviation. The background value from the control employing no phage in detection was subtracted. In the right panel, an illustration showing the assay construction is presented
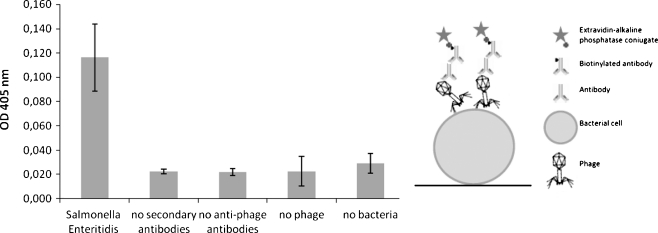
When we attempted to attach bacteria directly to the wells in the plate (107cells/well), and then added a lysate of bacteriophage O1 (108 pfu/well), followed by the addition of anti-O1 serum, we were able to detect the bacteria, whereas the signals of the controls were relatively weak (Fig. 2). To assess the specificity of the assay, we tested the detection of different strains of Salmonella by means of the ELISA test using phages to attach to bacterial cells. The results are summarized in Table 2.
Fig. 2.
Detection of Salmonella enterica ser. Enteritidis directly adsorbed to ELISA plate wells using phage O1 and anti-phage serum. In the left panel, the average signal generated in complete assay and in various controls is shown. The presented values are average results from four experiments, with error bars representing the standard deviation. The background value from the control employing no phage in detection was subtracted. In the right panel, an illustration showing the assay construction is presented
Table 2.
Detection of different Salmonella enterica strains by using indicated phages as capture agents in ELISA
| Phage | Signala | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S. London | S. Panama | S. Heidelberg | S. Anatum | S. Tennessee | S. Reading | No secondary antibodies | No primary antibodies | No bacteria | No phage | |
| O1 | 0.362 ± 0.068 | 0.104 ± 0.071 | 0.258 ± 0.202 | 0.533 ± 0.164 | 0.042 ± 0.020 | 0.083 ± 0.070 | 0.013 ± 0.004 | 0.011 ± 0.010 | 0.011 ± 0.006 | 0.052 ± 0.074 |
| T4D | 0.154 ± 0.057 | 0.036 ± 0.018 | 0.037 ± 0.031 | 0.112 ± 0.102 | 0.050 ± 0.035 | 0.038 ± 0.018 | 0.008 ± 0.014 | 0.014 ± 0.004 | 0.013 ± 0.014 | 0.006 ± 0.012 |
| P1vir | 0.054 ± 0.040 | 0.023 ± 0.017 | 0.034 ± 0.020 | 0.090 ± 0.063 | 0.049 ± 0.014 | 0.041 ± 0.022 | 0.003 ± 0.009 | 0.004 ± 0.009 | 0.016 ± 0.014 | 0.036 ± 0.067 |
| λcIb2 | 0.053 ± 0.028 | 0.026 ± 0.011 | 0.028 ± 0.012 | 0.068 ± 0.048 | 0.065 ± 0.061 | 0.034 ± 0.012 | 0.005 ± 0.017 | 0.004 ± 0.015 | 0.022 ± 0.031 | 0.004 ± 0.020 |
a The presented values are mean results from four experiments ± standard deviation
We found that the signals generated during the detection of various bacterial strains were different. As expected, among bacteriophages used for testing, phage O1 was the most effective in the detection of Salmonella strains. However, some Salmonella enterica strains, e.g., Salmonella enterica ser. Tennessee, generated weak signals, indicating that they were not recognized efficiently by this phage. Interestingly, the highest signals were generated by Salmonella enterica ser. Anatum (Table 2), which is immune to phage O1 when tested by a plaque test (data not shown). This suggests that this bacteriophage can adsorb efficiently on Salmonella enterica ser. Anatum, but is not able to complete its lytic development in this strain. This observation clearly shows that the use of phages may create unexpected positive signals, which could be even beneficiary in some cases. Thus, while constructing a detection test, more rigorous experiments than the sole ability of phage to form plaques should be employed, in order to identify properly the specificity of phage binding.
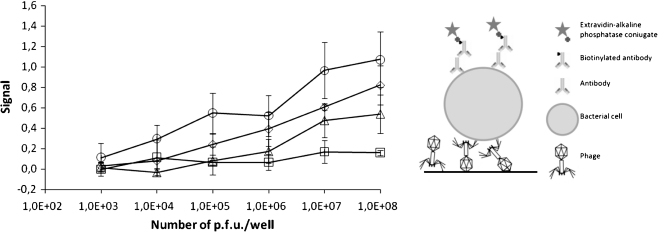
To optimize the method and to determine its sensitivity in respect to the number of phages added to each well, we performed a series of tests with various amounts of phage lysates. The results are summarized in Fig. 3. Generally, the results showed that the highest signals were obtained at the highest phage densities. Moreover, they showed that it is possible to detect bacterial strains when as few as 104 phage particles are deposited in a single well.
Fig. 3.
Detection of Salmonella enterica ser. Enteritidis (107/well) using different numbers of phage particles per well as a capture agent. In the left panel, the signal strength in relation to phage number used is presented for phages O1 (circles), T4 (diamonds), P1 (triangles), and λ (squares). The presented values are mean results of four experiments, with error bars representing the standard deviation. The background value from the control employing no phage in detection was subtracted. In the right panel, an illustration showing the assay construction is presented
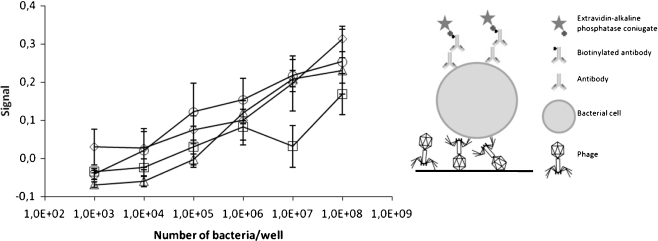
To assess the sensitivity of the detection of bacteria, we performed the assay using the optimal number of phages, 108 pfu/well, and different numbers of bacterial cells (Fig. 4). In this assay, 105 bacterial cells were detected unambiguously when phages T4 or O1 were employed.
Fig. 4.
Detection of different numbers of Salmonella enterica ser. Enteritidis cells using phage particles as a capture agent. In the left panel, the signal strength in relation to phage number used is presented for phages O1 (circles), T4 (diamonds), P1 (triangles), and λ (squares). The presented values are mean results of four experiments, with error bars representing the standard deviation. The background value from the control employing no phage in detection was subtracted. In the right panel, an illustration showing the assay construction is presented
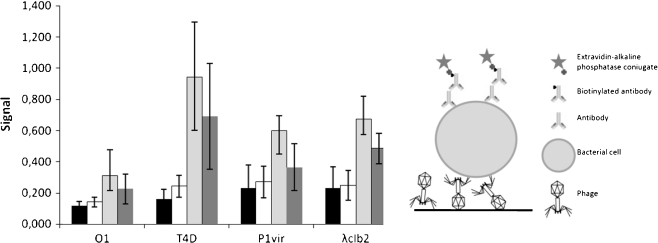
We employed the same bacteriophages to detect different strains of E. coli. First, we tested the efficiency of plating (e.o.p.) of purified phages (Table 3). The differences in e.o.p. values apparently resulted from differences in bacteriophage development in the strains used. Phage O1 formed no plaques on E. coli strains employed in this assay. This phage is known to form plaques on very few E. coli strains and is reported to be specific to the genus Salmonella [15]. One strain, E. coli O157:H7 86-24, appeared to be insensitive to all four phages. However, in ELISA tests, this strain generated the strongest signals (Fig. 5), which indicated efficient phage adsorption, and, thus, immobilization of the bacteria in the wells.
Table 3.
Results of titration of phage lysates on bacterial strains used in this study
| Phage | pfu/ml determined on Escherichia coli strainsa | |||
|---|---|---|---|---|
| E. coli B | E. coli C1a | E. coli 8624 | E. coli MG1655 | |
| O1 | <101 | <101 | <101 | <101 |
| λcIb2 | 4.0 × 105 | 1.2 × 1010 | <101 | 8.0 × 1010 |
| T4D | 1.2 × 10c11 | 2.4 × 109 | <101 | 2.0 × 1011 |
| P1vir | 4.0 × 106 | 8.0 × 109 | <101 | 1.2 × 108 |
apfu, plaque forming units
Fig. 5.
Detection of different E. coli strains (B: black bars, C1a: white bars, 86-24: pale gray bars, MG1655: dark gray bars) using different phage strains. The presented values are mean results of four experiments, with error bars representing the standard deviation. The background value from the control employing no phage in detection was subtracted. In the right panel, an illustration showing the assay construction is presented
Discussion
The results presented in this report suggest that the concept of using bacteriophages instead of primary or secondary antibodies in ELISA for the specific detection of bacterial strains may be an effective alternative to classical immuno-detection procedures. The availability of easy methods of phage isolation from natural sources and the relatively low number of bacteriophages necessary for the generation of unambiguous signals indicate that it is reasonable to use phages in this kind of assay. The sensitivity of the assay was also relatively high; nevertheless, we believe that it may be further improved when more advanced platforms are used for signal detection.
On the other hand, a proper selection of bacteriophage strains is not possible without detailed tests of their host range. Our results showed that the ability of generating plaques is not always a sufficient feature to choose a proper phage as a recognition agent, and, also, it is not sufficient to clearly define the range of host strains on which the phage may adsorb.
The recognition of bacterial cells by bacteriophage particles is based on the presence of phage receptors on the cell surface. As almost every structure present on the bacterial cell surface can be a potential receptor [19], it is possible to isolate phages varying in the range of bacterial strains which can be recognized. Owing to such a feature of the phage host recognition, it may be possible to construct assays with very generic or very specific recognition patterns, or tests that combine both types of recognition. This might be important if the bacterial phenotype is tightly connected to antigenic specificity, like Shiga toxin production and the O157:H7 serotype of E. coli [20]. This approach would be similar to the phage typing of bacteria but would differ in that typing by a plaque test would consume much more time than identification by the ELISA test. Moreover, while the plaque test is strongly affected by different ways of phage exclusion, the ELISA assay is affected only by the presence or absence of a phage receptor on the bacterial surface.
The sensitivity of the test described in this report was estimated to be 106 cells/ml. It is worth mentioning that the sensitivity of other ELISA tests detecting intact cells show similar or lower sensitivities [21]. As the use of bacteriophages instead of antibodies did not decrease the sensitivity of the assay, and the selection and production of bacteriophages is relatively easy, cheap, and does not involve any work done on live animals, we consider it to be an attractive alternative to traditionally used antibodies. One issue which should be considered in laboratories preparing the assays, is that bacteriophages may be produced and disseminated accidentally and cause false test results owing to phage contamination [22]. However, it is easy to inactivate bacteriophages using UV light prior to the coating of ELISA plates or after coating. When proper coating conditions are used, phage re-activation due to multiple infection can be easily avoided. This assumption is strongly supported by the fact that a relatively low number of phage particles deposited in a single well was enough to provide good detection sensitivity. Such conditions, due to the relatively low density of phages on the surface, minimize the possibility of high multiplicity of infection during bacterial capture. Thus, the use of phages as a recognition agent in the ELISA test may be a safe, sensitive, and easy alternative to standard antibody-based tests.
Acknowledgments
This work was supported by the Ministry of Science and Higher Education (Poland) and the European Union within the European Regional Development Fund, through grant Innovative Economy (POIG.01.01.02-00-008/08).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Horz H-P, Scheer S, Huenger F, Vianna ME, Conrads G. Selective isolation of bacterial DNA from human clinical specimens. J Microbiol Methods. 2008;72:98–102. doi: 10.1016/j.mimet.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Lv LL, Liu BC, Zhang CX, Tang ZM, Zhang L, Lu ZH. Construction of an antibody microarray based on agarose-coated slides. Electrophoresis. 2007;28:406–413. doi: 10.1002/elps.200600310. [DOI] [PubMed] [Google Scholar]
- 3.Balasubramanian S, Sorokulova IB, Vodyanoy VJ, Simonian AL. Lytic phage as a specific and selective probe for detection of Staphylococcus aureus—a surface plasmon resonance spectroscopic study. Biosens Bioelectron. 2007;22:948–955. doi: 10.1016/j.bios.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Shabani A, Zourob M, Allain B, Marquette CA, Lawrence MF, Mandeville R. Bacteriophage-modified microarrays for the direct impedimetric detection of bacteria. Anal Chem. 2008;80:9475–9482. doi: 10.1021/ac801607w. [DOI] [PubMed] [Google Scholar]
- 5.Jepson CD, March JB. Bacteriophage lambda is a highly stable DNA vaccine delivery vehicle. Vaccine. 2004;22:2413–2419. doi: 10.1016/j.vaccine.2003.11.065. [DOI] [PubMed] [Google Scholar]
- 6.Rabsch W. Salmonella typhimurium phage typing for pathogens. Methods Mol Biol. 2007;394:177–211. doi: 10.1007/978-1-59745-512-1_10. [DOI] [PubMed] [Google Scholar]
- 7.Głośnicka R, Dera-Tomaszewska B. Comparison of two Salmonella enteritidis phage typing schemes. Eur J Epidemiol. 1999;15:395–401. [PubMed] [Google Scholar]
- 8.Gourmelon M, Caprais MP, Ségura R, Le Mennec C, Lozach S, Piriou JY, Rincé A. Evaluation of two library-independent microbial source tracking methods to identify sources of fecal contamination in French estuaries. Appl Environ Microbiol. 2007;73:4857–4866. doi: 10.1128/AEM.03003-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehndiratta PL, Bhalla P, Ahmed A, Sharma YD. Molecular typing of methicillin-resistant Staphylococcus aureus strains by PCR-RFLP of SPA gene: a reference laboratory perspective. Indian J Med Microbiol. 2009;27:116–122. doi: 10.4103/0255-0857.45363. [DOI] [PubMed] [Google Scholar]
- 10.Kurlenda J, Grinholc M, Krzysztoń-Russjan J, Wiśniewska K. Epidemiological investigation of nosocomial outbreak of staphylococcal skin diseases in neonatal ward. Antonie Van Leeuwenhoek. 2009;95:387–394. doi: 10.1007/s10482-009-9318-7. [DOI] [PubMed] [Google Scholar]
- 11.Hendrix RW, Duda RL. Bacteriophage lambda PaPa: not the mother of all lambda phages. Science. 1992;258:1145–1148. doi: 10.1126/science.1439823. [DOI] [PubMed] [Google Scholar]
- 12.Sasaki I, Bertani G. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J Gen Microbiol. 1965;40:365–376. doi: 10.1099/00221287-40-3-365. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsh EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 14.Paton JC, Rogers TJ, Morona R, Paton AW. Oral administration of formaldehyde-killed recombinant bacteria expressing a mimic of the Shiga toxin receptor protects mice from fatal challenge with Shiga-toxigenic Escherichia coli. Infect Immun. 2001;69:1389–1393. doi: 10.1128/IAI.69.3.1389-1393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welkos S, Schreiber M, Baer H. Identification of Salmonella with the O-1 bacteriophage. Appl Microbiol. 1974;28:618–622. doi: 10.1128/am.28.4.618-622.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz M, Le Minor L. Occurrence of the bacteriophage lambda receptor in some enterobacteriaceae. J Virol. 1975;15:679–685. doi: 10.1128/jvi.15.4.679-685.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tétart F, Repoila F, Monod C, Krisch HM. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J Mol Biol. 1996;258:726–731. doi: 10.1006/jmbi.1996.0281. [DOI] [PubMed] [Google Scholar]
- 18.Murooka Y, Harada T. Expansion of the host range of coliphage P1 and gene transfer from enteric bacteria to other gram-negative bacteria. Appl Environ Microbiol. 1979;38:754–757. doi: 10.1128/aem.38.4.754-757.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Letellier L, Boulanger P, Plançon L, Jacquot P, Santamaria M. Main features on tailed phage, host recognition and DNA uptake. Front Biosci. 2004;9:1228–1239. doi: 10.2741/1333. [DOI] [PubMed] [Google Scholar]
- 20.Gould LH, Bopp C, Strockbine N, Atkinson R, Baselski V, Body B, Carey R, Crandall C, Hurd S, Kaplan R, Neill M, Shea S, Somsel P, Tobin-D’Angelo M, Griffin PM, Gerner-Smidt P, Centers for Disease Control and Prevention (CDC) Recommendations for diagnosis of shiga toxin-producing Escherichia coli infections by clinical laboratories. MMWR Recomm Rep. 2009;58:1–14. [PubMed] [Google Scholar]
- 21.Kim JW, Jin LZ, Cho S-H, Marquardt RR, Frohlich AA, Baidoo SK. Use of chicken egg-yolk antibodies against K88+ fimbrial antigen for quantitative analysis of enterotoxigenic Escherichia coli (ETEC) K88+ by a sandwich ELISA. J Sci Food Agric. 1999;79:1513–1518. doi: 10.1002/(SICI)1097-0010(199908)79:11<1513::AID-JSFA396>3.0.CO;2-0. [DOI] [Google Scholar]
- 22.Muniesa M, Blanch AR, Lucena F, Jofre J. Bacteriophages may bias outcome of bacterial enrichment cultures. Appl Environ Microbiol. 2005;71:4269–4275. doi: 10.1128/AEM.71.8.4269-4275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]