Abstract
Proteomic studies have identified a plethora of lysine acetylated proteins in eukaryotes and bacteria. Determining the individual lysine acetyltransferases responsible for each protein acetylation mark is crucial for elucidating the underlying regulatory mechanisms, but has been challenging due to limited biochemical methods. Here, we describe the application of a bioorthogonal chemical proteomics method to profile and identify substrates of individual lysine acetyltransferases. Addition of 4-pentynoyl-coenzyme A, an alkynyl chemical reporter for protein acetylation, to cell extracts, together with purified lysine acetyltransferase p300, enabled the fluorescent profiling and identification of protein substrates via Cu(I)-catalyzed alkyne-azide cycloaddition. We identified several known protein substrates of the acetyltransferase p300 as well as the lysine residues that were modified. Interestingly, several new candidate p300 substrates and their sites of acetylation were also discovered using this approach. Our results demonstrate that bioorthogonal chemical proteomics allows the rapid substrate identification of individual protein acetyltransferases in vitro.
Protein acetylation is a ubiquitous, highly regulated, and reversible posttranslational modification, with important functions in transcriptional and epigenetic regulation.1,2 In addition to the prominent roles of acetylation on histones and histone-associated proteins, protein acetylation also regulates the activity of many cytosolic proteins, involved in different biological activities.3 Lysine acetylation, the most prevalent form of protein acetylation, is catalyzed by lysine acetyltransferases (KATs), which transfer the acetyl group from acetyl-coenzyme A (acetyl-CoA) onto the primary ε-amine of lysine residues. The opposing reaction, the removal of the acetyl group, is mediated by lysine deacetylases (KDACs). Various biochemical and analytical methods have allowed the identification of thousands of acetylated proteins in eukaryotes and more recently also in various prokaryotes.3–5 However, the underlying enzyme-substrate pairs for most of these acetylated proteins are still unknown. To fully understand the importance of these posttranslational modifications and their embedded roles in cellular signaling networks, as well as their particular roles in different diseases, it is crucial to connect individual acetyltransferases with their cognate substrates and sites of modification.
We recently developed bioorthogonal chemical reporters for the detection and identification of acetylated proteins, based on alkynyl-acetate and alkynyl-acetyl-CoA.6 Introduction of alkynyl chemical reporter groups, either by metabolic labeling or in vitro, allows the subsequent bioorthogonal chemical tagging of alkyne-modified proteins with fluorophores or affinity reagents via Cu(I)-catalyzed alkyne-azide cycloaddition (CuAAC).6 In particular, 4-pentynoyl-CoA proved to be an efficient chemical reporter to monitor lysine acetylation on histones and histone peptides catalyzed by the KAT p300 in vitro.6 In this study, we evaluated the utility of 4-pentynoyl-CoA for the identification of KAT p300 substrates from in vitro labeled cell extracts by bioorthogonal chemical proteomics.
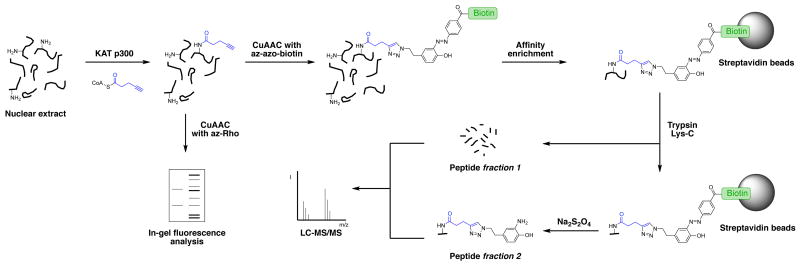
The mammalian KAT p300 exerts transcriptional regulatory function through interactions with the basal transcriptional RNA polymerase complex and different transcription factors.7 Furthermore, p300 acetylates various lysine residues on different histone proteins and non-histone substrates, such as the tumor suppressor protein p53.8 Mutations and aberrations in the p300 gene are associated with congenital diseases and different acquired pathologies.9 Some of these mutations abrogate the acetyltransferase activity of p300, indicating the importance of the downstream acetylation marks for proper cellular function.7 The full complement of p300 substrates in different cellular contexts is still unknown. The combination of 4-pentynoyl-CoA with recombinant p300, added to nuclear cellular fractions (Scheme 1), should allow fluorescent detection of modified substrates as well as their identification by affinity enrichment and mass spectrometry (MS).
Scheme 1.
Labeling of p300 substrates with 4-pentynoyl-CoA chemical reporter in nuclear extracts. The lysine acetyltransferase (KAT) p300 is added to one sample of nuclear extract and omitted in the negative control sample. KAT p300 transfers the 4-pentynoyl reporter group from 4-pentynoyl-CoA onto lysine target residues on its cognate protein substrates. Cu(I)-catalyzed alkyne-azide cycloaddion (CuAAC) is subsequently used to tag all labeled proteins with either azido-rhodamine (az-Rho) for in-gel fluorescence analysis, or azido-azo-biotin (az-azo-biotin) for mass spectrometry-based protein identification. The biotin moiety allows affinity enrichment of 4-pentynoyl-CoA labeled proteins on streptavidin agarose beads. Captured proteins are digested with trypsin and Lys-C on beads and the resulting peptide fraction 1 is collected for LC-MS/MS analysis. The remaining peptides, still bound to the affinity beads, contain the sites of modification. These peptides (fraction 2) are then eluted with sodium dithionite (Na2S2O4), yielding the characteristic product of the azo-bond reduction, which can be identified by LC-MS/MS analysis.
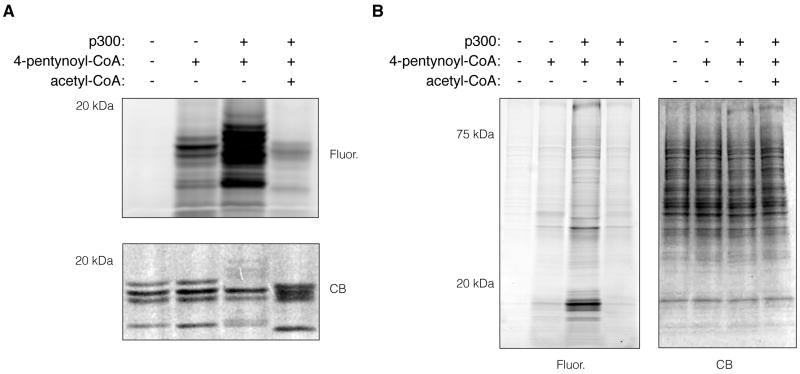
Initially, we set out to demonstrate modification of extracted core histones with the 4-pentynoyl-CoA reporter by p300 in vitro. p300 KAT has been shown to modify lysine residues on the N-terminal tails of histones H2B, H3, and H4 in vitro.7 Extracted core histones from HeLa cells were incubated with 50 μM 4-pentynoyl-CoA and 125 ng p300 for 2 h at 30 °C. The reaction products were then subjected to CuAAC with azido-rhodamine (az-Rho)10 and analyzed by in-gel fluorescence scanning. Concurrent addition of p300 and 4-pentynoyl-CoA resulted in a selective fluorescence signal in the molecular weight region of core histones (Fig. 1a). Even though we could observe limited intrinsic chemical reactivity of 4-pentynoyl-CoA, the addition of p300 resulted in a significant increase in fluorescence labeling of core histones. Pre-incubation of core histones with acetyl-CoA in the presence of p300 and subsequent addition of 4-pentynoyl-CoA greatly diminished the signal intensity (Fig. 1a). These results further demonstrate that 4-pentynoyl-CoA can be used in combination with purified KATs, such as p300, to specifically visualize protein acetylation in vitro.
Figure 1.
In-gel fluorescence analysis of p300-specific labeling of extracted core histones and nuclear extractes with 4-pentynoyl-CoA. A) In-gel fluorescence analysis of p300-catalyzed 4-pentynoylated core histone proteins. p300-catalyzed acylation reactions were carried out with 5 μg extracted core histones, 50 μM 4-pentynoyl-CoA (lanes 2-4) and 125 ng p300 (lanes 3-4) in 50 mM, pH 7.9 Tris buffer (+ 10% glycerol) for 2 h incubation time at 30 °C. In lane 4, 500 μM acetyl-CoA was pre-incubated with core histones in the presence of p300 for 10 min followed by the addition of 50 μM 4-pentynoyl-CoA and co-incubation for additional 2 h. The crude reaction products were subjected to CuAAC with az-Rho for 1 h followed by separation on SDS-PAGE and in-gel fluorescence scanning. B) In-gel fluorescence analysis of the p300-catalyzed 4-pentynoylated proteins in HeLa cell nuclear extract. p300-catalyzed acylation reactions were carried out with 10 μg HeLa cell nuclear extract, 50 μM 4-pentynoyl-CoA (lanes 2-4) and 250 ng p300 (lanes 3-4) in 50 mM, pH 7.9 Tris buffer (+ 10% glycerol) for 2 h incubation time at 30 °C. In lane 4, 500 μM acetyl-CoA was pre-incubated with nuclear extract in the presence of p300 for 10 min followed by the addition of 50 μM 4-pentynoyl-CoA and co-incubation for additional 2 h. The crude reaction products were subjected to CuAAC with az-Rho for 1 h followed by separation on SDS-PAGE and in-gel fluorescence scanning. Lanes are counted consecutively from left to right. Coomassie blue (CB), Fluorescence (Fluor.).
Due to the nuclear localization of most known p300 substrates, we decided to use total nuclear extracts from HeLa cells for the identification of new candidate p300 substrates. Nuclear extracts were prepared as previously described and incubated with 50 μM 4-pentynoyl-CoA and 250 ng p300 for 2 h at 30 °C.11 In-gel fluorescence profiling revealed that addition of the KAT p300 in the presence of 4-pentynoyl-CoA labels discrete protein sets compared to the negative control, which did not contain p300 (Fig. 1b). Intense protein labeling in the low molecular weight region is most likely due to modification of histone proteins. Interestingly, distinctly labeled proteins or sets of proteins could be observed throughout the entire molecular weight continuum, indicating the presence of many non-histone p300 substrates (Fig. 1b). Pre-incubation of cellular extracts with acetyl-CoA decreased the signal intensity to background levels (Fig. 1b), demonstrating the acetylation specificity of the chemical reporter and CuAAC-mediated fluorescence profiling.
To identify the proteins in nuclear extracts specifically modified by p300 we employed a bioorthogonal chemical proteomics protocol. In addition to fluorescence profiling, CuAAC also allows the modification of labeled proteins with Na2S2O4-cleavable azobenzene-biotin tags (azido-azo-biotin) for affinity enrichment with streptavidin beads and selective protein elution for MS-based protein identification.12 We recently described a facile protocol based on this strategy, which includes on-bead digestion and iterative recovery of two distinct peptide fractions for downstream MS analysis (Scheme 1).12 This protocol allows the identification of captured proteins through the recovery of peptides generated by on-bead digestion and the identification of the individual sites of modification by selective elution of the remaining peptides with Na2S2O4. We applied this protocol to identify possible nuclear targets of p300 and their individual sites of modification. Optimized reaction conditions for the analysis of HeLa cell nuclear extracts were established (Suppl. Fig. 1) and applied to the final proteomic experiment using 100 μM 4-pentynoyl-CoA, 125 ng p300 per 10 μg of cell lysate and 2 h labeling at 30 °C. Small fractions of the negative control and the experimental sample were reacted with az-Rho to analyze the labeling efficiency by in-gel fluorescence (Suppl. Fig. 2). The majority of the crude reaction products were reacted with azido-azo-biotin and subsequently precipitated to remove excess biotin tag. The protein samples were then solubilized, reduced, alkylated and loaded onto streptavidin beads for affinity capture. After extensive washing, specifically bound proteins were digested on beads with the endoproteases Lys-C and trypsin. The resulting peptide fractions (fraction 1) were collected for LC-MS/MS analysis. Peptides still bound to the beads, theoretically containing the modified lysine residues, were subsequently eluted with Na2S2O4 and also collected for LC-MS/MS analysis (fraction 2).
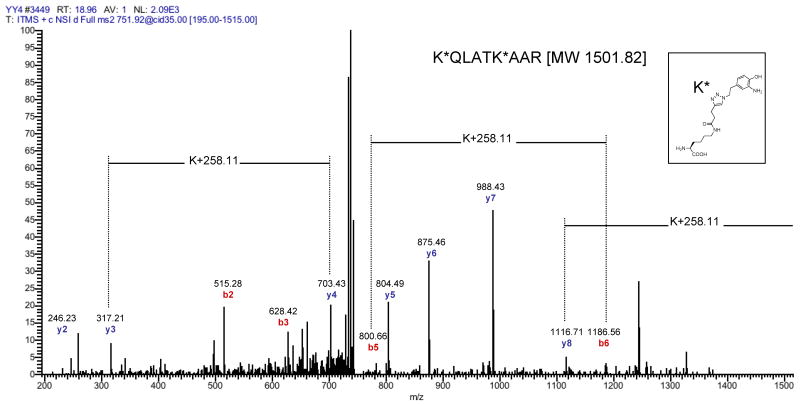
The combined proteomic analysis of fraction 1 and fraction 2 afforded the identification of 23 proteins (Suppl. Table 1c) that satisfied our filter criteria, of which 15 are known acetylated proteins. Furthermore, 3 proteins (nucleophosmin, histone H4, and histone H3.2) identified in this data set, are known substrates of p300. Examined separately, fraction 1 yielded 17 protein identifications (Suppl. Table 1a), whereas 11 proteins were identified in fraction 2 (Suppl. Table 1b). This difference is to be expected, since fraction 2 contains the minor subset of informative peptides per protein, due to the preceding recovery of fraction 1. Four proteins were identified in both peptide fractions. While detection of a protein in fraction 1 is already a strong indicator for a covalent modification of this protein by p300, due to the affinity enrichment process, direct evidence for the covalent modification with the reporter group is preferable. Interestingly, all accepted peptides in fraction 2 contained the characteristic mass of the covalent lysine modification resulting from the chemical reporter. Table 1 displays all peptides and corresponding proteins whose MS/MS spectra contained at least one informative differential b- or y-ion pair indicative of the 4-pentynoylated lysine residue. The site of modification can be assessed by MS/MS analysis, due to a characteristic mass shift of 258.11 Da, resulting from the mass added by the chemical reporter and the triazol-5-ethyl-2-aminophenol moiety of the cleaved azido-azo-biotin tag (Fig. 2). Collectively, our results demonstrate that the acetylation reporter 4-pentynoyl-CoA in combination with purified p300 enables rapid fluorescence profiling and identification of lysine acetylated proteins from cell extracts.
Table 1.
Peptides identified in fraction 2 with at least one diagnostic differential b- or y-ion pair for the 4-pentynoylated lysine residue K*. Number (#) of total spectra equals sum of all acquired spectra of all peptides containing a modified lysine K* (see Fig. 2) for a certain protein. −p300, sample without p300; + p300, sample with p300.
| Protein Name | # of Total Spectra | Peptides with 4-pentynoylation site K* | |
|---|---|---|---|
| - p300 | + p300 | ||
| Nucleolin | 0 | 23 | GATPGK*ALVATPGK TVTPAK*AVTTPGK |
| Histone H3.2 | 0 | 14 | K*QLATK K*QLATK*AAR |
| Isoform 2 of Protein SET | 0 | 14 | K*ELNSNHDGADETSEK |
| Activated RNA polymerase II transcriptional coactivator p15 | 0 | 13 | ALSSSK*QSSSR |
| Isoform 1 of DNA polymerase zeta catalytic subunit | 0 | 4 | ISLPHPMEIGESLDGTLK* RK*VNYETEDSESSFVTHNSK |
| Isoform Long of Antigen KI-67 | 0 | 3 | ISLGK*VGVK LTPSAGK*AMHTPK |
| Isoform 1 of Transcription factor BTF3 | 0 | 1 | IGGK*GTAR |
Figure 2.
MS/MS analysis of a H3.2 peptide identified in fraction 2 containing two lysine residues modified by p300. The identified H3.2 peptide has the sequence KQLATKAAR. Both lysines carried the indicated covalent modification (K*), resulting from the modification with the 4-pentynoyl reporter group, the subsequent formation of the triazol-azobenzene-biotin product by CuAAC, and the final reduction of the azo-bond by Na2S2O4. The resulting species introduces a mass shift of 258.11 Da relative to the unmodified lysine.
The identification of specific protein substrates of individual enzymes is still a major challenge in elucidating posttranslational mechanisms of cellular regulation. For protein acetylation studies, our preliminary studies demonstrate that bioorthogonal chemical reporters can be used with purified enzymes to profile and identify enzyme specific substrates as well as their sites of modification. The bioorthogonal chemical proteomics strategy described here provides a complementary approach to allele-specific chemical reporters (bump-and-hole) or protein array profiling for substrate identification.13,14 We envision this bioorthogonal chemical approach could be applied to other KATs or enzyme-families and combined with quantitative mass spectrometry methods for large-scale substrate profiling and identification studies.
Supplementary Material
Acknowledgments
We congratulate Professor Carolyn Bertozzi on the 2011 Tetrahedron Young Investigator Award in Bioorganic and Medicinal Chemistry. We thank Paul Dossa for providing az-Rho reagent, Dr. Janet Ascano, Dr. Sohail Malik and Professor Robert Roeder for recombinant p300 and the Rockefeller Proteomics Resource Center for mass spectrometry analysis. This work was funded by grants from Irma T. Hirschl/Monique Weill-Caulier Trust, Ellison Medical Foundation and NIH/NIGMS (1R01GM087544).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Allfrey VG, Faulkner R, Mirsky AE. Proc Natl Acad Sci U S A. 1964;51:786. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kouzarides T. Cell. 2007;128:693. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Science. 2009;325:834. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y, Hang HC. Chembiochem. 2011;12:314. doi: 10.1002/cbic.201000558. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning Z, Zeng R, Xiong Y, Guan K, Zhao S, Zhao G. Science. 2010;327:1004. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y, Ascano JM, Hang HC. J Am Chem Soc. 2010;132:3640. doi: 10.1021/ja908871t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalkhoven E. Biochem Pharmacol. 2004;68:1145. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 8.An W, Kim J, Roeder RG. Cell. 2004;117:735. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Giles RH, Peters DJ, Breuning MH. Trends Genet. 1998;14:178. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 10.Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. J Am Chem Soc. 2009;131:4967. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 11.Dignam JD, Lebovitz RM, Roeder RG. Nucleic Acids Res. 1983;11:1475. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y, Grammel M, Raghavan AS, Charron G, Hang HC. Chem Biol. 2010;17:1212. doi: 10.1016/j.chembiol.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Nature. 2003;425:859. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 14.Lin Y, Lu J, Zhang J, Walter W, Dang W, Wan J, Tao S, Qian J, Zhao Y, Boeke JD, Berger SL, Zhu H. Cell. 2009;136:1073. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.