Abstract
Programmed Cell Death 4 (PDCD4) has been described as a tumor suppressor, with high expression correlating with better outcomes in a number of cancer types. Yet a substantial number of cancer patients with high PDCD4 in tumors have poor survival, suggesting that oncogenic pathways may inhibit or change PDCD4 function. Here, we explore the significance of PDCD4 in breast cancer and identify Protein Arginine Methyltransferase 5 (PRMT5) as a cofactor that radically alters PDCD4 function. Specifically, we find that co-expression of PDCD4 and PRMT5 in an orthotopic model of breast cancer causes accelerated tumor growth and that this growth phenotype is dependent on both the catalytic activity of PRMT5 and a site of methylation within the N-terminal region of PDCD4. In agreement with the xenograft model, elevated PDCD4 expression was found to correlate with worse outcome within the cohort of breast cancer patients whose tumors contain higher levels of PRMT5. These results reveal a new cofactor for PDCD4 that alters its tumor suppressor functions and point to the utility of PDCD4/PRMT5 status as both a prognostic biomarker and a potential target for chemotherapy.
Keywords: Breast Cancer, PDCD4, PRMT5, Arginine Methylation, Prognostic Biomarkers
Introduction
Although an active area of research where significant advances have been achieved, breast cancer is a leading cause of mortality among women (1). Breast cancer is a heterogeneous disease that can be difficult to stratify into distinct categories with precise outcomes, especially as improved detection methods allow for diagnosis of early subclinical disease (2). The dilemma of unknown/unquantifiable risk assessments can lead to over-treatment, accompanied by unwanted side effects, or under-treatment, which risks increased recurrence (3). Increasingly, biomarkers, such as hormone receptors, are used to enhance outcome predictions and refine treatment plans. Expanding the arsenal of prognostic biomarkers will help reduce guesswork in treatment plans, leading to better overall survival and improved quality of life.
Programmed Cell Death 4 (PDCD4) is a promising biomarker that correlates with better outcomes in lung, colon, ovarian and esophageal cancer (4–7). PDCD4 is expressed at lower levels in invasive breast carcinoma compared with ductal carcinoma in situ or normal samples, indicating there may be loss of PDCD4 during disease progression (8). Experimental models substantiate a tumor suppressor role for PDCD4. In a mouse epithelial cell line, PDCD4 expression reduced phorbol ester-induced transformation (9) and, in transformed cells, PDCD4 expression suppressed anchorage-independent cell growth (10). Transgenic mice expressing epidermal PDCD4 were resistant to chemically induced skin tumors (11), while PDCD4 knockout mice developed B-cell lymphoma (12).
Here, we find that PDCD4 expression in breast cancer correlates with better survival. Yet, as with many biomarkers, there are limitations in using PDCD4 to predict outcome: subsets of patients whose tumors contain elevated PDCD4 mRNA still experienced poor clinical outcome, indicating the presence of mechanisms that abrogate or change PDCD4 function. Pursuing such a mechanism, we have found a novel PDCD4-interacting partner, PRMT5, which post-translationally methylates PDCD4. Together, these proteins cause a pro-growth tumor phenotype in an orthotopic breast cancer model. Moreover, we have found that PRMT5 levels are significant in determining long term survival in PDCD4-upregulated breast cancer.
Materials and Methods
Constructs
Open reading frames for human PDCD4, PRMT5 and the PDCD4 truncations were cloned into pDONR221. PRMT5cd was created by mutation of amino acids glycine 367 and arginine 368 to alanines (13). PDCD4, PDCD4 point mutants and truncations were cloned into pGEX-4T. PRMT5 and PRMT5cd were cloned into the pcDNA3.1 nV5/DEST (Invitrogen). PDCD4 and PDCD4mm were cloned into the pMIG vector (Addgene plasmid 9044, William Hahn); PRMT5 and PRMT5cd were cloned into pMSCVpuro (Clontech) vector.
Recombinant protein production
BL21/RIL cells (Novagen) transduced with PDCD4-pGEX4T or parental vector were induced using 0.1mM IPTG for 1–3 hrs. Pellets were resuspended and sonicated in 1×PBS, 0.4mM PMSF, 5μg/ml leupeptin and aprotinin, 0.1% deoxycholate. GST proteins were purified following standard protocol.
Recombinant PDCD4 pull-downs in cell lysates
5–10 μg of GST-proteins were incubated with glutathione resin (GE Healthcare) in binding buffer (20mM Hepes pH7.6, 100 mM KCl, 0.5mM EDTA, 0.25% TritonX100, 20% glycerol) at room temperature for 1 hr. GST-glutathione conjugates were washed and incubated with 100–300 μg of cell lysate for 1 hr at RT. Pull-downs assessing the eIF4A1 interaction and the pleural effusion samples used methylation buffer (1mM DTT, 0.25% TritonX100, 5% glycerol in 1X PBS).
Preparation of ecotropic retrovirus and MCF7e infections
To prepare virus, HEK293T cells were cotransfected with pCL-Eco plasmid and pMIG or pMSCVpuro in a 1:3 ratio. One day after transfection, the medium was exchanged with fresh media. Medium was collected 48 hrs after transfection and filtered through a 0.45 micron filter. Virus was diluted 1:4 into MCF7 media (DME:F12 1:1, 10% FBS, 10mg/L insulin) and mixed with polybrene (8 μg/ml). This virus mixture was overlaid on 20–50% confluent MCF7e cells, which stably express an ecotropic receptor (a kind gift from Dr. Alex Swarbrick, Garvan Institute, Sydney, Australia). Virus media was exchanged for MCF7 media at 24 hrs. pMIG-transduced cells were selected by FACS for GFP-positive cells and pMSCVpuro-transduced cells were selected for 2 weeks with 2 μg/ml puromycin. All generated cell lines were found to be insulin independent and subcultured in DME:F12 1:1, 10% FBS.
Transplantation of transduced MCF7e cells and tumor growth
1×106 cells were diluted into 10 μL of matrigel (BD Bioscience) and kept on ice. Three-week-old recipient NOD/SCID mice were injected with the cell suspension as described (14). 30 mg beeswax pellets containing 15 μg of β-estradiol were implanted subcutaneously into the interscapular region of recipient mice during the same surgical procedure. Tumors were measured using calipers at 14–21 day intervals. Mice were sacrificed by CO2 asphyxiation and visible tumors collected. For immunoblots, tumor tissue was homogenized in methylation buffer plus 5 μg/ml leupeptin and aprotinin and 400 μM PMSF. Lysates were clarified by centrifugation at 20k × g for 15 minutes at 4°C and resulting supernatants were separated by PAGE. Upon sacrifice of mice, lungs, spleen, and liver were dissected and visually inspected for macro-metastases. Micro-metastases were assessed by detection of GFP positive cells in tissues using an Olympus MVX10 dissecting scope with a UV light source.
V5-Immunoprecipitation
HEK293 cells transfected with catalytically active or dead PRMT5 pcDNA3.1 nV5/EXP vectors were lysed in methylation buffer and incubated with V5 antibody (Invitrogen) conjugated to protein A beads. 1 mg of lysate was incubated with prebound beads for a minimum of 2 hours at 4°C. IPs were washed 3 times with methylation buffer prior to use in further reactions.
Human Pleural Effusion (PE) lysate preparation
Pleural effusion samples were collected from consented breast cancer patients under an approved IRB protocol. Cells were pelleted from the pleural fluid, and if red blood cells were present, the pellet was resuspended in 135mM NH4Cl, 17 mM Tris pH7.4 and incubated at 37°C for 5 minutes to remove red blood cells. The remaining cells were pelleted, washed twice with HBSS, resuspended in freezing medium and stored in liquid nitrogen. Cells were thawed on ice and lysed by dounce homogenization in methylation buffer with protease inhibitors.
Methyltransferase reactions
0.5–5 μg of GST tagged protein was incubated with PE lysate, HEK293 cell lysate or immunoprecipitated V5-PRMT5 in methylation buffer and 0.5–2 μCi 3H-SAM (Perkin Elmer). Reactions were incubated at 30°C for 1 hr and terminated by addition of SDS sample buffer. Tritiated proteins were separated by SDS-PAGE, and transferred to PVDF. For autoradiography, blots were treated with En3hance Spray (Perkin Elmer) according to manufacturer’s recommendations and exposed to film at −80°C for 6 hrs-5 days.
Antibodies
For immunoblots, α-PDCD4 antibody (Abcam) was used at 1:2000 while α-PRMT5 antibody (Abcam) was used at 1:1000 and cell lysates were loaded at 2–5 micrograms protein per lane. Anti-V5 antibody (2F11F7 Invitrogen) was used at 1:2000, anti-tubulin (YL1/2, Accurate chemical and Scientific Corp.) at 1:2000 and anti-β-actin (AC-74, Sigma) at 1:2000 for immunoblots.
Gene expression and statistical analysis
Gene expression and clinical outcome information were obtained from two independent publicly available datasets (15–17). Clinical outcomes from the Pawitan study (16) was obtained from data published in the Ivshina study (17). Data for PRMT5 and PDCD4 were extracted from normalized expression data for each breast tumor sample, and patients were divided into groups based on expression of the two genes. Each dataset was analyzed separately. See further information in supplemental material. Kaplan-Meier survival curves were generated using the software WINSTAT FOR EXCEL (R. Fitch Software, Staufen, Germany), and p values were calculated by log-rank analysis. p values <0.05 were considered significant.
Cell lines
MCF7e cells were a kind gift of Alex Swarbrick (Garvan Institute, Australia), MDA-MB-453 cells were a gift from Biogen-Idec, DU4475 cells were obtained from ATCC. The cell lines were not further authenticated.
Results
Analysis of PDCD4 as a prognostic marker in breast cancer
To determine the relationship between PDCD4 expression in breast cancer and patient survival, we evaluated a previously published microarray dataset. This dataset was obtained from tumors less than 5cm collected from 295 breast cancer patients under the age of 55 at the Netherlands Cancer Institute (15). We stratified this dataset into two groups based on PDCD4 levels, with the “high” group representing those above the median. High expression correlated significantly with better probability of survival (p=0.0014) (Fig. 1). Nevertheless, a significant fraction of patients within the high PDCD4 cohort (~35%, which extrapolates to over 38,000 new patients per year in the U.S. alone (1)) did not appear to gain benefit from this elevated expression. One explanation for this could be the presence of interacting partners of PDCD4 that inactivate or change its role as a tumor suppressor.
Figure 1. Analysis of PDCD4 as a prognostic marker in breast cancer.

Microarray data from tumors (15) was analyzed with respect to PDCD4 expression. Patients with tumors expressing PDCD4 mRNA above the median (“high”, dark grey) had a significantly higher probability of survival than the other patients (“low”, light grey).
Discovery of an interaction between PDCD4 and PRMT5 reveals a potential pathway of regulation in breast cancer
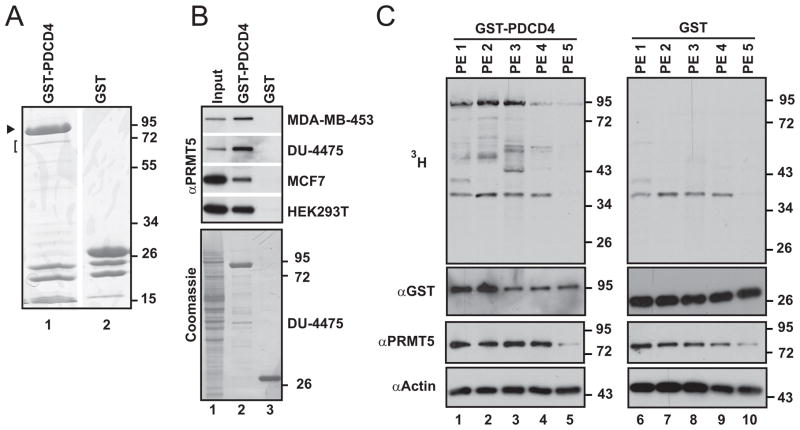
To look for new regulatory factors of PDCD4, we took a biochemical approach to identify binding partners. We used Xenopus laevis egg extract, which has the advantage of limited manipulation to disrupt protein complexes and retains a very high concentration of proteins. A 72 kD protein was consistently recovered from egg extract with recombinant PDCD4 (Fig. 2A). When analyzed by mass spectrometry, this protein was identified as Protein Arginine Methyltransferase 5 (PRMT5; Fig. S1). To confirm that PDCD4 also interacts with human PRMT5, lysates were made from a panel of cell lines (representing both estrogen receptor positive and estrogen receptor negative breast cancers), and interacting proteins were isolated on an analytical scale. Immunoblotting the proteins retained with GST-PDCD4 confirmed the interaction with human PRMT5 (Fig. 2B).
Figure 2. PRMT5 is a protein partner of PDCD4.
(A) Coomassie-stained gel of proteins associated with GST-PDCD4 (lane 1; GST-PDCD4 indicated with arrowhead), compared to background levels of proteins that associate with GST (lane 2), following incubation with Xenopus laevis egg extract. The gel was sliced (bracket) to recover a 72 kD band. Analysis by mass spectrometry identified PRMT5 in this gel slice. (B) Upper panel: immunoblot to detect PRMT5 among proteins associated with GST-PDCD4 (lane 2) or GST (lane 3), following incubation with lysates from the human cell lines indicated. 8.5–17% of input material is shown in lane 1. Lower panel: Coomassie stain of immunoblot (DU-4475 samples), showing recovery of GST-tagged proteins (lanes 2, 3). (C) Autoradiograph of a panel of pleural effusion lysates supplemented with 3H-SAM and GST-PDCD4 (left panel) or GST (right panel), indicating that PDCD4 can be targeted for methylation in the context of breast cancer.
PRMT5 is a type II methyltransferase and catalyzes addition of a methyl group to both terminal nitrogens of arginine residues (18). Like other post-translational modifications, methylation can alter protein function, localization, and/or binding partners (18). To assess whether PRMT5 has the potential to be a regulatory factor in the context of breast cancer, we examined a set of clinical breast cancer specimens for protein expression. Again, we surveyed both estrogen receptor positive and estrogen receptor negative tumors. This analysis found that PRMT5 is expressed in breast cancer and can vary in level as well as whether it is co-expressed with PDCD4 (Fig. S2). To assess whether these proteins interact and have the potential to directly influence each other in the context of breast cancer, breast cancer cells were isolated directly from patients, as pleural effusions. When GST-PDCD4 was incubated with lysates of these cells, PRMT5 was again found to associate (data not shown).
The discovery of a partnership between PRMT5 and PDCD4 raised the question of whether PDCD4 is a target of methylation. To probe the potential for this modification to occur in the context of breast cancer, lysates of tumor cells isolated from fresh pleural effusions were incubated with GST-PDCD4 (Fig. 2C, lanes 1–5) or GST (lanes 6–10) along with tritiated S-adenosylmethionine (SAM) as the methyl donor. Specific labeling of GST-PDCD4 was detected, indicating that PDCD4 is a target of protein methyltransferases present in these lysates. GST-PDCD4 was modified to varying degrees in this assay, underscoring the possibility that the extent to which PDCD4 is targeted by this type of pathway in breast cancer is variable and, in turn, may impact the role of PDCD4 to different extents, depending on the individual context.
PDCD4 and PRMT5 synergistically enhance tumor growth
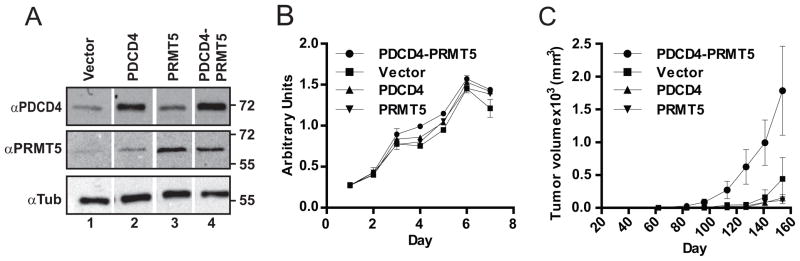
Over-expression of PRMT5 transforms NIH3T3 cells and has been found to be upregulated in leukemia, lymphoma and gastric cancer (13, 19, 20). The presence of PRMT5 in breast cancer, its potential to modulate protein activity, and its reported attributes as a pro-tumor factor prompted us to test whether PRMT5 influences the role of PDCD4 in a tumor context. To do so, MCF7e breast cancer cells, (MCF7 cells that ectopically express the mouse ecotropic receptor enabling infection by murine-specific retroviruses) were engineered to express elevated levels of PDCD4 and PRMT5 alone or in concert (Fig. 3A). There were no gross phenotypic changes as a result of increased expression of these proteins and all four cell lines had similar doubling times under normal tissue culture conditions (Fig. 3B).
Figure 3. Co-expression of PDCD4 and PRMT5 enhances tumor growth in xenograft model.
(A) MCF7e cells expressing PDCD4 and PRMT5 or empty constructs, as indicated, were assessed by immunoblot. Tubulin levels were tracked as a loading control. (B) Cell growth in tissue culture was measured for 7 days using a WST assay (BioVision). (C) The cell panel was transplanted orthotopically and tumor volume was monitored over a 22 week period, with mean numbers and standard error graphed. See supplementary methods for additional information on xenograft tumor growth.
To assess tumor growth, cells were transplanted orthotopically into NOD/SCID mice that also received estrogen supplementation (see Materials and Methods). Increasing levels of either PDCD4 or PRMT5 alone in the context of MCF7e cells did not significantly alter the growth rate of the tumor (Fig. 3C). However, the PDCD4-PRMT5 co-expressing cells exhibited significantly faster growth as tumors than singly-expressing PDCD4, PRMT5 or control cell lines (Fig. 3C). Analysis of lungs, spleen, and liver showed no significant difference in metastasis between cell lines. Immunohistochemical analysis of harvested tumors showed the expected elevated levels of PDCD4 and PRMT5 expression (Fig. S3). No significant difference between PDCD4-PRMT5 tumors compared to vector control tumors was found in analysis of activated caspase 3 and Ki67 (Fig. S4). In general, however, more necrosis, edema, and vasculature were observed in PDCD4-PRMT5 tumors relative to vector-expressing tumors by H&E stain (Fig. S5).
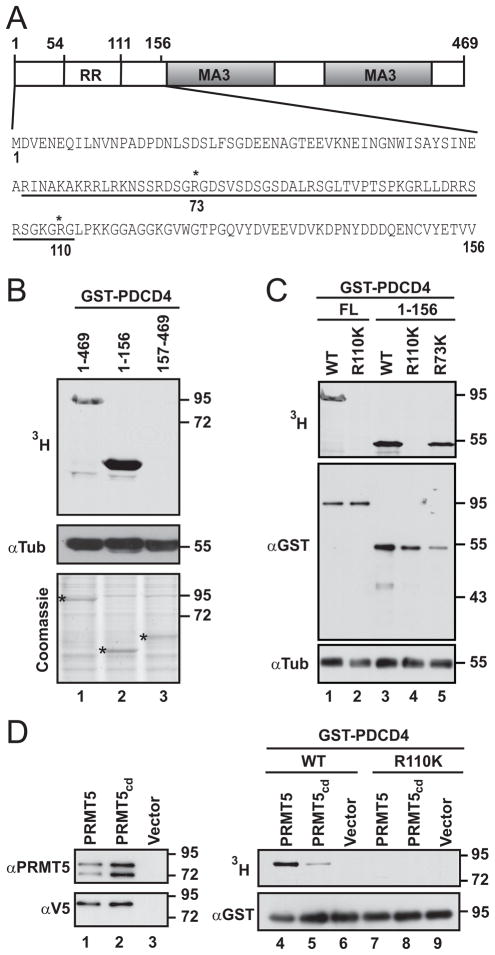
PDCD4 is methylated in the N-terminal domain at Arginine110 and is a target of PRMT5
To narrow down the region of methyl modification seen in Figure 2C, we fragmented PDCD4 into two domains. We noted that the N-terminal region contains an arginine rich (RR) subdomain (Fig. 4A). The second region contains the MA3 domains responsible for eIF4A binding (Fig. 4A) (21–23). Purified GST-fusion proteins of full-length and both truncation mutants were incubated with cell lysate as an enzyme source and supplemented with 3H-SAM. Only full-length protein and the N-terminal fragment were labeled (Fig. 4B). To further map the methylation site of PDCD4, we took a candidate approach. Within the arginine-rich region of PDCD4, R73 and R110 are flanked by glycines, resembling canonical methylation sites (Fig. 4A). These arginines were mutated to lysine, conserving the amino acid charge but disrupting potential arginine methyl acceptor sites. Mutation of R73 had negligible effect, whereas mutation of R110 abolished methylation (Fig. 4C).
Figure 4. PDCD4 can be methylated in its N-terminal domain and is a target of PRMT5.
(A) Schematic of PDCD4, with arginine rich (RR) region and MA3 regions indicated. N-terminal sequence is shown with the RR region underlined. (B) Upper panel: autoradiograph of reactions containing purified GST-PDCD4 (lane 1) or domain fragments (lanes 2, 3) along with HEK293 cell lysate and 3H-SAM. Middle panel: immunoblot for tubulin, tracking lysate level in methylation reactions. Bottom panel: total protein in each reaction detected by Coomassie staining; * indicates recombinant protein present in each reaction. (C) Arg110 and/or Arg73 were mutated to lysines in full length (FL) PDCD4 or the N-terminal region. These proteins, and wild-type (WT) counterparts, were incubated with cell lysates supplemented with 3H-SAM. Top panel: autoradiography. Middle panel: immunoblot to detect GST fusion proteins. Lower panel: immunoblot for tubulin, to track levels of cell lysate. (D) V5-PRMT5, the catalytically dead mutant (V5-PRMT5cd), or control vector were transiently expressed. Left panels are immunoblots of material immunoprecipitated with V5 antibody and probed with PRMT5 (upper panel) or V5 (lower panel) antibody. GST-PDCD4 (lanes 4–6) or GST-PDCD4-R110K (lanes 7–9) were incubated with immunoprecipitates shown in lanes 1–3 along with 3H-SAM. Methylation was monitored by autoradiography (upper panel). The presence of recombinant PDCD4 in all reactions was confirmed by immunoblot with anti-GST (lower panel).
To determine if PRMT5 itself methylates PDCD4, wild-type or catalytically dead PRMT5 (PRMT5cd) were transiently expressed and then immuno-isolated (Fig. 4D). These enzyme sources were incubated with purified GST-PDCD4 and 3H-SAM. Labeled GST-PDCD4 was detected in reactions using wild-type PRMT5 (Fig. 4D, lane 4), demonstrating that PDCD4 can be targeted for methylation by PRMT5. Mutation of R110 again resulted in the absence of methylation (Fig. 4D, lane 7), confirming that this is the acceptor site for methylation by PRMT5. PDCD4 was labeled at greatly reduced levels in reactions using PRMT5cd; this residual activity is likely due to the ability of mutant PRMT5 to homo-oligomerize with its endogenous counterpart ((24) and Fig. 4D, lanes 1 and 2).
The catalytic activity of PRMT5 and PDCD4-R110 are necessary for enhanced tumor growth
To determine if PRMT5 enzymatic activity is necessary to promote tumor growth in our breast cancer model system, MCF7e cell lines were generated expressing PRMT5cd alone or in conjunction with wild-type PDCD4 (Fig. S6A). These cells were transplanted orthotopically and assessed for tumor growth over a 22 week period. As controls, vector only and wild-type PDCD4-PRMT5 cells were also re-transplanted. The new injection of the PDCD4-PRMT5 cell line tracked well with the previous PDCD4-PRMT5 tumor growth rate, underscoring their synergistic effect (Fig. 5A). Tumor measurements from the second experiment with vector control and PDCD4-PRMT5 were combined with the respective data from the first set to assess growth rates at week 22 and a clear difference in tumor growth was seen between these cell lines (Fig. 5B). The PRMT5cd-PDCD4 cells, however, did not show an accelerated growth rate compared to the vector control (Fig. 5A, B), indicating that the enzymatic activity of PRMT5 is necessary for enhanced tumor growth due to co-expression with PDCD4.
Figure 5. The catalytic activity of PRMT5 and arginine 110 of PDCD4 are necessary for synergistic tumor cell growth.
(A) Mean tumor growth of PDCD4mm-PRMT5 and PDCD4-PRMT5cd cell-lines is graphed along with repeats of vector only and PDCD4-PRMT5 (denoted “−2”). Data from the first tumor growth experiment (denoted “−1”) with these two cell-lines (Figure 3) is shown for comparison. (B) Compiled comparison of tumor volume at 22 weeks, with the mean and standard error indicated. The only cell line that showed statistically different growth than the vector control cell line were the PDCD4-PRMT5 expressing cells, as indicated. See supplementary methods for additional information on xenograft tumor growth. (C) Representative tumors from panel A were assessed for construct expression by immunoblot.
To determine whether PDCD4 is the relevant target of PRMT5 in this context or whether PRMT5 is working through a parallel pathway, MCF7e cell lines were generated expressing the methyl mutant PDCD4mm (PDCD4 with R110K mutation) with or without wild type PRMT5 and then assessed for tumor growth (Fig. S6 and Fig. 5A, B). Tumor growth of PDCD4mm with PRMT5 was not significantly different from the control cell line (Fig. 5B). Tumor growth properties in these cohorts are not explained by loss of expression of exogenous constructs, as confirmed at the level of both protein (Fig. 5C) and RNA sequence (Fig. S6). This indicates that methylation of PDCD4, and R110 in particular, is necessary for the enhanced tumor growth observed with co-expression of both wild type PDCD4 and PRMT5.
Two important properties of PDCD4 are its ability to interact with eIF4A and rapid degradation in response to cell proliferation signals (21, 25, 26). Binding of eIF4A is mediated by two MA3 domains that are distal to the methylation site in PDCD4 (Fig. 4A) (27, 28). To determine if mutation of this methylation site alters eIF4A binding, both GST-PDCD4 and GST-PDCD4mm were incubated in cell lysate and then recovered on a glutathione matrix. These samples were probed by immunoblot for eIF4A1, which was found at equal levels (Fig. S7A). Therefore, at the level of methylation that takes place under these assay conditions, an influence on the association of eIF4A is not detectable. We next examined whether increased PRMT5 expression alters the stability of PDCD4. To do so, the MCF7e lines expressing exogenous PDCD4, PDCD4 plus PRMT5, and PDCD4mm plus PRMT5 were stimulated with PMA to trigger S6K activation and with cycloheximide to stop new protein synthesis. We then tracked the stability of exogenous PDCD4 over a short time course by probing immunoblots with anti-V5. PRMT5 expression did not appear to alter the stability of PDCD4 (Fig. S7B, upper and middle panels). Consistent with this, we did not observe a significant difference between wild-type PDCD4 and the methyl mutant (Fig. S7B, middle and lower panels).
Clinical data indicate that PRMT5 levels impact the probability of survival when tumors express PDCD4
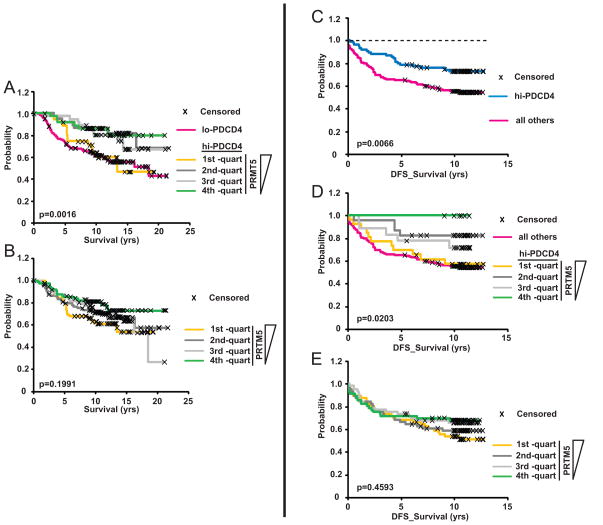
Our observations that simultaneous expression of elevated PDCD4 and PRMT5 causes accelerated tumor growth in an orthotopic mouse model raised the question of whether this combination of markers would be useful in a clinical setting. To address this, we stratified patients with high tumor levels of PDCD4 into quartiles based on PRMT5 expression. Each one of these cohorts extrapolates to approximately 25,000 patients a year in the U.S. (1). We found that the top quartile –in which tumors highly express both PDCD4 and PRMT5– had poor outcomes, similar to the low PDCD4 cohort. In contrast, with decreases in PRMT5 expression, the probability of survival increased significantly (p=0.0016). The bottom quartile –high for PDCD4 and low for PRMT5– had remarkably better outcome than the top quartile (Fig. 6A), with a 20-year survival of 80% vs 43%. Similar quartile analysis of PRMT5 alone showed a trend in improved probability of survival with decreasing PRMT5, but was not significant (Fig. 6B). This suggests that the combination of PRMT5 and PDCD4 as biomarkers for outcome is potentially useful: high levels of PDCD4 are protective, unless PRMT5 is also highly expressed.
Figure 6. Expression of PRMT5, in conjunction with PDCD4, improves prediction of survival in two independent datasets.
Breast cancer mRNA expression data was analyzed with respect to both PDCD4 and PRMT5 using the two independent data sets (Fig. 6A, B (15); Fig. 6C-E (17)). (A) The “high” PDCD4 cohort of patients (see Fig. 1, (15)) were stratified into quartiles based on PRMT5 expression, with the first quartile containing the highest levels of PRMT5 expression. (B) Patients from the entire dataset were stratified into quartiles based on PRMT5 expression alone. (C–E) Analysis of a second dataset of mRNA expression in breast cancer (17). (C) This dataset was stratified into thirds based on PDCD4 expression levels. Patients in the “high” expression cohort (top one-third) had greater probability of disease-free survival (DFS) compared to all others. (D) The “high” PDCD4 cohort was further stratified into quartiles based on PRMT5 expression; less PRMT5 expression corresponded to increased probability of disease-free survival. (E) Patients stratified into quartiles based on tumor expression of PRMT5 alone did not show significant differences in survival.
To test the reproducibility of these observations, we used another, independent breast cancer dataset (17). We selected patients from this cohort based on availability of both clinical follow-up and gene expression data. This resulted in a set of 224 tumors, which we then stratified for PDCD4 expression, with “high” defined as those in the top third of expression levels. Patients in this high expression group again had a significantly higher probability of longer disease-free survival (p= 0.0066) (Fig. 6C). Yet, within the high PDCD4 expression group there was still a >25% probability of relapse. We then further stratified the PDCD4 high group by PRMT5 levels (each sub-group extrapolates to greater than 15,000 patients a year in the U.S. (1)) and, strikingly, lower levels of PRMT5 proved to similarly correspond to better outcome in this context (p=0.0203; Fig. 6D), whereas quartile analysis of PRMT5 alone did not show a correspondence to outcome (Fig. 6E). This data confirms that considering PRMT5 levels in the context of PDCD4 could be an effective strategy toward an improved prognostic tool in breast cancer.
Discussion
PDCD4 shows promise as a biomarker, with prognostic attributes in a number of cancers (4–7). Here, we further find that PDCD4 expression is informative with regard to survival of breast cancer patients, where increased levels correlate with better outcome in two large scale clinical evaluations. PDCD4 expression alone, however, has limitations in stratifying cancer patient outcomes as ~30% of patients with tumors expressing relatively high PDCD4 levels have poor survival. This could be due to a potential discrepancy between mRNA and protein levels (26, 29), to environmental factors, or to interacting regulatory pathways.
The discovery here of PRMT5 as an interacting partner opened a new level of regulation to consider. Neither PDCD4 nor PRMT5 affected tumor cell growth when expressed alone in our model system. Although this was somewhat surprising, it may indicate that in a particularly aggressive or advanced tumor context, elevated expression of either protein alone is insufficient to alter tumor properties. In the case of PDCD4, this may be due at least in part to endogenous levels of PRMT5. Importantly, the orthotopic tumor model revealed that, together, elevated PDCD4 and PRMT5 expression significantly enhanced tumor growth. This result suggests that PRMT5 does not just negate a PDCD4 tumor suppressor function, but works synergistically to promote tumor growth. This pro-growth effect occurred only in the tumor context, not in tissue culture, hinting that the combination of proteins activates a pathway that enhances the ability to establish a productive tumor microenvironment. The similar numbers of cells staining for activated caspase-3 and Ki67 in tumors expressing elevated levels of both PDCD4 and PRMT5 versus tumors with vector alone suggests that alteration in apoptosis or proliferation does not account for the larger tumor size in the former case. It will be important to investigate these features at earlier points in tumorigenesis. Changes in early events coordinated between cancer and host cells may accelerate the growth of PDCD4-PRMT5 tumors. The increase in necrosis, edema, and vasculature is consistent with the larger size of the PDCD4-PRMT5 xenograft tumors, but further investigation is needed to determine whether any of these features are critical to the altered tumor growth properties conferred by elevated levels of PDCD4 and PRMT5.
The change in PDCD4 function that occurs in the presence of elevated PRMT5 pointed to a role for post-translational modification. We found that indeed PDCD4 is methylated and further, a PDCD4 methyl mutant expressed with wild type PRMT5 or a catalytically dead PRMT5 expressed with wild type PDCD4 failed to promote tumor growth. This demonstrates that methylation of PDCD4 by PRMT5 is critical for enhanced tumor growth. The methylation target residue, R110, lies near a S6 Kinase 1 site (S67) reported to regulate PDCD4 stability (26, 30). Levels of ectopic expression of PDCD4 did not appear to be influenced by the presence of PRMT5 or mutation of R110 (Figs. 3 and 5). Furthermore, under conditions that stimulate PDCD4 degradation, the methyl mutant did not enhance or decrease stability indicating that methylation of PDCD4 does not help coordinate proteasome targeting.
PDCD4 is thought to exert its tumor suppressor role by regulating translation (21, 25), fitting with a growing theme of translation initiation as a node of regulation in cancer (31). Interestingly, PRMT5 has been shown to influence eIF4E levels (32) and was recently shown to influence ribosomal stability by methylation of ribosomal subunit RPS10 (33). Methylation of PDCD4 may impact its role in translation regulation, although we did not observe a difference in eIF4A1 binding activity when the methylation site was mutated. Another interesting possibility is that, conversely, methylated PDCD4 changes PRMT5 function or specificity. In addition to the recent connection to translation, PRMT5 is known to be involved in transcription (13, 20, 34), in efficient assembly of the spliceosome (35), and in modulation of p53-dependent cell cycle arrest (32, 36). Methylated PDCD4 potentially accelerates tumor growth by altering these or other functions downstream of PRMT5 (32, 36).
By integrating biochemical and tumor model data, we have found that an elevated level of PRMT5 in conjunction with PDCD4 reverses the tumor suppressive properties of PDCD4. The finding that combined expression analysis of PDCD4 and PRMT5 is a powerful prognostic indicator for outcome in breast cancer suggests that these factors could be used as rational, activity-based biomarkers to aide in decisions about how aggressively to treat a breast cancer patient. In the future, chemical inhibition of PRMT5 methyltransferase activity could be used to abrogate the synergism between PDCD4 and PRMT5, potentially unmasking PDCD4 tumor suppressor function in cancers that express both proteins. Finally, although our focus here has been on breast cancer, PDCD4 plays a tumor suppressor role in a wide spectrum of cancers (4–6), raising the possibility that its connection to PRMT5 will be of broad prognostic, and perhaps therapeutic, value.
Supplementary Material
Acknowledgments
Financial Support: We acknowledge funds from the NIH (R01 GM61275, KSU), a Leukemia & Lymphoma Society Scholar Award (1438-06, KSU), a DoD Breast Cancer Research Program Era of Hope Scholar Award (#08-1-0109, AW), and Huntsman Cancer Foundation (AW, KU) for support of this work. MF was supported by the Multidisciplinary Cancer Research Training Grant (T32 CA093247).
We are grateful to Tyler Jarrett and Yoko DeRose for help with immunohistochemistry, we thank Krishna Parsawar and Chad Nelson for input on mass spectrometry and Guoying Wang for processing the pleural effusion samples and IHC. We also acknowledge shared resources (Mass spectrometry, DNA sequencing, DNA synthesis, Comparative Oncology) supported by P30 CA042014 awarded to Huntsman Cancer Institute.
Footnotes
Conflicts of Interest: Authors have no conflicts of interest.
Author contributions: MP: designed and performed research, analyzed data, wrote paper; MF: designed and performed research, analyzed data, edited paper; RF: assessed protein expression and histology in clinical samples and mouse tumor IHC; AW: designed research and analyzed data, edited paper; KU: designed research and analyzed data; wrote paper.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Burke HB. Outcome prediction and the future of the TNM staging system. J Natl Cancer Inst. 2004;96:1408–9. doi: 10.1093/jnci/djh293. [DOI] [PubMed] [Google Scholar]
- 3.Duffy MJ, Crown J. A personalized approach to cancer treatment: how biomarkers can help. Clin Chem. 2008;54:1770–9. doi: 10.1373/clinchem.2008.110056. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Knosel T, Kristiansen G, Pietas A, Garber ME, Matsuhashi S, et al. Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J Pathol. 2003;200:640–6. doi: 10.1002/path.1378. [DOI] [PubMed] [Google Scholar]
- 5.Fassan M, Cagol M, Pennelli G, Rizzetto C, Giacomelli L, Battaglia G, et al. Programmed cell death 4 protein in esophageal cancer. Oncol Rep. 2010;24:135–9. doi: 10.3892/or_00000838. [DOI] [PubMed] [Google Scholar]
- 6.Mudduluru G, Medved F, Grobholz R, Jost C, Gruber A, Leupold JH, et al. Loss of programmed cell death 4 expression marks adenoma-carcinoma transition, correlates inversely with phosphorylated protein kinase B, and is an independent prognostic factor in resected colorectal cancer. Cancer. 2007;110:1697–707. doi: 10.1002/cncr.22983. [DOI] [PubMed] [Google Scholar]
- 7.Wei NA, Liu SS, Leung TH, Tam KF, Liao XY, Cheung AN, et al. Loss of Programmed cell death 4 (Pdcd4) associates with the progression of ovarian cancer. Mol Cancer. 2009;8:70. doi: 10.1186/1476-4598-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wen YH, Shi X, Chiriboga L, Matsahashi S, Yee H, Afonja O. Alterations in the expression of PDCD4 in ductal carcinoma of the breast. Oncol Rep. 2007;18:1387–93. [PubMed] [Google Scholar]
- 9.Yang HS, Jansen AP, Nair R, Shibahara K, Verma AK, Cmarik JL, et al. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene. 2001;20:669–76. doi: 10.1038/sj.onc.1204137. [DOI] [PubMed] [Google Scholar]
- 10.Yang HS, Knies JL, Stark C, Colburn NH. Pdcd4 suppresses tumor phenotype in JB6 cells by inhibiting AP-1 transactivation. Oncogene. 2003;22:3712–20. doi: 10.1038/sj.onc.1206433. [DOI] [PubMed] [Google Scholar]
- 11.Jansen AP, Camalier CE, Colburn NH. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res. 2005;65:6034–41. doi: 10.1158/0008-5472.CAN-04-2119. [DOI] [PubMed] [Google Scholar]
- 12.Hilliard A, Hilliard B, Zheng SJ, Sun H, Miwa T, Song W, et al. Translational regulation of autoimmune inflammation and lymphoma genesis by programmed cell death 4. J Immunol. 2006;177:8095–102. doi: 10.4049/jimmunol.177.11.8095. [DOI] [PubMed] [Google Scholar]
- 13.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–45. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rijnkels M, Rosen JM. Adenovirus-Cre-mediated recombination in mammary epithelial early progenitor cells. J Cell Sci. 2001;114:3147–53. doi: 10.1242/jcs.114.17.3147. [DOI] [PubMed] [Google Scholar]
- 15.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 16.Pawitan Y, Bjohle J, Amler L, Borg AL, Egyhazi S, Hall P, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–64. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 18.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JM, Sohn HY, Yoon SY, Oh JH, Yang JO, Kim JH, et al. Identification of gastric cancer-related genes using a cDNA microarray containing novel expressed sequence tags expressed in gastric cancer cells. Clin Cancer Res. 2005;11:473–82. [PubMed] [Google Scholar]
- 20.Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, Sif S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. Embo J. 2007;26:3558–69. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang HS, Cho MH, Zakowicz H, Hegamyer G, Sonenberg N, Colburn NH. A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol Cell Biol. 2004;24:3894–906. doi: 10.1128/MCB.24.9.3894-3906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang JH, Cho YH, Sohn SY, Choi JM, Kim A, Kim YC, et al. Crystal structure of the eIF4A-PDCD4 complex. Proc Natl Acad Sci U S A. 2009;106:3148–53. doi: 10.1073/pnas.0808275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loh PG, Yang HS, Walsh MA, Wang Q, Wang X, Cheng Z, et al. Structural basis for translational inhibition by the tumour suppressor Pdcd4. Embo J. 2009;28:274–85. doi: 10.1038/emboj.2008.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rho J, Choi S, Seong YR, Cho WK, Kim SH, Im DS. Prmt5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J Biol Chem. 2001;276:11393–401. doi: 10.1074/jbc.M008660200. [DOI] [PubMed] [Google Scholar]
- 25.Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, et al. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol. 2003;23:26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1-and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–71. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 27.LaRonde-LeBlanc N, Santhanam AN, Baker AR, Wlodawer A, Colburn NH. Structural basis for inhibition of translation by the tumor suppressor Pdcd4. Mol Cell Biol. 2007;27:147–56. doi: 10.1128/MCB.00867-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki C, Garces RG, Edmonds KA, Hiller S, Hyberts SG, Marintchev A, et al. PDCD4 inhibits translation initiation by binding to eIF4A using both its MA3 domains. Proc Natl Acad Sci U S A. 2008;105:3274–9. doi: 10.1073/pnas.0712235105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 30.Schmid T, Jansen AP, Baker AR, Hegamyer G, Hagan JP, Colburn NH. Translation inhibitor Pdcd4 is targeted for degradation during tumor promotion. Cancer Res. 2008;68:1254–60. doi: 10.1158/0008-5472.CAN-07-1719. [DOI] [PubMed] [Google Scholar]
- 31.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10:254–66. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 32.Scoumanne A, Zhang J, Chen X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 2009;37:4965–76. doi: 10.1093/nar/gkp516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren J, Wang Y, Liang Y, Zhang Y, Bao S, Xu Z. Methylation of ribosomal protein S10 by protein-arginine methyltransferase 5 regulates ribosome biogenesis. J Biol Chem. 2010;285:12695–705. doi: 10.1074/jbc.M110.103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, et al. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol. 2003;23:7475–87. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meister G, Fischer U. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. Embo J. 2002;21:5853–63. doi: 10.1093/emboj/cdf585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10:1431–9. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.