Abstract
Two base excision repair glycosylase (BER) transition state (TS) mimics, (3R, 4R)-1-benzyl (hydroxymethyl) pyrrolidin-3-ol (1NBn) and (3R, 4R)-(hydroxymethyl) pyrrolidin-3-ol (1N), were synthesized using an improved method. Several BER glycosylases that repair oxidized DNA bases, bacterial formamidopyrimdine glycosylase (Fpg), human OG glycosylase (hOGG1) and human Nei-like glycosylase 1 (hNEIL1) exhibit exceptionally high affinity (Kd ~ pM) with DNA duplexes containing the 1NBn and 1N nucleotide. Notably, comparison of the Kd values of both TS mimics relative to an abasic analog (THF) in duplex contexts paired opposite C or A suggest that these DNA repair enzymes use distinctly different mechanisms for damaged base recognition and catalysis despite having overlapping substrate specificities.
Keywords: Base excision repair, DNA glycosylase, pyrrolidine analogs, transition state analogs, 8-oxo-7, 8-dihydro-2-deoxyguanosine, hOGG1, Fpg, Nei, hNEIL1
Chemical tools developed to study enzymes that act upon oligosaccharides provide inspiration for the use of similar strategies with functionally similar enzymes that act on nucleic acids. 1 Indeed, DNA glycosylases initiate base excision repair (BER) at damaged bases within DNA by catalyzing N-glycosidic bond cleavage2,3 in a manner mechanistically analogous to oligosaccharide hydrolysis catalyzed by a large and diverse class of glycoside hydrolases.4 Chemically modified sugars have played key roles in elucidating the structural and mechanistic properties of glycoside hydrolases and hold promise as powerful tools to visualize and tag glycosidases in cellular contexts.5–7
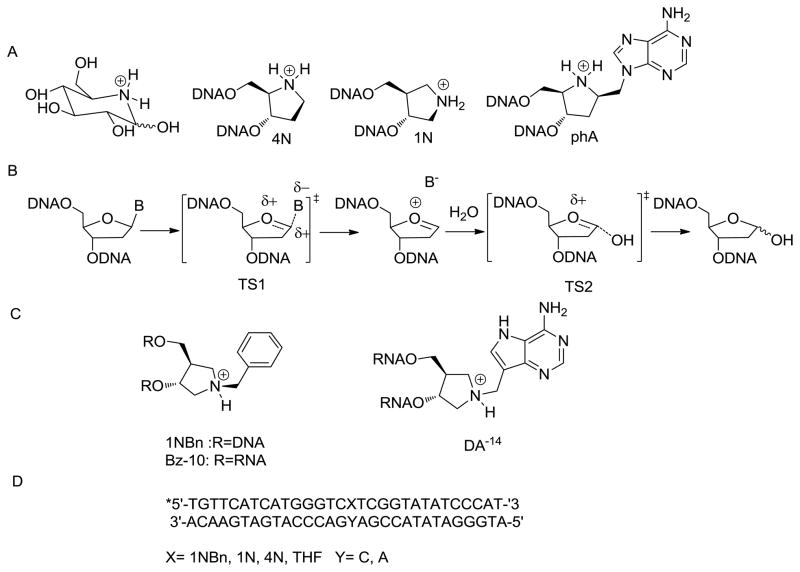
There is a particularly rich history of the use of azasugars as high affinity inhibitors of glycosidase enzymes and many of these inhibitors have shown promise as pharmaceuticals.5 The efficacy of inhibitors such as Nojirimycin (Figure 1A) has been attributed to mimicry of the glycosidase “oxacarbenium ion-like” transition state providing for favorable electrostatic interactions within the active site.5 The pyrrolidine analogs, (3R, 4R)-3-hydroxymethyl-2-(hydroxymethyl) pyrrolidinium (4N) and (3R, 4R)-(hydroxymethyl) pyrrolidin-3-ol (1N) (Figure 1A) incorporated within DNA duplexes have been found to have subnanomolar affinity for several DNA glycosylases.8–10 Kinetic isotope effects studies of Uracil-DNA glycosylase (UDG) and the adenine glycosylase MutY indicate an SN1 mechanism of base hydrolysis that proceeds through two oxacarbenium ion-like transition states (TS1 and TS2, Figure 1B).11,12 The pyrrolidines 1N and 4N mimic the positive charge on the 2′-deoxyribose oxacarbenium ion and may more closely approximate TS2.13 Pyrrolidine homonucleosides (Figure 1C) that contain an appended base or base mimic would be expected to more closely mimic the first oxacarbenium ion-like TS (TS1). These TS mimics incorporated into RNA have been shown to be potent inhibitors of ricin A-chain (RTA) that hydrolyzes adenosine within ribosomal RNA.14–17 Consistent with similar mechanisms for base excision, an adenine pyrrolidine homonucleotide, phA incorporated into a DNA duplex (Figure 1B) has been used for binding studies with MutY.8
Figure 1.
A. Nojirimycin and aza-ribose transition state mimics. B. SN1 mechanism for monofunctional DNA glycosylases (such as uracil-DNA glycosylase) based on KIE studies.3 Note that the enzymes studied herein are bifunctional glycosylase/lyase enzymes that utilize an amine nucleophile (Lys or N-terminal proline) rather than a water molecule in the base displacement process. C. Structure of 1NBn, Bz-10 and DA-14. D. Sequence context of the 30-base pair duplex used in this study.
Among the myriad of modified DNA bases, oxidative modifications are particularly mutagenic and are contributors to human diseases, including cancer and neurological disorders.18–20 A cadre of DNA glycosylases play crucial roles in excising oxidatively damaged DNA bases as the first step in BER.20,21 In mammalian cells, the human OG glycosylase (hOGG1) removes primarily 8-oxo-7,8-dihydroguanine (OG) and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG), while the human endonuclease III enzyme (hNTH) removes oxidized thymines, such as thymine glycol (Tg). The human nei-like (NEIL1) glycosylase has a broad substrate specificity in removing damaged pyrimidines, such as Tg and 5-hydroxyuracil (5-OHU) as well as the oxidized purines FapyG, guanidinohydantoin (Gh) and spiroiminodihydantoin (Sp) but not OG.20,22–26 In contrast, in Escherichia coli, the formamidopyrimidine glycosylase (Fpg) is able to remove a broad array of oxidized bases, primarily those derived from purines, including OG, FapyG, and the hydantoins27,28 while the Nei glycosylase removes primarily oxidized pyrimidines (e.g. thymine glycol), Gh, Sp and 4,6-diamino-5-formamidopyrimidine (FapyA).29 An additional important feature of these oxidized-base specific glycosylases is the ability to catalyze associated β-(hOGG1) or β– and δ-lyase (hNEIL1, Fpg, Nei) strand scission reactions.
Due to the overlapping but distinct substrate specificity of this group of DNA glycosylases, we reasoned that each glycosylase may exhibit distinct binding preferences for pyrrolidine analogs that mimic TS1 versus TS2. An inhibitor of RTA that attracted our attention was Bz-10 (Figure 1C). Bz-10 was reported to exhibit a Ki of 0.099 μM, establishing it as a far better inhibitor than DA-14 (Figure 1C, Ki value of 0.280 μM) which has a 9-deazaadenyl substituent and closely resembles adenine. The N1 atom of Bz-10 has a pKa around 8.5 making it cationic at physiological pH. 15 Herein, we report modified and improved syntheses of 1N and 1NBn phosphoramidites and evaluate the utility of these analogs within duplex DNA as TS mimics for the BER glycosylases, Fpg, hOGG1, NEIL1 and Nei.10,30 An expected feature of TS mimicry is high enzyme affinity and therefore we determined dissociation constants (Kd) of this group of BER glycosylases with a 30 bp duplex containing centrally located synthetic transition state mimics (1N, 4N, 1NBn) relative to the product analog (THF) using electrophoretic mobility shift assays (EMSA).31–34
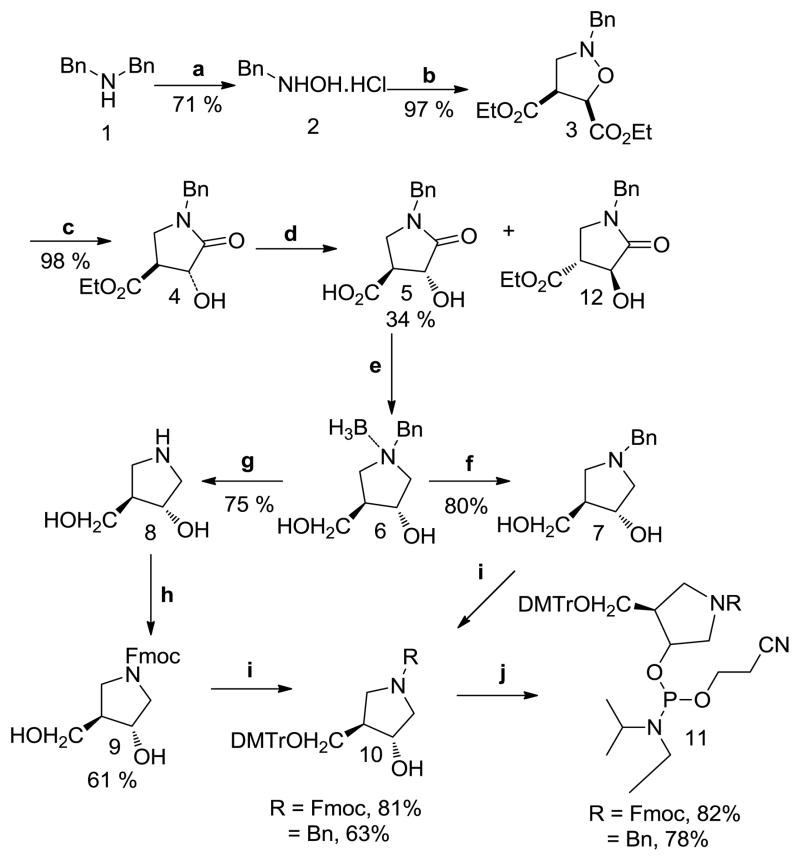
1NBn (7) has recently been published as an intermediate product in the synthesis of (3R,4R)-N-tert-butoxycarbonyl-4-hydroxymethyl.35 We modified this procedure to start from an economical precursor (1) and provide higher yielding syntheses of 1NBn (7), 1N (8) and the corresponding phosphoramidite monomers (Scheme 1). Briefly, dibenzylamine (1) was used to make the N-benzylhydroxylamine (2).36 The reaction time was lengthened for steps c–d to obtain lactam (5): the cleavage of the N-O bond of isoxazolidine (3) and the hydrolytic resolution of the racemic ester (4) using the immobilized form of lipase B of Candida antarctica (Cal-B).35 The reduction of the lactam (5) to benzylamine (6) was accomplished using borane dimethyl sulfide.37 The latter reaction produced a stable amine-borane complex (6) as confirmed by HRMS analysis and a signal at −9.6 ppm in 11BNMR, which is known to arise from a tertiary-amine-borane complex (Supporting Information (SI)).38 Decomplexation to generate 1NBn (7) was accomplished by stirring with methanol for 24 h in the presence of 10% Pd/C catalyst under H2 gas atmosphere. The increased time of methanolic cleavage resulted in tandem decomplexation and debenzylation to generate 1N (8). HRMS, 1HNMR, and 13CNMR for 1N (8) and additional X-ray crystallography for intermediates (5), (12), and 1NBn (7) (SI, Figure 2) confirmed the identity and absolute stereochemistry of the products. Fmoc protection of 1N was performed using the procedure of Filichev et. al.39 Tritylation (10) and phosphitylation (11) for both nucleosides were carried out with minor modifications of the previous methods17 to provide an overall yield of the 1NBn and 1N phosphoramidites of 9 % and 4%, respectively. 31PNMR (CDCl3) indicated formation of the phosphoramidite linkage with peaks at ~148–9 ppm for both 1NBn and IN phosphoramidite (11) (SI). These phosphoramidites were incorporated into oligonucleotides with >95% efficiency using standard automated phosphoramidite synthesis and were deprotected using standard conditions. HRMS analysis of an 11-nucleotide fragment containing 1NBn [d(5′TGTCCA1NBnGTCT-3′)] gave a strong molecular ion peak at the expected molecular weight of m/z 3261.77 (SI) indicating that 1NBn is stable under the conditions used for the DNA synthesis and deprotection. The 1NBn nucleoside was also generated from 1N by reductive amination. 40
Scheme 1.
Reagents and conditions: (a) 1.) Na2WO4•2H20, H2O2, MeOH, rt 18 h; 2.) 20% HCl; (b) diethymaleate, CH2O, EtOH, reflux, 2.5h; (c) Zn, AcOH, 8 h; (d) Cal-B lipase, pH 7.5, 30°C,16–24h; (e) 1.) BH3.DMS, THF, reflux, 11h; 2.) H2O; (f) MeOH, 10% Pd/C, H2, 24h, rt, 80%; (g) MeOH, 10% Pd/C,10 days, rt, 75%; (h) FmocCl, dioxane, 10 % aq. NaHCO3, rt; (i) DMTrCl, Hunig’s base, DMAP, DCM, rt, 1h; (j) ClPNi-Pr2(OCH2CH2CN), DCM, iPr2NEt, rt, 2 h
Electrophoretic mobility shift assays (EMSA) were used to measure the relevant dissociation constants of Fpg, hOGG1, hNEIL1 and Nei in a manner analogous to previous reports from our laboratory with MutY.33 Briefly, these experiments entailed titration of the relevant enzyme into a solution of 5′-[32P-phosphate]-end-labeled DNA duplex and analysis on native PAGE to separate and detect both the free DNA duplex and protein-DNA complex. Quantitation via storage phosphor autoradiography and fitting of the data to a one binding isotherm provide the Kd values listed in Table 1. Indeed, inspection of these Kd values reveal distinct differences between the various enzymes with respect to their preference for the TS mimics over the abasic site analog, the influence of the base opposite and relative nonspecific and specific DNA binding affinity. Interestingly, Fpg exhibits the highest affinities while Nei exhibits the lowest affinities for all of the DNA duplexes examined.
Table 1.
Dissociation constants (Kd, nM)a for various DNA repair glycosylases
| Central base pair | Fpg | hOGG1d | hNEIL1 | Nei |
|---|---|---|---|---|
| 1NBn:C | <0.005b | <0.005b (72%), 32 (20%) | 0.2 ± 0.1 | 32 ± 12 |
| 1N:C | <0.005 b | <0.01 (36%), 5 ± 2 (50%) | 0.2 ± 0.1 | 8 ± 3 |
| 4N:C | <0.005 b | <0.01 (50%), 5 ± 2 (20%) | 0.1 ± 0.05 | NDe |
| THF:C | <0.005 b | <0.005 b | 1 ± 1 | 24 ± 8 |
| 1NBn:A | ~ 0.02c | 400 ± 300 | 2 ± 1 | 22 ± 4 |
| 1N:A | ~ 0.01c | 500 ± 400 | 4 ± 1 | 9 ± 4 |
| THF:A | ~0.05c | 400 ± 200 | 4 ± 3 | 8 ± 1 |
| G:C | >150f | >1200 f | 44 ± 5 f | >2000 f |
Dissociation constant measured were performed at 25 °C, 100 mM NaCl and pH 7.5. The DNA duplex concentration was 5 pM for all experiments except those with X:A-duplexes in binding titrations with hOGG1 where 20 pM was used. Errors reported in dissociation constants are standard deviation of the average of at least 3 trials. The concentrations of enzyme are active enzyme concentrations.
Upper limit estimation; too tight to measure accurately, Kd is estimated to be lower than the [DNA] used.
Tight apparent Kd estimated from the plot. The data fits better using a two-site binding isotherm with the second weaker binding capacity representing 20% for 1N and 1NBn, and 50% for THF duplex and providing for a similar Kd value with all three duplexes of approximately 6 nM.
The data with hOGG1 fits best using a two-site binding isotherm that provides two Kd values with relative capacities indicated in closed parentheses. The percentages of the two binding fractions differ between duplexes with the 1NBn giving the highest fraction of the high affinity Kd (see SI for representative data).
ND- not determined
G:C represents non-specific binding. The lower limit estimates Kd are based on the highest enzyme concentration used that still provided less than 50% bound DNA.
Remarkably, Fpg shows exceptionally high affinity for all of the TS analogs (1N, 4N and 1NBn) and the THF product analog when paired with C in the 30 bp duplex. Using the lowest concentration of the analog-containing duplex that may be practically detected via storage phosphor autoradiography (5 pM duplex), the DNA duplex was observed completely bound at all concentrations of Fpg used (SI). On this basis of the DNA concentration used in the EMSA, an upper limit for the Kd of 5 pM was estimated. Although Fpg and hNEIL1 have significant sequence homology, hNEIL1 exhibits a significantly reduced affinity for all of the TS analogs and the THF-containing duplexes. Moreover, like Fpg, hNEIL1 exhibits a 5-fold higher affinity for the duplexes containing positively charge analogs 1N, 4N, 1NBn over the uncharged THF product analog when the opposite base is C (Table 1). In contrast, the bacterial Nei enzyme exhibited weak affinity for all the analogs and did not show a significant preference for positively charged TS analogs over the THF analog.
Surprisingly, hOGG1, a functional human homolog of bacterial Fpg in preventing mutations associated with OG, has a very high affinity for both THF:C and 1NBn:C, with Kd values that are similar to those for Fpg with the same duplexes (Table 1). Curiously, the hOGG1 binding titration curves with the duplexes containing the 1N, 4N and 1NBn analogs paired with C are biphasic and are best fit with a two-site binding isotherm yielding two distinct Kd values. Notably, the 1NBn:C duplex exhibited the largest fraction with the extremely tight Kd compared to the corresponding 1N and 4N containing DNA. This behavior does not appear to be due to the presence of ds and ss DNA in the initial DNA duplex sample after annealing (SI). Indeed, EMSA analysis of hOGG1 binding to the 30 nt ss oligonucleotide containing 1N and 1NBn-containing DNA provided Kd values that are significantly higher (76 ± 12 and 83 ± 15 nM, respectively) than either of the Kd values observed with duplex DNA. Moreover, if the presence of ss DNA is a factor, the fraction representing binding to the duplex (the tighter Kd) would be expected to follow the duplex stabilities with THF >1N >1NBn (Chu, A., David, S. S., unpublished results). The fact that hOGG1 binds 1NBn more efficiently than 1N suggests that the two-site binding data reflects an enzyme-dependent effect on the DNA duplex, such as hOGG1 providing stability for the duplex form. An alternative intriguing possibility is that the biphasic curves are capturing two mutually exclusive DNA binding sites on the enzyme. Indeed, hOGG1 been shown to have two base-binding sites.20 Further investigation of this interesting feature of hOGG1 binding with these TS mimics is in progress using alternative binding methods.
Interestingly, the EMSA data with Fpg fit more accurately to a two-site binding isotherm in the case of the analogs paired with the A-containing duplex, with the proportion representing the tighter Kds being greater with the 1N and 1NBn analog compared to THF. Comparison of the tighter Kd values indicates a reduction in binding affinity of Fpg for the TS analogs paired with A relative to C, consistent with the known ability of Fpg to select against removal of OG paired with A.28,41 Notably, these results also show that by pairing with A, there is a slightly higher preference of Fpg for the presence of the positively charged 1N and 1NBn analogs over the neutral THF analog consistent with published data that indicated an 8-fold tighter binding of Fpg to a DNA duplex containing a central 4N:T over the corresponding THF:T containing duplex.10 Similarly, the relative affinity of hNEIL1 for the 1N and 1NBn TS analog containing duplexes when paired with A is less than for C, as was observed with Fpg, indicating a similar preference for C opposite the lesion, TS or product analog site. In contrast, Nei glycosylase shows a slight preference for base pair context with A, similar to the base specificity that has been reported previously.29
Of this group of glycosylases, hOGG1 is distinct in showing a marked preference for the uncharged product analog (THF) over the positively charged abasic/TS mimic (1N, 4N). Moreover, when the analogs were paired with A, the high affinity and specificity of hOGG1 for the 1NBn and uncharged THF analog is completely lost. Consistent with these results, cleavage of an abasic site by hOGG1 is dependent on the opposite base.42 The reduced binding affinity observed when the analogs were base paired to A is also consistent with the large degree of selectivity of hOGG1 for removal of OG or FapyG when paired with C over A, especially compared to Fpg.43 The results with the 1NBn:C duplex show that the presence of a base mimic (Bn) and C are needed for high affinity. This suggests that both the damaged base and its opposite partner C are utilized in TS stabilization. Indeed, recent structural studies of an inactive hOGG1 mutant (Q315F) bound to an OG:C duplex shows that the OG lesion is almost completely and properly inserted into the OG binding pocket.44 This suggests that there are strict steric requirements for positioning of the base within the active site to reach the proper TS required for hOGG1 mediated base excision. The ability of the simple benzyl substituent to provide for improved recognition of the positively charged azaribose suggests that further elaboration of pyrrolidine nucleotides with base-like moieties and screening may provide for derivatives with even higher affinity for hOGG1 over other glycosylases.
Overall, the results suggest that even though the BER glycosylases studied herein have similar and overlapping substrate specificities, there are distinct differences in the damaged base recognition and excision process within this group. This may be related to their biological function and the particular demand for accuracy versus efficiency when removing damaged bases. The different features of the base excision reaction coordinate for each enzyme are suggested by the distinct recognition patterns for the series of TS and product analogs studied herein. This data indicates that 1NBn, in addition to 4N and 1N, is a useful TS mimic for studying the catalytic mechanism of BER glycosylases and may serve as general inhibitors of these enzymes. These new chemical tools will provide avenues for additional structural studies of DNA glycosylases as well as potential chemical biology tools to identify and probe BER glycosylases and associated protein partners in a cellular milieu. Moreover, such studies provide a starting point for developing high affinity inhibitors of BER glycosylases that may have chemotherapeutic applications.
Supplementary Material
Figure 2.
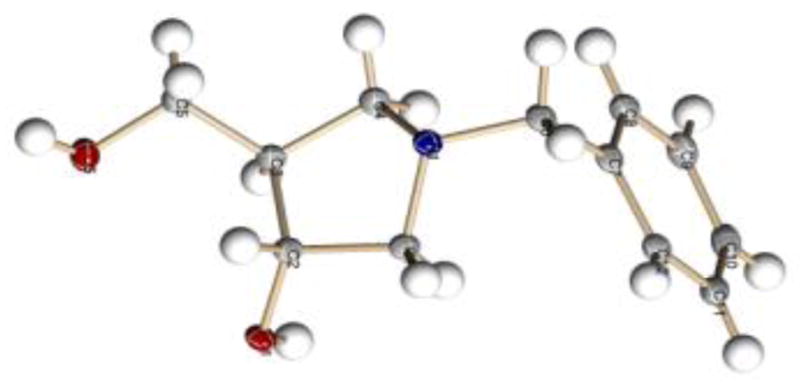
X-ray crystal structure of 1NBn (7) CCDC no: 825694.
Acknowledgments
We thank Dr. Sheng Cao, Dr. Micheal Leipold and Ms. Paige McKibbin for providing the purified enzymes used in this work. We also thank Jennifer Mason for useful discussions on the synthetic work. Andrew Ferreira and Dr. Jerry Dallas of the UCD NMR facility helped obtain the 11BNMR. This work was supported by grants from the National Cancer Institute of the National Institutes of Health (CA67985 and CA090689). We also acknowledge NSF (#0840444) for the department dual source diffractometer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bertozzi CR, Kiessling LL. Science. 2001;291:2357. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- 2.David SS, Williams SD. Chem Rev. 1998;98:1221. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 3.Berti PJ, McCann JAB. Chem Rev. 2006;106:506. doi: 10.1021/cr040461t. [DOI] [PubMed] [Google Scholar]
- 4.Henrissat B, Davies G. Curr Opin Struct Biol. 1997;7:637. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 5.Vocadlo DJ, Davies GJ. Curr Opin Chem Biol. 2008;12:539. doi: 10.1016/j.cbpa.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Volcadlo DJ, Bertozzi CR. Angew Chemie Int Ed. 2004;2004:5338. doi: 10.1002/anie.200454235. [DOI] [PubMed] [Google Scholar]
- 7.Witte MD, Kallemeign WW, Aten J, Li K-Y, Strijland A, Donker-Koopman WE, van den Nieuwendijk AMCH, Bleijlevens B, Kramer G, Florea BI, Hooibrink B, Hollak CEM, Ottehnhoff R, Boot RG, van der Marel GA, Voerkleeft HS, Aerts JMFG. Nat Chem Biol. 2010;6:907. doi: 10.1038/nchembio.466. [DOI] [PubMed] [Google Scholar]
- 8.Deng L, Scharer OD, Verdine GL. J Am Chem Soc. 1997;119:7865. [Google Scholar]
- 9.Jiang YL, Drohat AC, Ichikawa Y, Stivers JT. J Biol Chem. 2002;277:15385. doi: 10.1074/jbc.M200634200. [DOI] [PubMed] [Google Scholar]
- 10.Schärer OD, Nash HM, Jiricny J, Laval J, Verdine GL. J Biol Chem. 1998;273:8592. doi: 10.1074/jbc.273.15.8592. [DOI] [PubMed] [Google Scholar]
- 11.Werner RM, Stivers JT. Biochemistry. 2000;39:14054. doi: 10.1021/bi0018178. [DOI] [PubMed] [Google Scholar]
- 12.McCann JAB, Berti PJ. J Am Chem Soc. 2008;130:5789. doi: 10.1021/ja711363s. [DOI] [PubMed] [Google Scholar]
- 13.Bianchet MA, Seiple LA, Jiang YL, Ichikawa Y, Amzel LM, Stivers JT. Biochemistry. 2003;42:12455. doi: 10.1021/bi035372+. [DOI] [PubMed] [Google Scholar]
- 14.Amukele TK, Schramm VL. Biochemistry. 2004;43:4913. doi: 10.1021/bi0498508. [DOI] [PubMed] [Google Scholar]
- 15.Roday S, Amukele T, Evans GB, Tyler PC, Furneaux RH, Schramm VL. Biochemistry. 2004;43:4923. doi: 10.1021/bi0498499. [DOI] [PubMed] [Google Scholar]
- 16.Sturm MB, Tyler PC, Evans GB, Schramm VL. Biochemistry. 2009;48:9941. doi: 10.1021/bi901425h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturm MB, Roday SVLS. J Am Chem Soc. 2007;129:5544. doi: 10.1021/ja068054h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neeley WL, Essigmann JM. Chem Res Toxicol. 2006;19:491. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 19.Delaney S, Delaney JC, Essigmann JM. Chem Res Toxicol. 2007;20:1718. doi: 10.1021/tx700273u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.David SS, O’Shea VL, Kundu S. Nature. 2007;447:941. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fortini P, Pascucci B, Parlanti E, D’Errico M, Simonelli V, Dogliotti E. Mutat Res. 2003;531:127. doi: 10.1016/j.mrfmmm.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Biochemistry. 2008;47:7137. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bandaru V, Sunkara S, Wallace SS, Bond JP. DNA Repair. 2002;1:517. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 24.Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. DNA Repair. 2005;4:41. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Bandaru V, Blaisdell JO, Wallace SS. In: Methods in Enzymology. Judith C, Paul M, editors. Vol. 408. Academic Press; 2006. p. 15. [DOI] [PubMed] [Google Scholar]
- 26.Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, Izumi T. J Biol Chem. 2002;277:30417. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 27.Tchou J, Bodepudi V, Shibutani S, Antoshechkin I, Miller J, Grollman AP, Johnson F. J Biol Chem. 1994;269:15318. [PubMed] [Google Scholar]
- 28.Leipold MD, Muller JG, Burrows CJ, David SS. Biochemistry. 2000;39:14984. doi: 10.1021/bi0017982. [DOI] [PubMed] [Google Scholar]
- 29.Dizdaroglu M, Burgess SM, Jaruga P, Hazra TK, Rodriguez H, Lloyd RS. Biochemistry. 2001;40:12150. doi: 10.1021/bi015552o. [DOI] [PubMed] [Google Scholar]
- 30.Erzberger JP, Barsky D, Schärer OD, Colvin ME, Wilson DM. Nucleic Acids Res. 1998;26:2771. doi: 10.1093/nar/26.11.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chmiel NH, Livingston AL, David SS. J Mol Biol. 2003;327:431. doi: 10.1016/s0022-2836(03)00124-4. [DOI] [PubMed] [Google Scholar]
- 32.Chmiel NH, Golinelli MP, Francis AW, David SS. Nucleic Acids Res. 2001;29:553. doi: 10.1093/nar/29.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chepanoske CL, Porello SL, Fujiwara T, Sugiyama H, David SS. Nucleic Acids Res. 1999;27:3197. doi: 10.1093/nar/27.15.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porello SL, Willaims SD, Kuhn H, Michaels ML, David SS. J Am Chem Soc. 1996;118:10684. [Google Scholar]
- 35.Clinch K, Evans GB, Furneaux RH, Lenz DH, Mason JM, Mee SPH, Tyler PC, Wilcox SJ. Organic & Biomolecular Chemistry. 2007;5:2800. doi: 10.1039/b708796a. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen TB, Martel A, Dhal R, Dujardin G. Synthesis. 2009;2009:3174. [Google Scholar]
- 37.Zheng J-F, Chen W, Huang S-Y, Ye J-L, Huang P-Q. Beilstein J Org Chem. 2007;3:41. doi: 10.1186/1860-5397-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zambrano V, Rassu G, Roggio A, Pinna L, Zanardi F, Curti C, Casiraghi G, Battistini L. Org Biomol Chem. 2010;8:1725. doi: 10.1039/b924721a. [DOI] [PubMed] [Google Scholar]
- 39.Filichev VV, Pedersen EB. Tetrahedron. 2001;57:9163. [Google Scholar]
- 40.Abdel-Magid AF, Carson KG, Harris BD, Maryanoff CA, Shah RD. J Org Chem. 1996;61:3849. doi: 10.1021/jo960057x. [DOI] [PubMed] [Google Scholar]
- 41.Leipold MD, Workman H, Muller JG, Burrows CJ, David SS. Biochemistry. 2003;42:11373. doi: 10.1021/bi034951b. [DOI] [PubMed] [Google Scholar]
- 42.Bjoras M, Luna L, Johnsen B, Hoff E, Haug T, Rognes T, Seeberg E. EMBO J. 1997;16:6314. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnamurthy N, Haraguchi K, Greenberg MM, David SS. Biochemistry. 2008;47:1043. doi: 10.1021/bi701919u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radom CT, Banerjee A, Verdine GL. J Biol Chem. 2006;282:9182. doi: 10.1074/jbc.M608989200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.