Abstract
Chronic myeloid leukemia in chronic phase (CML-CP) cells that harbor oncogenic BCR-ABL1 and normal ABL1 allele often become resistant to the ABL1 kinase inhibitor imatinib. Here we report that loss of the remaining normal ABL1 allele in these tumors, which results from cryptic interstitial deletion in 9q34 in patients who did not achieve a complete cytogenetic remission during treatment, engenders a novel unexpected mechanism of imatinib resistance. BCR-ABL1-positive Abl1−/− leukemia cells were refractory to imatinib as indicated by persistent BCR-ABL1 -mediated tyrosine phosphorylation, lack of BCR-ABL1 protein degradation, increased cell survival and clonogenic activity. Expression of ABL1 kinase, but not a kinase-dead mutant, restored the anti-leukemic effects of imatinib in ABL1-negative CML cells and in BCR-ABL1-positive Abl1−/− murine leukemia cells. The intracellular concentration of imatinib and expression of its transporters were not affected, while proteins involved in BCR-ABL1 degradation were downregulated in Abl1−/− cells. Furthermore, twelve genes associated with imatinib resistance were favorably deregulated in Abl1−/− leukemia. Taken together, our results indicate that loss of the normal ABL1 kinase may serve as a key prognostic factor that exerts major impact on CML treatment outcomes.
Introduction
BCR-ABL1 results from t(9;22)(q34;q11) reciprocal translocation or variants generating the Philadelphia chromosome (Ph), which initiates CML-CP. The second (wild-type) ABL1 allele remains intact on the non-rearranged homologue of chromosome 9 and CML-CP cells at early stages express both forms of the ABL1 kinase, oncogenic BCR-ABL1 and normal ABL1 (1).
ABL1 and BCR-ABL1 can exert opposite effects on a variety of cellular functions (2). For example, BCR-ABL1 can act upstream and downstream of cytochrome c to inhibit apoptosis. In contrast, ABL1 kinase may facilitate apoptosis by stimulation of p73, p53, and caspase 9.
Tyrosine kinase inhibitor (TKI) imatinib revolutionized the treatment of BCR-ABL1-positive leukemias (3). The incidence of a continuous complete cytogenetic remission (CCyR) in CML-CP patients treated for 12 months with the drug was 66% (4). Mutations within the kinase domain of BCR-ABL1, over-expression of LYN kinase, loss of p53 and BCR-ABL1 amplification were implicated in the lack of achieving CCyR (5).
Here we demonstrate that loss of expression of normal ABL1 kinase due to cryptic deletion in remaining normal chromosome 9 [del(9q34)] reduced the sensitivity of BCR-ABL1 leukemia cells to imatinib and may contribute to drug resistance in CML patients.
MATERIALS AND METHODS
Chromosome and whole genome analysis of CML-CP samples
Bone marrow cells (BMCs) of CML-CP patients who failed to achieve CCyR within 12 months of TKI treatment were obtained after informed consent and analyzed at presentation and at 3-monthly intervals by G-banding and dual color/dual fusion probe fluorescent in situ hybridization (D-FISH) with a range of bacterial artificial chromosome (BAC) probes to detect the loss of normal 9q34 (6). DNA from BMCs of patients 1 and 3 was also analyzed by array comparative genomic hybridization (aCGH) using DNA Analytics (105K Agilent) and Formatter software (7). All genome addresses are derived from NCBI36/hg18 (March 2006) of the Human Genome. Additional information about the patients and cytogenetic and molecular results are described in Supplementary Materials and Supplementary Table 1.
Cells
Abl1+/− mice were kindly obtained from Dr. A.J. Koleske (Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT, USA) and bred to obtain −/− and +/+ littermates. Animal studies were approved by the Institutional Animal Care and Use Committee at Temple University. p210BCR-ABL1 –positive growth factor-independent leukemia cells were generated by retroviral infection of Abl1+/+ and Abl1−/− BMCs with pMIG-BCR-ABL1-IRES-GFP retroviral construct as previously described (8). CML-BP LAMA84R and KCL22 cell lines were described before (9, 10). LAMA84R and KCL22 cells, and GFP+ BCR-ABL1 –positive Abl1−/− leukemia cells were infected with pKI retroviral construct encoding YFP-ABL1 fusion protein or kinase-dead YFP-ABL1(K290R) mutant (kindly obtained from Dr Koleske). GFP, YFP and YFP/GFP -positive cells were sorted and expanded in growth factor-free conditions.
Sensitivity to imatinib
Cells were treated with imatinib (Novartis Pharma, Basel, Switzerland) and evaluated by clonogenic assay as described before (8). TKI-resistant BCR-ABL1 kinase mutations were not detected in cells used for these experiments.
Imatinib retention
Radiolabeled drug uptake was performed using 14C-labeled imatinib (Novartis) as previously described with modifications (11). Briefly, 2 × 106 cells were incubated with 1.6μM 14C-labeled imatinib (3,052 MBq/mg) at 37°Cfor 2 hours. After incubation, cells were washed twice with ice-cold phosphate-buffered saline and incubated in culture medium at 37°C for another 15 minutes. Cell pellet was then solubilized in 50 μl of distilled water and radioactivity was counted using β-counter (Perkin Elmer, Waltham MA, USA).
Protein expression
Total cell lysates were analyzed by Western blotting using primary antibodies recognizing ABL1, Abcb1, CHIP and tubulin (Calbiochem, San Diego, CA, USA), phosphotyrosine (Upstate, Lake Placid, NY, USA), Bag1, Cbl and GFP (Santa Cruz Biotechnology, Santa Cruz, CA, USA), Oct-1 (Novus Biologicals, Littleton, CO, USA), Abcg2, HSP90 and cathepsin B (Abcam Inc., Cambridge, MA, USA), and Hsc70 (Enzo Life Sciences International, Inc., Plymouth Meeting, PA, USA) as described before (8).
Genome-wide expression array
Affymetrix Mouse gene 1.0ST array containing 28,815 probe sets (Affymetrix, Santa Clara, CA, USA) was used to measure mRNA expression levels. Affymetrix arrays were processed by Partek Genomic Suite at the Penn Molecular Profiling Facility - Bioinformatics Group (University of Pennsylvania, Philadelphia, PA, USA) to determine whether a given transcript was present and if there were consistently significant differences between BCR-ABL1 –positive Abl−/− and BCR-ABL1 –positive Abl+/+ BMC based on three separate experiments. Genes were considered to have a significant differential expression between the two groups when displayed a False Discovery Rate (FDR) not exceeding 5% and a cut-off value > 1-fold (up-regulated or down-regulated).
Results and Discussion
Using G-banding, D-FISH, and aCGH we detected that three CML patients who initially failed to achieve CCyR within 12 months of TKI treatment acquired a cryptic deletion in 9q34 region in the normal chromosome 9 [del(9q34)] resulting in the loss of normal ABL1 allele (Figure 1 and 2, Supplementary Table 1 and Figure 1). Two of these are among 21 CML–CP patients without CCyR on first line TKI we analyzed in the lab. Importantly, in addition to the cryptic loss at 9q34.1 all three patients showed karyotype evolution – from the presence of a second Ph (Figure 2A) to multiple numerical and structural aberrations (Supplementary Table 1).
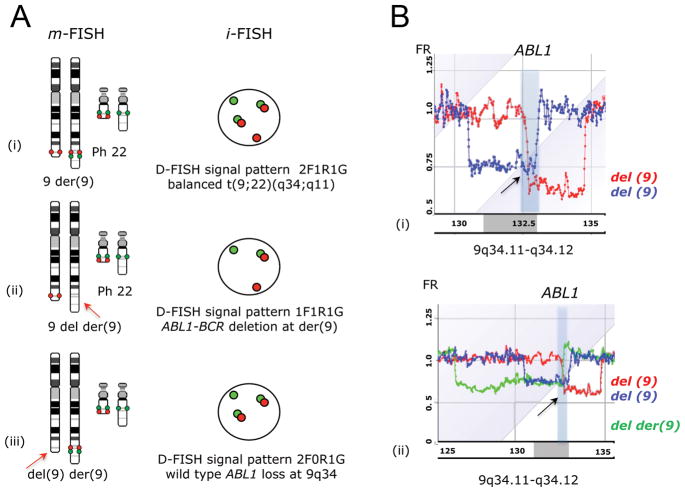
Figure 1. Deletions of the normal ABL1 allele in CML-CP patients.
(A) The classical and variants of t(9;22)(q34;q11) are usually balanced and reciprocal translocations, however the translocation may be unbalanced. (i) balanced t(9;22)(q34;q11) with two fusion signals on Ph and der(9), one red signal and one green signal on the normal 9q34 and 22q11 respectively (2F1R1G), (ii) t(9;22)(q34;q11) accompanied by the loss of ABL1-BCR signal at der(9) (1F1R1G), and (iii) t(9;22)(q34;q11) accompanied by the loss of wild-type ABL1 at del(9) (2F0R1G). (B) aCGH results: (i) the 9q34-qter region of patient 1 showing a 2.6Mb genome loss including the entire ABL1 together with proximally flanking sequences leading to 2F0R1G aberrant FISH signal pattern (blue) and a 1.8Mb loss in patient 3 encompassing sequences downstream of the ABL1 breakpoint (red), and (ii) added for comparison the ‘classic’ der(9) deletion (graph in green) involves the 5′ ABL1 and sequences proximal to the ABL1 breakpoint thus resulting in aberrant 1F1R1G pattern.
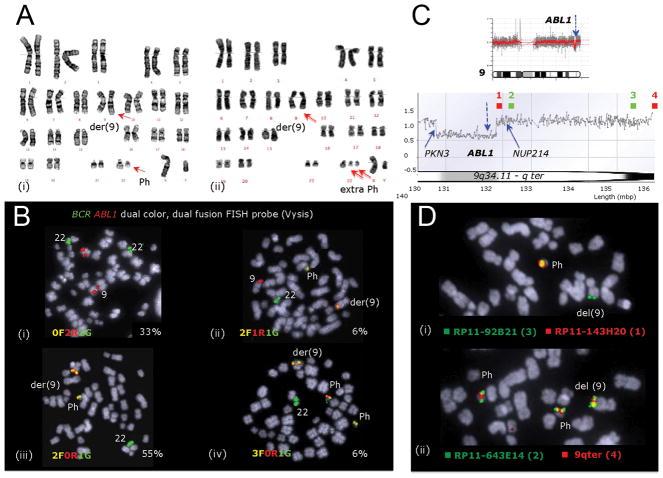
Figure 2. Analysis of del(9) causing a loss of the wild-type ABL1 allele during imatinib treatment in patient #1.
(A) G-banding analysis identified (i) a t(9;22)(q34;q11) and (ii) an additional Ph in 20% of cells. (B) FISH analysis showed that (i) 33% of the cells were BCR-ABL1 negative (0F2R2G), (ii) 6% were BCR-ABL1 positive with fusion products on both the Ph and der(9) chromosome and signals from the normal 9q34 and 22q11 regions (2F1R1G) as expected, and (iii) 55% had an abnormal 2F0R1G signal pattern indicating cryptic loss of the ABL1 signal at 9q34 [del(9)] and (iv) 6 % had in addition to del(9) also an extra Ph. (C) Chromosomes 9 aCGH profile indicates the loss at 9q34.1 (top, blue arrow) with a close-up of 9q34.1-qter region (bottom), showing that the breakpoints fall within the PKN3 gene and downstream of ABL1. The locations of BAC probes (1–4) are shown in green and red. (D) FISH mapped the missing ABL1 sequences to del(9) affecting the wild-type allele of ABL1 and confirmed the location of the distal breakpoint between (i) BACs RP11-143H20 [note the missing red signal from del(9)] and (ii) RP11-643E14 within a 128 Kb region containing NUP214 gene.
In patient 1 FISH analysis revealed an aberrant signal pattern due to a missing ABL1 signal, which was mapped to the morphologically normal chromosome 9 indicating a cryptic deletion (Figure 1A(iii) and 1B). In patient 2 the deletion was detectable by G-banding and assessed as del(9q31?;q34) (Supplementary Table 1). In patient 3 the loss of ABL1 was revealed by aCGH and confirmed by FISH mapping (Figure 1B, Supplementary Figure 1). aCGH results for the 9q34 region in patient 1 (blue) and patient 3 (red) show the extent of the cryptic deletions (Figure 1B(i)). The common loss is defined proximally by the ABL1 breakpoint in patient 3 and distally by the telomeric breakpoint in patient 1 (arrow in Figure 1B(i), Figure 2C, D, and Supplementary Figure 1). The estimated size of the common genome loss is 567Kb, which includes ABL1 exons a2 to a11 together with downstream sequences encompassing the LAMC3 and NUP214 genes. These deletions differ significantly from the deletions at der(9), where the genome loss involves only ABL1 exons 1a and 1b and spans towards the centromere (Figure 1A(ii) and 1B(ii)).
The observed loss of the wild allele of ABL1 is an evolutionary event as evidenced by the presence of Ph(+) cells with and without ABL1 deletion in patient 1 (Figure 2B). Furthermore, it is the ABL1 deficient cell clone that sustains the disease progression by acquiring a second copy of Ph (Figure 2A, B, D). Our observation combined with other report that inhibition of Abl1 kinase compromises genomic stability suggests that loss of ABL1 not only decreases imatinib sensitivity but also promotes accumulation of chromosomal aberrations (12).
Cryptic deletions in 9q34 causing the loss of normal ABL1 allele may be under-reported in CML-CP patients probably because they would be missed unless either D-FISH or aCGH have been used for monitoring therapy response. In concordance, similar del(9q34) was found in several CML-BP cell lines (Supplementary Table 2). To detect loss of the ABL1 signal from the normal 9 homologue, FISH using a BCR-ABL1-ASS tricolor dual fusion translocation probe could be recommended (Supplementary Figure 2), which produces unique signals for the translocation products and the normal non-rearranged loci at 9q34 and 22q11. Importantly, identification of del(9q34) using any of the two double fusion BCR-ABL1 probe sets is as reliable on interphase cells as on chromosome preparations. In contrast, the popular ES-FISH probe creates in BCR-ABL1 positive cell with ABL1 loss a signal pattern (1R1G1F) that cannot reliably differentiate the ‘wild’ ABL1 allele from the ABL1-BCR fusion (Supplementary Figure 2) thus misreporting del(9q34) as deletions at der(9).
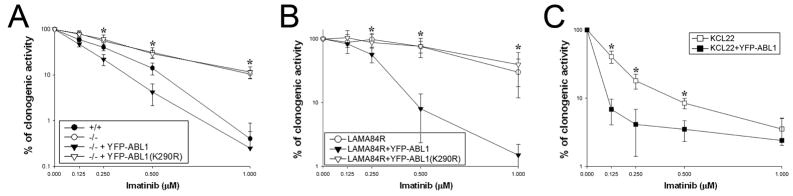
To prove that loss of ABL1 directly contributes to imatinib resistance, BCR-ABL1 was expressed in Abl1−/− and +/+ BMCs. The absence of Abl1 reduced the sensitivity of BCR-ABL1 leukemia cells to imatinib whereas expression of YFP-ABL1 fusion kinase, but not the kinase-dead YFP-ABL1(K290R) mutant, in BCR-ABL1 –positive Abl1−/− leukemia cells restored anti-leukemia effect of the drug (Figure 3A). YFP-ABL1, but not YFP-ABL1(K290R), also increased imatinib sensitivity in drug-resistant CML-BP cell line LAMA84R (ABL1-negative, Supplementary Table 2) (Figure 3B). Since BCR-ABL1 gene amplification and overexpression of the multidrug resistance P-glycoprotein was observed in LAMA84R cells, loss of ABL1 may collaborate with other genetic/epigenetic abnormalities to promote drug resistance in CML-BP (9). Moreover, expression of YFP-ABL1 kinase in KCL22 CML-BP cells (relatively low ABL1 expression (10)) increased their sensitivity to imatinib suggesting that increased BCR-ABL1: ABL1 ratio observed during the course of disease can also limit the effect of imatinib (Figure 3C) (13).
Figure 3. ABL1 kinase regulates imatinib sensitivity of BCR-ABL1 leukemias.
(A) BCR-ABL1 –positive Abl1−/− leukemia cells (−/−), BCR-ABL1 –positive Abl1+/+ leukemia cells (+/+) and BCR-ABL1 –positive Abl1−/− leukemia cells reconstituted with YFP-ABL1 and YFP-ABL1(K290R), (B) LAMA84R cells and these transfected with YFP-ABL1 and YFP-ABL1(K290R), and (C) KCL22 cells and these transfected with YFP-ABL1 were incubated with imatinib and clonogenic cells were counted. Results represent mean percentages ± s.d. of control untreated cells; *p<0.05 in comparison to other group(s) as determined by two-tailed Student t test.
The fact that ABL1 kinase may regulate the sensitivity of CML cells to imatinib is rather unexpected because BCR-ABL1 and ABL1 kinases are equally sensitive to imatinib in vitro (3). However, inhibition of intracellular ABL1 kinase usually requires higher concentration of the drug in comparison to BCR-ABL1 kinase; in addition ABL1 may work in a kinase-independent manner (14, 15). Moreover, imatinib-induced inhibition of BCR-ABL1 kinase is associated with release of ABL1 from the complex with 14-3-3 sigma, which promotes ABL1 relocation to the nucleus (triggers p73-dependent apoptosis), to the mitochondrial membranes (causes the loss of mitochondrial membrane potential) and to the complex with caspase 9 (activates caspase cascade) (2, 16). In summary, the presence of ABL1 kinase may exert a significant impact on anti-CML effect of imatinib. This speculation is supported by the observation that expression of YFP-ABL1 fusion kinase, but not its kinase-dead K290M mutant restored sensitivity to imatinib in BCR-ABL1 –positive Abl1−/− leukemia cells and LAMA84R (ABL1-negative) CML-CP cells (Figure 3A and B).
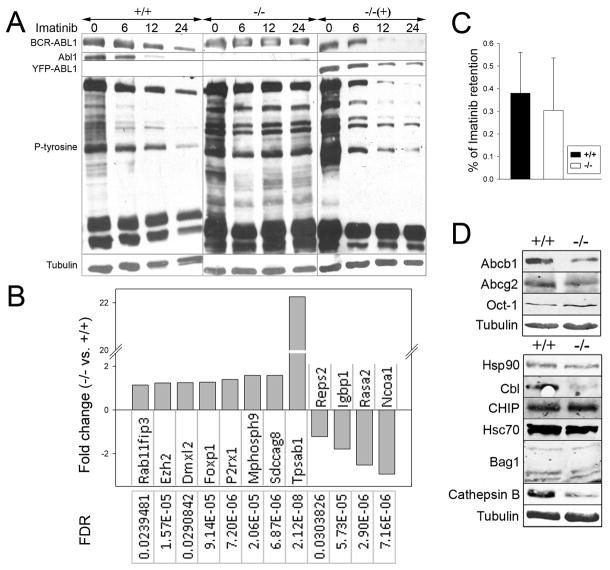
In Abl1−/− leukemia cells, imatinib displayed reduced capability to inhibit BCR-ABL1 kinase-mediated tyrosine phosphorylation and to induce BCR-ABL1 protein degradation in comparison to Abl1+/+ counterparts and Abl1−/− leukemia reconstituted with YFP-ABL1 (Figure 4A). Moreover, genome-wide array confirmed imatinib-resistant signature of BCR-ABL1 –positive Abl1−/− cells by detecting deregulated expression of 12 genes previously reported in imatinb-resistant CML patients (Figure 4B) (17, 18). Intracellular retention of imatinib, and expression of drug importer Oct-1 and exporters Abcb1 and Abcg2, appear unaffected by Abl1 (Figure 4C, D), but the impact of Abl1 on metabolism of imatinib cannot be excluded. On the other hand, >10-fold downregulation of Cbl E3 ligase, which induce ubiquitin-dependent degradation of “mature” BCR-ABL1 protein, and/or >3-fold downregulation of cathepsin B, which cleaves BCR-ABL1 may be responsible for lack of degradation of BCR-ABL1 protein in imatinib-treated Abl1−/− cells (Figure 4D) (19, 20). Hsc70, Bag1 and E3 ligase CHIP responsible for degradation of “immature” BCR-ABL1 protein, and chaperone protein Hsp90 protecting BCR-ABL1 from proteasomal degradation, are not affected by Abl1 (Figure 4D) (19). Downregulation of BCR-ABL1 in imatinib-treated CD34+ CML-CP cells was implicated in regulating their sensitivity to the drug (20).
Figure 4. Imatinib-resistant phenotype of BCR-ABL1 –positive Abl1−/− leukemia cells.
BCR-ABL1 –positive Abl1−/− leukemia cells (−/−), BCR-ABL1 –positive Abl1+/+ leukemia cells (+/+) and BCR-ABL1 –positive Abl1−/− leukemia cells reconstituted with YFP-ABL1 (−/− (+)) were used. (A) Western analysis of the total cell lysates from cells incubated with 1μM imatinib for 0, 6, 12 and 24 hrs. (B) Statistically significant (FDR<0.05) fold-changes (>1) of the expression of indicated genes in BCR-ABL1-positive Abl1−/− versus BCR-ABL1-positive Abl1+/+ samples. (C) Intracellular retention of imatinib; results represent mean percentages ± s.d. of total C14-imatinib. (D) Western blots of total cell lysates to detect imatinib transporters (upper box) and proteins involved in BCR-ABL1 degradation (lower box).
Altogether, it can be postulated that loss of expression of ABL1 kinase may contribute to imatinib resistance in CML-CP patients which do not achieve CCyR during 12 months on imatinib and eventually progress to CML-BP. ABL1 loss in CML-CP can be achieved by interstitial deletion in chromosome 9 [del(9q34)] causing a loss of normal ABL1 allele (this report), which may be combined with epigenetic silencing of the alternative ABL1 promoter retained in t(9;22) (13). Therefore, detection of del(9q34) may serve as an important prognostic factor and have a significant impact on CML treatment.
Supplementary Material
Acknowledgments
Financial support: Supported by the grants from NIH/NCI CA123014 (T. Skorski), Leukemia and Lymphoma Research Fund 05098 (E.N.), 1M19/NK1W/2009 (T. Stoklosa) and 1M19/WB1/2010 (E.G-M.) from Medical University of Warsaw.
We thank H. Mazzullo, J. Howard-Reeves, D. Brazma, C. Grace and Katia Gancheva for their contribution to the cytogenetic and aCGH analysis as well and L. Foroni for the BCR-ABL1 mutation analysis. We also thank Dr. J. Tobias for help with statistical analysis of the microarray data.
References
- 1.Diamond J, Goldman JM, Melo JV. BCR-ABL, ABL-BCR, BCR, and ABL genes are all expressed in individual granulocyte-macrophage colony-forming unit colonies derived from blood of patients with chronic myeloid leukemia. Blood. 1995;85:2171–5. [PubMed] [Google Scholar]
- 2.Colicelli J. ABL tyrosine kinases: evolution of function, regulation, and specificity. Sci Signal. 2010;3:re6. doi: 10.1126/scisignal.3139re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Druker BJ, Tamura S, Buchdunger E, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–6. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 4.Kantarjian H, Shah NP, Hochhaus A, et al. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2010;362:2260–70. doi: 10.1056/NEJMoa1002315. [DOI] [PubMed] [Google Scholar]
- 5.Quintas-Cardama A, Kantarjian HM, Cortes JE. Mechanisms of primary and secondary resistance to imatinib in chronic myeloid leukemia. Cancer Control. 2009;16:122–31. doi: 10.1177/107327480901600204. [DOI] [PubMed] [Google Scholar]
- 6.Virgili A, Brazma D, Reid AG, et al. FISH mapping of Philadelphia negative BCR/ABL1 positive CML. Mol Cytogenet. 2008;1:1–13. doi: 10.1186/1755-8166-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nacheva EP, Brazma D, Virgili A, et al. Deletions of immunoglobulin heavy chain and T cell receptor gene regions are uniquely associated with lymphoid blast transformation of chronic myeloid leukemia. BMC Genomics. 2010;11:41. doi: 10.1186/1471-2164-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slupianek A, Poplawski T, Jozwiakowski SK, et al. BCR/ABL stimulates WRN to promote survival and genomic instability. Cancer Res. 2011;71:842–51. doi: 10.1158/0008-5472.CAN-10-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahon FX, Deininger MW, Schultheis B, et al. Selection and characterization of BCR-ABL positive cell lines with differential sensitivity to the tyrosine kinase inhibitor STI571:diverse mechanisms of resistance. Blood. 2000;96:1070–9. [PubMed] [Google Scholar]
- 10.Bueno MJ, Perez de Castro I, Gomez de Cedron M, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 11.Thomas J, Wang L, Clark RE, Pirmohamed M. Active transport of imatinib into and out of cells: implications for drug resistance. Blood. 2004;104:3739–45. doi: 10.1182/blood-2003-12-4276. [DOI] [PubMed] [Google Scholar]
- 12.Fanta S, Sonnenberg M, Skorta I, et al. Pharmacological inhibition of c-Abl compromises genetic stability and DNA repair in Bcr-Abl-negative cells. Oncogene. 2008;27:4380–4. doi: 10.1038/onc.2008.68. [DOI] [PubMed] [Google Scholar]
- 13.Zion M, Ben-Yehuda D, Avraham A, et al. Progressive de novo DNA methylation at the bcr-abl locus in the course of chronic myelogenous leukemia. Proc Natl Acad Sci U S A. 1994;91:10722–6. doi: 10.1073/pnas.91.22.10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Mishra N, Raina D, Saxena S, Kufe D. Abrogation of the cell death response to oxidative stress by the c-Abl tyrosine kinase inhibitor STI571. Mol Pharmacol. 2003;63:276–82. doi: 10.1124/mol.63.2.276. [DOI] [PubMed] [Google Scholar]
- 15.Sawyers CL, McLaughlin J, Goga A, Havlik M, Witte O. The nuclear tyrosine kinase cAbl negatively regulates cell growth. Cell. 1994;77:121–31. doi: 10.1016/0092-8674(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 16.Mancini M, Corradi V, Petta S, et al. A New Non-Peptidic Inhibitor of the 14-3-3 Docking Site Induces Apoptotic Cell Death in Chronic Myeloid Leukemia Sensitive or Resistant to Imatinib. J Pharmacol Exp Ther. 2010;2010:1. doi: 10.1124/jpet.110.172536. [DOI] [PubMed] [Google Scholar]
- 17.Frank O, Brors B, Fabarius A, et al. Gene expression signature of primary imatinib-resistant chronic myeloid leukemia patients. Leukemia. 2006;20:1400–7. doi: 10.1038/sj.leu.2404270. [DOI] [PubMed] [Google Scholar]
- 18.de Lavallade H, Finetti P, Carbuccia N, et al. A gene expression signature of primary resistance to imatinib in chronic myeloid leukemia. Leuk Res. 2009;34:254–7. doi: 10.1016/j.leukres.2009.09.026. [DOI] [PubMed] [Google Scholar]
- 19.Tsukahara F, Maru Y. Bag1 directly routes immature BCR-ABL for proteasomal degradation. Blood. 2010;2010 doi: 10.1182/blood-2009-10-249623. in press. [DOI] [PubMed] [Google Scholar]
- 20.Puissant A, Colosetti P, Robert G, Cassuto JP, Raynaud S, Auberger P. Cathepsin B release after imatinib-mediated lysosomal membrane permeabilization triggers BCR-ABL cleavage and elimination of chronic myelogenous leukemia cells. Leukemia. 2010;24:115–24. doi: 10.1038/leu.2009.233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.