Abstract
Biomimetic scaffolds have been proposed as a means to facilitate tissue regeneration by multipotent stromal cells (MSCs). Effective scaffold colonization requires a control of multiple MSC responses including survival, proliferation, differentiation, and migration. As MSC migration is relatively unstudied in this context, we present here a multi-level approach to its understanding and control, integratively tuning cell speed and directional persistence to achieve maximal mean free path (MFP) of migration. This approach employs data-driven computational modeling to ascertain small molecule drug treatments that can enhance MFP on a given materials substratum. Using poly(methyl methacrylate)- graft-poly(ethylene oxide) polymer surfaces tethered with epidermal growth factor (tEGF) and systematically adsorbed with fibronectin, vitronectin, or collagen-I to present hTERT-immortalized human MSCs with growth factor and extracellular matrix cues, we measured cell motility properties along with signaling activities of EGFR, ERK, Akt, and FAK on 19 different substrate conditions. Speed was consistent on collagen/tEGF substrates, but low associated directional persistence limited MFP. Decision tree modeling successfully predicted that ERK inhibition should enhance MFP on collagen/tEGF substrates by increasing persistence. Thus, we demonstrated a two-tiered approach to control MSC migration:materials-based “coarse-graining” complemented by small molecule“fine-tuning”.
Keywords: mesenchymal stem cells, cell migration, extracellular matrix, epidermal growth factor, computational modeling
1. Introduction
Multipotent stromal cells (MSCs) derived from bone marrow comprise heterogeneous populations of self-renewing “mesenchymal” stem cells as well as their further differentiated downstream progenitors[1]. MSCs hold promise for bone tissue engineering applications because of their ability to home to injury sites and to differentiate along mesodermal lineages to become osteocytes, chondrocytes, and adipocytes and aid in bone and cartilage repair and regeneration[2–4]. One key challenge to the therapeutic success of MSCsis the colonization of MSCs in biomimetic scaffolds to aid tissue integration[5,6]. Toward this end, a variety of growth factors and biomaterials have been investigated for their effects on MSC survival and proliferation[7–11]. In particular, epidermal growth factor (EGF) has been found to promote MSC survival and proliferation without biasing the stem cells down any particular lineage, thus retaining their multipotent differentiation potential[12,13]. Moreover, EGF immobilized to a biomaterials substrate (tEGF for ‘tethered EGF’) promoted MSC spreading and survival under Fas ligand antagonism, whereas soluble EGF did not, indicating a protective advantage for the growth factor-tethered scaffold to MSCs under anticipated pathophysiological stress conditions[14]. Despite migration being vital for vigorous MSC infiltration of scaffolds, to enable population of regions beyond initial seeding sites, little information is available for EGF-related MSC migration behavior that could aid in integrative scaffold design. This motivates our work to characterize and understand MSC migration to improve biomaterials scaffolds development.
Cell migration is a delicately coordinated biophysical process governed by a variety of extracellular molecular and mechanical cues via intracellular biochemical signals[15]. Membrane protrusions at the cell front adhere to the substrate, followed by contractile forces that translocate the cell body, and ultimately adhesion disassembly at the cell rear allows for detachment and productive forward motion. Migrating cells cycle through these biophysical processes with each step mediated by underlying signaling pathways downstream of stimulatory cues such as extracellular matrix (ECM) proteins and growth factors including EGF[16]. Little is known about MSC signaling control, although some hints might be obtained from extensive studies in other adherent mesenchymal cells such as fibroblasts. EGF receptor (EGFR) is expressed at moderate levels (approximately 10,000 #/cell) in fibroblasts and its activation induces a signaling network involving multiple pathways including PLCγ, ERK, and PI3K as well as additional propagated signals further downstream, which cooperatively regulate lamellipod protrusion, cytoskeletal contractile force generation, and dynamic adhesion/deadhesion [17]. PLCγ is involved in lamellipod protrusion and ERK modulates adhesion dynamics via multiple mechanisms [18,19]. Downstream of PI3K, ROCK and MLCKare involved in contractile force generation[20] while Akt influences actin cytoskeleton reorganization[21]. Concomitantly, integrinmediated adhesion to extracellular matrix (ECM) and affiliated mechanical forces generate signals that synergize with these pathways. The integrin adhesion complex-associated focal adhesion kinase (FAK) plays a central role in integrating crosstalk with growth factor receptors qualitatively and quantitatively[22,23]. Since the overall migration behavior produced by these various pathways depends on the quantitative relationships among them, and as MSCs express substantially fewer EGFR[21] than most of the cell types for which EGF-elicited motility signaling has been investigated, analogous studies dedicated to MSC behavior is clearly motivated.
To date, most studies of MSC migration employ transmembrane assays (e.g.,[21,24]), which are unable to capture vital characteristics of cell motility processes. In this new study, we track migration via live single-cell video microscopy to quantitatively analyze MSC migration in terms of two independent properties: translational speed and directional persistence [25]. A foundational premise is that the greatest product of translational speed and directional persistence will facilitate most vigorous infiltration of biomaterials scaffold; this product can be termed ‘mean free path’ (MFP), and it essentially characterizes the average distance a cell moves before changing direction significantly[25]. Using a poly(methyl methacrylate)-grafted-poly(ethylene oxide) copolymer surface as a model of biomimetic scaffolds, we systematically present fibronectin, vitronectin, and collagen-Ias alternative ECM cues along with tEGF for migration of hTERT-immobilized MSCs to ascertain their respective effects on cell speed and persistence. Furthermore, to gain basic insights concerning key signaling pathways controlling migration speed and persistence, we measured EGFR, ERK, Akt, and FAK phosphoprotein activities in MSCs, hypothesizing that one or more of these might exhibit significant predictive capacity for migration under the various conditions. Finally, in order to analyze these multi-variate data most usefully, we apply a ‘systems biology’ computational modeling technique previously shown to be useful for predictive understanding of fibroblast migration governed integratively by ECM and EGF cues[26,27].
2. Materials and methods
2.1. Cells and cell culture
Human telomerase reverse transcriptase immortalized multi-potent stromal cells (hTERTMSCs) were used as a model cell system for MSCs and were a generous gift from Dr. Junya Toguchida[28]. Cells were routinely cultured in Dulbecco’s modified eagle’smedium (DMEM) supplemented with 10% fetal bovine serum (Gemini Bio-Products, West Sacramento, CA), 1% Lglutamine, 1% penicillin-streptomycin, 1% sodium pyruvate, and 1% non-essential amino acids (all from Invitrogen, Carlsbad, CA). Cells were cultured at 37°C with 5% CO2, split when they were ~90% confluent, and used between passages 10 to 25 for all experiments.
2.2. Surface preparation for presenting growth factor and ECM cues
Polymer-coated glass coverslips were prepared and modified with tEGF as described previously[14] and further adsorbed with ECM proteins (Fig. 1). The base polymer was a poly(methyl methacrylate)-graft-poly(ethylene oxide) (PMMA-g-PEO) comb copolymer consisting of a hydrophobic PMMA backbone off of which extend PEO sidechains. By altering the total weight percent (wt%) of PEO during synthesis, the density of the sidechains can be systematically controlled. We synthesized two polymers as previously described by the Griffith laboratory[29,30]: Comb 1 and Comb 2, with 33 wt% PEO and 22 wt% PEO respectively. Comb 1 with its higher density PEO sidechains is resistive to cell adhesion, whereas Comb 2 allows for protein adsorption and subsequent cell adhesion. Furthermore, Comb 1 allows for the presentation of tEGF in a locally dense concentration suitable for receptor dimerization[31]. PEO sidechain ends in Comb 1 were activated with 4-nitrophenyl chloroformate (NPC) to react and covalently link EGF, and a 60:40 ratio blend of Comb 1 to Comb 2was used to achieve optimal tEGF presentation while still allowing for protein and cell adhesion. The mixed polymer blend was dissolved to 20 mg/mL in toluene and spin coated to glass coverslips. Murine EGF (Peprotech, Rocky Hill, NJ) was coupled to the activated PEO sidechains by incubation with a solution of 25 ug/mL EGF in 0.1 M phosphate buffer, pH 8.7, at room temperature for 20 hours. Control surfaces were incubated for the same period of time with phosphate buffer without EGF. The tethered surfaces were then blocked with 0.1 M Tris buffer, pH 9, at room temperature for 2 hours to react any remaining NPC, followed by phosphate buffered saline (PBS) rinses to achieve approximately 5,000–7,000 tethered EGF molecules per um2. ECM proteins were coated onto the surfaces via non-specific adsorption. Surfaces were incubated at room temperature for 2 hours with solutions of stock fibronectin, vitronectin, or collagen-I diluted to the target concentration in PBS, then blocked with 1% bovine serum albumin (BSA) for 1 hour at room temperature, sterilized with UV light for 30 minutes and used immediately.
Figure 1.
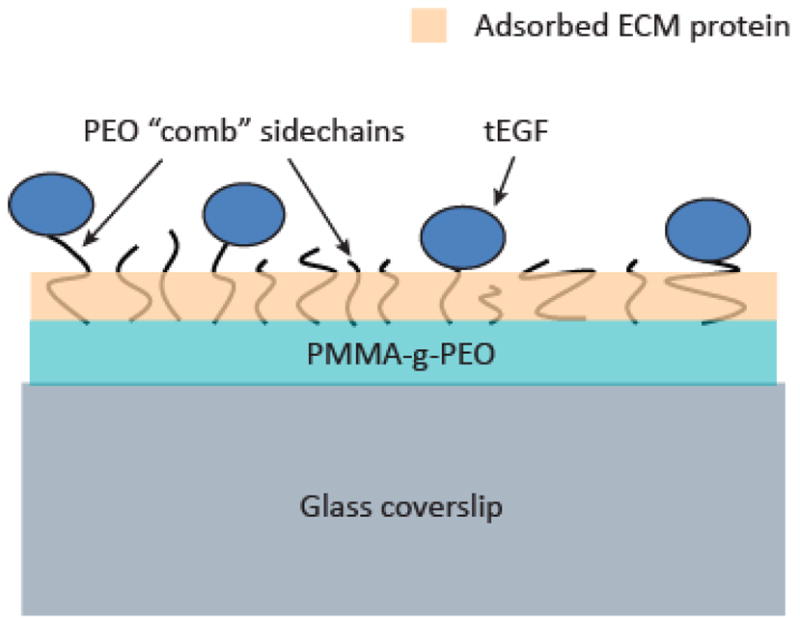
Illustration of PMMA-g-PEO polymer materials modification to provide growth factor and ECM cues influencing MSC migration. Solvent-dissolved polymer was spincoated onto glass coverslips, and the PEO sidechains reacted to tether EGF (tEGF). Varying concentrations of fibronectin, vitronectin, or collagen were adsorbed onto the polymer surface.
2.3. Cell migration tracking and quantification
Substrates were glued to DeltaT (Bioptechs) imaging dishes with minimal epoxy, and MSCs sparsely seeded(~4,000 cells/cm2) in 2 mL of media.18 hours post-seeding, the plates were sealed with a vacuum grease-lined cover glass lid and placed in a heated stage insert for a Ludl 99S008 motorized stage on a Zeiss Axiovert 35 microscope.15 randomly selected fields of cells, with five to ten cells per field, were tracked by recording an image for each field every 10 minutes for seven hours. On the order of one hundred cells were tracked for each replicate of each condition, typically yielding approximately two hundred cells for each condition aggregated across replicates. Individual cell coordinates were determined using Visible (Reify Corporation, Cambridge, MA), which differentiates cells from the background by comparing time-adjacent images locating clusters of changing pixels of a threshold size and assigning coordinates to each cluster. Extracting these coordinates to Excel (Microsoft Corp., Redmond, WA), we removed those associated with cells dividing, colliding, and/or exiting the field of view and calculated average speed for the remaining time points. Using the Persistent Random Walk model with overlapping time interval sampling [32], implemented in Mat lab (Natick, MA), we fitted for each cell’s persistence time. For EGFR and ERK inhibition studies, AG1478 (EMD Chemicals, Gibbstown, NJ) and U0126 (Selleck Chemicals, Houston, TX) were respectively added to the media at the time of seeding.
2.4. Phosphoprotein measurements
Each substrate was placed in individual wells of a 12-well plate and seeded with~31,000 cells/cm2 (80,000 cells per well) in 1 mL of media and 10 uM AG1478 to inhibit EGFR and minimize tEGF signaling during the attachment and spreading phases. Cells were allowed to attach and spread for 5 hours after which the media was replaced with normal media without EGFR inhibitor, thereby starting the tEGF stimulation. Cells were lysed and collected 0, 5, 15, 30, 60, and 180 minutes after tEGF treatment. At each time point, substrates were transferred to new 12-well plates, washed, lysed with proprietary cell lysis buffer (Bio-Rad Laboratories, Hercules, CA), collected and centrifuged for 10 minutes at 13,000 rpm at 4°C. The supernatant was collected and stored at −80°C until use, with a small amount removed to quantify protein concentrations via bicinchoninic acid (BCA) assay (Pierce, Rockford, IL).
Phosphorylated EGFR (Tyr), phosphorylated ERK 1/2 (Thr202/Tyr204, Thr185/Tyr187), and phosphorylated Akt (Ser473) were measured via the Bio-Plex suspension array system using Bio-Rad Phosphoprotein Kits (Bio-Rad Laboratories, Hercules, CA). Bio-Plex uses the Luminex xMAP platform, high-throughput, multiplexible, and quantitative bead-based assay to quantify protein. Antibodies immobilized to differentially dyed fluorescent beads bind the target proteins in cell lysates, one dye per target, and quantifies phosphoprotein activity via a detection antibody specific to the phosphorylation site. Briefly, 10 ug of total lysate protein were diluted with proprietary lysis buffer and assay buffer to a final protein concentration of 200 ug/mL and incubated with the appropriate beads in filter plates (Millipore, Billerica, MA) overnight. Unbound protein was aspirated away by vacuum filtration, trapping the beads in the wells of the plate. Biotinylated detection antibodies for specific phosphorylation sites were incubated with the beads for 30 minutes, followed by 10 minutes incubation with streptavidin phycoerythin (Strep-PE), fluorescently tagging the detection antibody. Fluorescence was quantified using the Bioplex 200 System reading at least 25 beads per condition per target protein.
Phosphorylated FAK (Tyr378) was measured via enzyme-linked immunosorbent assay (ELISA) kits (Millipore, Billerica MA) following manufacturer protocols.10 ug of cell lysate protein per condition were diluted in ELISA diluent to a total volume of 50 uL and loaded into 96-well plates pre-coated with capture antibody, and a detection antibody specific for the Tyr378 site added to each well. The plate was incubated at room temperature with constant agitation for three hours, washed with vendor-supplied buffer, and then incubated with 100 uL of HRP-conjugated antibody against the phosphospecific antibody for 30 minutes at room temperature with constant agitation. After washing, the plates were incubated with 100 uL of TMB detection reagent, protected from light, and 100 uL of stop solution added after appropriate color development (5–10 minutes). Each well’s absorbance was read at 450 nm, and fluorescence was normalized to a standard curve generated with increasing concentrations of vendor-supplied pFAK.
2.5. Decision tree modeling
We employed decision tree modeling to analyze our ‘cue-signal-response’ data set following our previous successful demonstration of this approach for fibroblast migration [26,27]. We used the Mat lab function ‘classregtree’; its underlying methodological details are outlined in an excellent recent review on decision tree modeling[33]. Our dataset consisted of measurements for:migration translational speed and directional persistence, and phosphorylation levels of EGFR, ERK, Akt, and FAK across 19 materials substrate conditions. Mean free path values were calculated as the product of speed and persistence for each cell. The temporal signaling profiles collected per phosphoprotein per condition over 6 time points were time-integrated to generate one comparable value across all conditions. Each set of19 migration speeds, persistence times, and mean free paths, along with each set of phosphoprotein time-integrated phosphoprotein signals were discretized into equal range bins of low, medium, and high. Tree models were generated via the Matlab function ‘classregtree’. The ‘classregtree’ function takes two inputs, a 19-by-4matrix X of time-integrated signals, and a vector Y of length 19 containing each of the migration responses individually. We examined results from multiple parameter settings corresponding to optional algorithmic variations; in the end, we found for this application that classification tree with minimum split criterion of 4 conditions gave most robust models across randomization trials. A best-fit tree model was generated for each type of migration response (speed, persistence, mean free path).
3. Results
3.1. MSCs migration responses on biomaterials surfaces
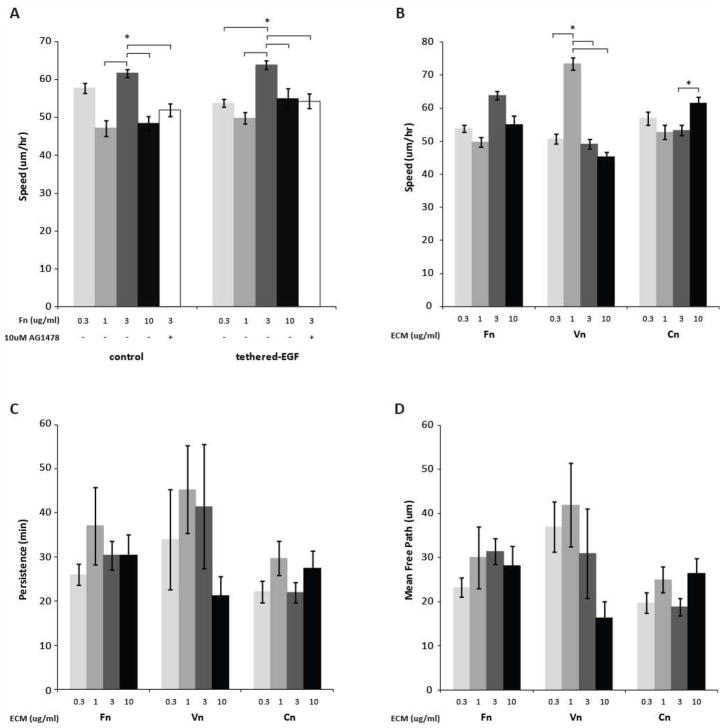
hTERT-MSCs were tracked to determine differential effects of tEGF and surface adhesiveness on cell migration (Fig. 2). Because of our previous experience with fibronectin effects on fibroblast migration, we extensively investigated MSC migration on fibronectin (Fn) adsorbed co-polymer substrates first (Fig. 2A). Within each set of four Fn concentrations on tEGF and control conditions, we observed the highest cell speeds on the intermediate 3 ug/mL Fn (Fn3) substrates with an average of 64 um/hr on Fn3 in the presence of tEGF (n ≈ 180 cells) and 62um/hr on Fn3 without tEGF (n ≈ 220 cells). Average speed was highest on intermediate Fn levels for both tEGF and control substrates as found previously with fibroblasts[16]. Peak speed was decreased by the EGFR kinase inhibitor AG1478 on both control and tEGF surface, with the latter effect likely due to autocrine EGFR ligands as we have previously found in primary MSCs[34].
Figure 2.
MSC migration speed and persistence are affected by tEGF PMMA-g-PEO surfaces with varying levels of ECM protein coating; bar color from lightest to darkest correspond to protein levels from lowest to highest. Sparsely seeded hTERT-MSCs on biomaterials surfaces were tracked via time-lapse image capture for 7 hours. (A) Cell speed on fibronectin-adsorbed polymer surfaces. (B) Cell speeds on tEGF polymer surfaces adsorbed with fibronectin, vitronectin, and collagen. (C) Migration persistence as fitted using the Persistent Random Walk model with overlapping intervals. (D) Mean free path (speed x persistence).* denotes statistical significance between conditions at p < 0.01; error bars show +/−SEM.
As along with Fn-binding integrins, MSCs also express integrins that bind vitronectin (Vn) and collagen-I (Cn)([21,35] and data not shown), both present in typical in vivo MSC environments, we additionally investigated the effects of these ECM proteins on MSC migration in the presence of tEGF. We found cell speed on Vn/tEGF to also show a higher maximum of about 73 um/hr (Fig. 2B), likewise at intermediate levels – in this case, 1 ug/mL (Vn1). In contrast to both the Fn and Vn influences, cell speed on Cn/tEGF was fairly consistent across levels (Fig. 2B). Cell persistence exhibited larger variability within any given condition (Fig. 2C); this situation derives from the quantity being derived as a fitted parameter in the Persistent Random Walk model. A more comprehensive migration parameter is that of cell mean free path (MFP), the product of speed and persistence. MSC MFP trends were found here to reflect those of speed and persistence, as expected (Fig. 2D). The most striking result is that MFP is consistently lowest and most independent of ECM level on Cn/tEGF surfaces compared to Fn/tEGF and Vn/tEGF. We thus next sought to account for these ECM effects in terms of intracellular signaling pathway activities elicited by the tEGF/ECM inputs.
3.2. MSCs phosphoprotein signaling and univariate correlation of signal to response
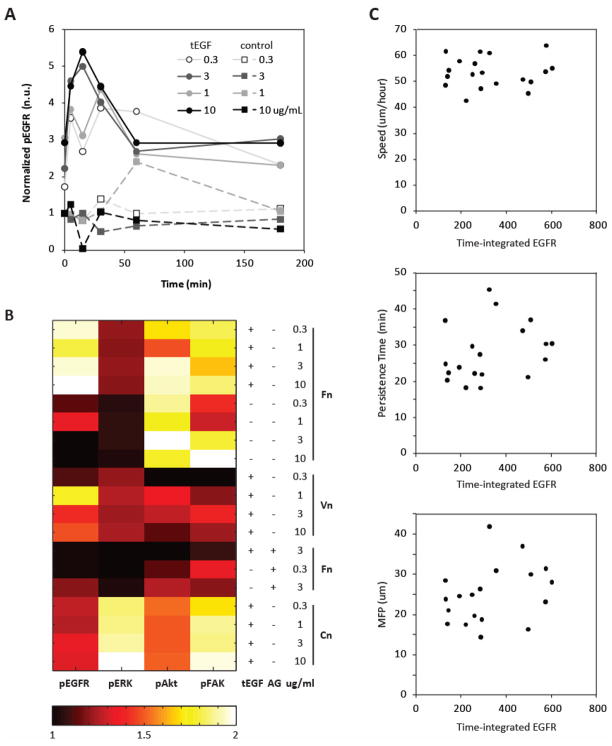
Fig. 3A shows a representative time course of EGFR activation on Fn-adsorbed tEGF substrates. Peak EGFR phosphorylation occurred 15 minutes after tEGF treatment on the most adhesive 10 ug/mL Fn substrate and was >5-fold that of the control (0.3 ug/mL Fn, no tEGF, time 0). We also observed a >2- fold activation of EGFR on control surfaces adsorbed with 1 ug/mL Fn, although peak activity was at 60 minutes, delayed from the tEGF substrates. Three hours after tEGF stimulation, EGFR phosphorylation on tEGF substrates was still sustained at levels ~3-fold that of the control on all levels of Fn adhesiveness.
Figure 3.
MSC signals a cross conditions. Lysates from cells on biomaterials surfaces were collected at 0, 5, 15, 30, 60, and 180 minutes after treatment and levels of EGFR, ERK, Akt, and FAK measured. (A) Representative signaling time-course of EGFR activity of cells on Fn-adsorbed substrates. (B) Heat map of normalized signals for 19substrate conditions. Raw signal measurements within each ECM group (Fn, Vn, Cn) were normalized to 0.3 ug/ml control and then integrated for each condition. All 19 integrated signals for each phosphoprotein were then normalized to that of Fn0.3 control, time 0. (C) Lack of univariate correlation of migration responses of Speed, Persistence, and MFP versus time-integrated EGFR.
To more robustly compare across various conditions, we time-integrated all signaling measurements across their individual time courses for each of 19 substrate conditions to present one signaling parameter per phosphoprotein per substrate. These are shown graphically in Fig. 3B, with the values for each phosphoprotein rescaled to be between 1 and 2. EGFR and Akt were highest on Fn-adsorbed tEGF substrates, whereas ERK and Akt were highest on Cn-adsorbed tEGF substrates. Signaling was relatively low on all Vn substrates and in cells treated with AG1478, though a dark block indicates lower activity relative to the other 18 substrates rather than a complete dearth of phosphorylation.
To investigate whether any single phosphoprotein is predictive of migration response, we plotted univariate correlations of migration speed, persistence, and MFP versus each of the four time integrated phosphoprotein signals. We saw no clear relationships for any of the responses; the representative plots for time-integrated EGFR are shown in Fig. 3C. This motivated a multi-variate ‘systems biology’ approach to relate all four phosphoproteins in an effort to predict MSC migration response from intracellular signaling.
3.3. Decision tree ‘signal-response’ modeling
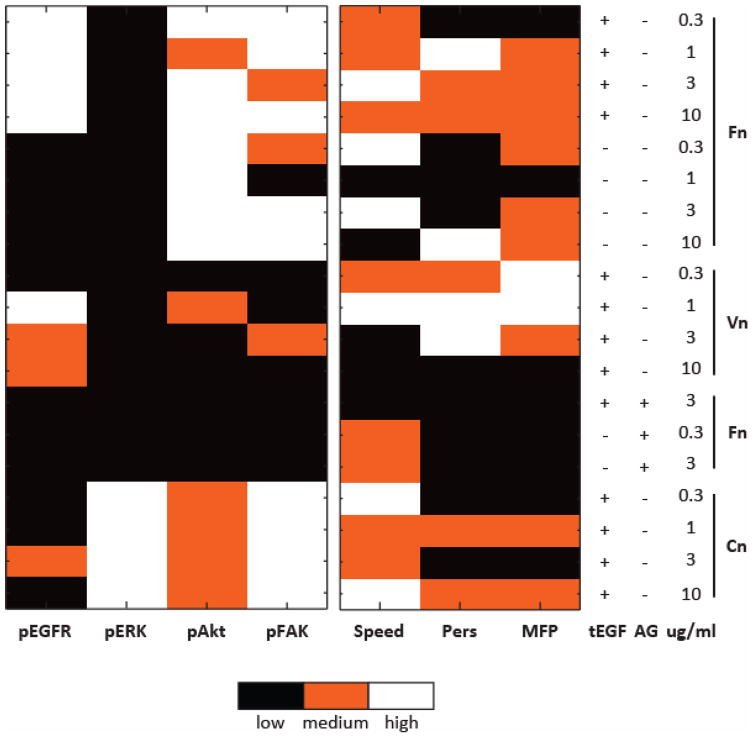
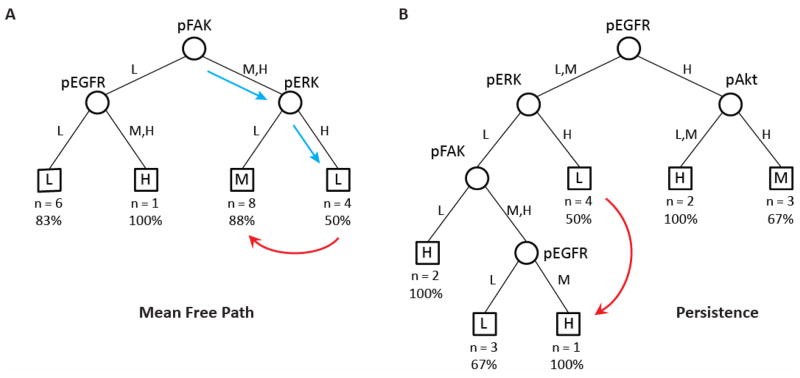
Decision tree modeling to classify dependences of a dependent variable, or ‘response’ (in our case here, migration properties – speed, persistence, mean free path) on quantitative combinations of multiple independent variables, or ‘signals’ (here, the four phosphoproteins) across many conditions is particularly well suited for problems in which the number of signals is relatively small[33] – precisely our situation at hand. Resulting trees have signals as nodes that classify to responses at the terminal tree leaves. To minimize over-fitting, we discretized our training data sets of signals and responses to the levels ‘Low’, ‘Medium’, and ‘High’ using range discretization (Fig. 4), and then generated binary trees that classify each of the migration responses of speed, persistence, and MFP using the set of time integrated signals. The MFP tree discerned EGFR, ERK, and FAK as dominant predictors (Fig. 5A). L, M, and H on the branches below each node show the discretized signaling levels that follow each branch, and the leaves show the discretized MFP class. For example, starting at the root predictor of FAK, we would follow the right branch if FAK were medium or high which leads to a new predictor of ERK (blue arrow). If ERK were high, then we would follow the right branch again (blue arrow), and our MFP tree predicts that cells with these signaling states would migrate with low MFP. Of the 19 substrates in our training set, 4 classified to this leaf (N = 4), and of those, 50% had low MFP. The split at ERK as a predictor generates a non-intuitive prediction: decreasing ERK from high to low should increase MFP from low to medium (red arrow). Applying this same hypothesis of decreasing ERK, the persistence tree predicts that lowering ERK would increase persistence only if FAK is not low and if EGFR is medium; otherwise, persistence would remain low. Taken together, these two trees predict that when FAK and EGFR are activated, inhibiting ERK would increase MSC migration MFP by increasing directional persistence. With ERK a critical signal in cell migration, this hypothesis that decreasing ERK would promote cell migration is a testable non-intuitive prediction.
Figure 4.
MSC signaling and migration response discretized to low, medium, and high for 19 substrate conditions to minimize model-overfitting. For each column of signal or response, the range of values between the column minimum and column maximum was evenly divided into three bins so that each bin contains an equal-sized range.
Figure 5.
Decision tree ‘signal-response’ models where signal nodes classify response leaves. Classification trees were generated via Mat lab using the discretized data from Figure 3C for: (A) cell mean free path; and (B) migration persistence.
3.4. Experimental test of decision tree model prediction: inhibition of ERK on Cn/tEGF substrates
We examined the four substrates that classified along the high ERK branch in both the MFP and persistence trees to determine their viability as hypothesis-testing conditions. For both trees, the four substrates were all four of the Cn-adsorbed tEGF conditions. The discretized data for these conditions (Table 1) show that FAK and ERK are high on all four, Akt medium on all four, and EGFR low except on 3 ug/mL Cn where EGFR is medium. Since our persistence tree predicted that decreasing ERK would only increase persistence if EGFR were medium, this makes Cn3 substrates with tEGF the ideal hypothesis testing condition.
Table 1.
Discretized levels of signal and response on collagen-adsorbed tEGF substrates. C0.3t indicates 0.3 ug/mL collagen adsorbed onto copolymer with tEGF.
| pEGFR | pAkt | pFAK | pERK | MFP | Pers | |
|---|---|---|---|---|---|---|
| C0.3t | L | M | H | H | L | L |
| C1t | L | M | H | H | M | M |
| C3t | M | M | H | H | L | L |
| C10t | L | M | H | H | M | M |
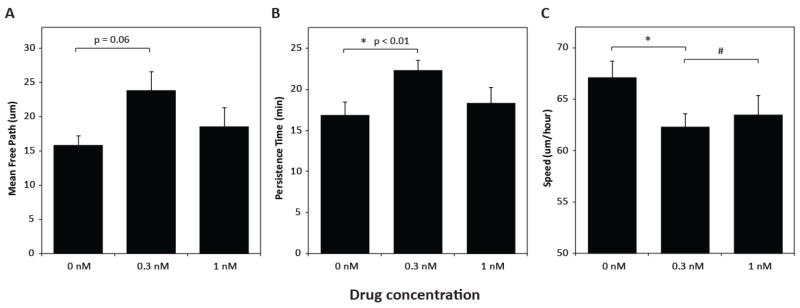
We tracked the migration of MSCs seeded on tEGF copolymer substrates adsorbed with 3 ug/ml Cn in the presence of U0216, a small molecule inhibitor of MEK, upstream of ERK. Partial ERK inhibition with 0.3 nM of U0126 (data not shown)significantly increased MSC persistence time by 30% from 17 minutes to 22 minutes (Fig. 6B) and increased MFP by 50%, from 16 um to 24 um (Fig. 6A). Speed was decreased mildly, by <10% (Fig. 6C), although it stayed within the range of values we observed on Cn/tEGF substrates in Fig. 2B -- indicating that MSCs still migrateat consistent speeds at this level of ERK inhibition. Essentially total inhibition of ERK using 1 nM of U0216 (data not shown) abrogated the effects; this is not surprising given that the decision tree model was trained on data in which substantive ERK activity was always found, due to signaling from ECM along with likely autocrine EGF stimulation[34].
Figure 6.
Successful test of decision tree model prediction that reducing ERK signal on Cn/tEGF substrates will enhance MSC migration persistence and mean free path.(A) mean free path, (B) persistence time, and (C) speed under partial and total ERK inhibition with MEK inhibitor U0126. * and # denote statistical significance with p < 0.01 and p < 0.05 respectively. Error bars show +/− SEM.
4. Discussion
We have quantitatively characterized the effects of tEGF and adsorbed ECM biomaterials cues on MSC migration using single-cell video microscopy, measuring cell translational speed and directional persistence behavior across different ECM proteins (fibronectin (Fn), vitronectin (Vn), collagen-I (Cn)) at a range of levels. We also calculated a derived quantity, the cell migration ‘mean free path’ (MFP): the product of speed and persistence. Conceptually, MFP can be interpreted as how far a cell is able, on the average, to travel from its original seeding site. For applications in biomaterials scaffold colonization, MFP can be appreciated to be more important than either speed or persistence individually -- slow but persistently moving cells may actually colonize a scaffold more vigorously than fast but erratically moving cells [25,36]. A key finding was that cell migration speed, directional persistence, and their product termed mean free path (MFP) exhibited categorically different behavior on Cn/tEGF surfaces compared to on Fn/tEGF and Vn/tEGF: these migration behaviors were relatively independent of Cn levels whereas they were strongly influenced by the level of Fn and of Vn, and were consistently lower on the Cn-coated materials than on the Fn- or Vn-coated materials (Fig. 2). One consequence might be that Cn/tEGF scaffolds could be considered inferior to Fn/tEGF and Vn/tEGF materials because the former would yield less vigorous MSC colonization. At the same time, the variable dependence of migration properties on ECM levels in the Fn/tEGF and Vn/tEGF materials could render them less attractive since predictability of colonization vigor would be less certain. An optimal design, therefore, could aspire to combine the reliable consistency of the Cn/tEGF surfaces with the potential higher velocity inherent in the Fn/tEGF or Vn/tEGF surfaces. In order to do so, we need to generate an underlying “design principle” that might hold validity across all the different materials conditions; as has been argued elsewhere, both for biomaterials [37,38] and for stem cell biology [39,40], a computational model comprehending the multi-variate intracellular signaling network state, downstream of ECM and growth factor cues, is an appealing approach.
We there fore measured the activity levels of four key phosphoproteins downstream of EGF and ECM stimuli: phosphorylated EGFR, phosphorylated ERK, phosphorylated Akt, and phosphorylated FAK. Each of these signals has been shown to be an important governor of EGF-induced fibroblast migration on ECM-coated substrata, so we hypothesized that they could be informative for our MSC situation. Consistent with our previous findings, we observed sustained EGFR activation on tEGF surfaces[34,38], a phenomenon thought to be caused by immobilized growth factors delaying and/or preventing receptor internalization and degradation upon ligand binding[31,41]. Because of the beneficial roles that tethered EGF has been demonstrated to play in promoting MSC survival, proliferation, and controlled differentiation previously[12,14,34,38], and now here for migration, this sustained EGFR signaling may offer a major advantage for continued control of MSCs for scaffold colonization.
Not surprisingly, no single phosphoprotein was predictive of MSC motility behavior (Fig.3). Since migration is an exquisitely coordinated process of distinct biophysical steps regulated by different signaling pathways, it follows that migration response is most likely governed by a quantitative combination of multiple signals. As in our previous work on fibroblasts [26,27], we found here that decision tree modeling can usefully ascertain quantitative combinations of EGFR, ERK, Akt, and FAK signals that classify MSC migration behavior into different levels of vigor (Fig. 4, Fig. 5). From these models emerged an attractive, non-intuitive hypothesis for gaining the best of Cn/tEGF motility consistency and Fn/tEGF or Vn/tEGF potential motility vigor: inhibiting ERK to increase MSC persistence and thus mean free path. We successfully tested this prediction (Fig. 6), providing proof-of-concept that “fine-tuning” an intracellular signaling pathway in a manner predictable from a multi pathway signal- response model generated from “coarse-graining” the cell signaling network using biomaterials scaffolds can produce a desirable result based on a molecular-level design principle.
5. Conclusion
We have successfully demonstrated in this study an application to controlling MSC migration on biomaterials scaffolds by means of a two-tiered approach: “coarse-graining” the biomaterials substrate to obtain reliable translational speeds, followed by “fine-tuning” using small molecule treatment to increase directional persistence -- together achieving a more vigorous migration mean free path.
Acknowledgments
This work was supported by the NIH Cell Migration Consortium U54-GM064346 (DAL, LGG), NIH grant R01-GM018336 (DAL, AW), and NIH grant R01-DE019523 (LGG). We are grateful to Linda Stockdale and Mary Rhoads for accomplishing the PMMA-g-PEO polymer synthesis and spin coating.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Phinney DG, Prockop DJ. Mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair-- current views. Stem Cells. 2007;25:2896–902. doi: 10.1634/stemcells.2007-0637. [DOI] [PubMed] [Google Scholar]
- 2.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 3.Prockop DJ. Repair of tissues by adult stem/progenitor cells (MSCs): controversies, myths, and changing paradigms. Molec Ther. 2009;17:939–46. doi: 10.1038/mt.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salem HK, Thiemermann C. Mesenchymal stem cells: current understanding and clinical status. Stem Cells. 2010;28:585–95. doi: 10.1002/stem.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shekaran A, Garcia AJ. Extracellular matrix-mimetic adhesive biomaterials for bone repair. J Biomed Mat Part A. 2011;96:261–72. doi: 10.1002/jbm.a.32979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Healy KE, Guldberg RE. Bone tissue engineering. J Musculoskel Neur Interactions. 2007;7:328–30. [PubMed] [Google Scholar]
- 7.Baba S, Inoue T, Hashimoto Y, Kimura D, Ueda M, Sakai K, et al. Effectiveness of scaffolds with pre-seeded mesenchymal stem cells in bone regeneration: assessment of osteogenic ability of scaffolds implanted under the periosteum of the cranial bone of rats. Dental Mat J. 2010;29:673–81. doi: 10.4012/dmj.2009-123. [DOI] [PubMed] [Google Scholar]
- 8.Gloria A, De Santis R, Ambrosio L. Polymer-based composite scaffolds for tissue engineering. J Appl Biomat Biomech. 2010;8:57–67. [PubMed] [Google Scholar]
- 9.Xu C, Su P, Chen X, Meng Y, Yu W, Xiang AP, et al. Biocompatibility and osteogenesis of biomimetic Bioglass-Collagen-Phosphatidylserine composite scaffolds for bone tissue engineering. Biomaterials. 2011;32:1051–8. doi: 10.1016/j.biomaterials.2010.09.068. [DOI] [PubMed] [Google Scholar]
- 10.Johnson AJW, Herschler BA. A review of the mechanical behavior of CaP and CaP/polymer composites for applications in bone replacement and repair. Acta Biomat. 2010;7:16–30. doi: 10.1016/j.actbio.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 11.Dozza B, Di Bella C, Lucarelli E, Giavaresi G, Fini M, Tazzari PL, et al. Mesenchymal stem cells and platelet lysate in fibrin or collagen scaffold promote non-cemented hip prosthesis integration. J Orthopaed Res. 2011;29:961–8. doi: 10.1002/jor.21333. [DOI] [PubMed] [Google Scholar]
- 12.Tamama K, Fan VH, Griffith LG, Blair HC, Wells A. Epidermal growth factor as a candidate for ex vivo expansion of bone marrow-derived mesenchymal stem cells. Stem Cells. 2006;24:686–95. doi: 10.1634/stemcells.2005-0176. [DOI] [PubMed] [Google Scholar]
- 13.Krampera M, Pasini A, Rigo A, Scupoli MT, Tecchio C, Malpeli G, et al. HB-EGF/HER-1 signaling in bone marrow mesenchymal stem cells: inducing cell expansion and reversibly preventing multilineage differentiation. Cell. 2005;106:59–66. doi: 10.1182/blood-2004-09-3645. [DOI] [PubMed] [Google Scholar]
- 14.Fan VH, Tamama K, Au A, Littrell R, Richardson LB, Wright JW, et al. Tethered epidermal growth factor provides a survival advantage to mesenchymal stem cells. Stem Cells. 2007;25:1241–51. doi: 10.1634/stemcells.2006-0320. [DOI] [PubMed] [Google Scholar]
- 15.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 16.Maheshwari G, Wells A, Griffith LG, Lauffenburger DA. Biophysical integration of effects of epidermal growth factor and fibronectin on fibroblast migration. Biophys J. 1999;76:2814–23. doi: 10.1016/S0006-3495(99)77435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wells A, Harms B, Iwabu A, Koo L, Smith K, Griffith LG, et al. Motility signaled from the EGF receptor and related systems. Methods Mol Biol. 2006;327:159–77. doi: 10.1385/1-59745-012-X:159. [DOI] [PubMed] [Google Scholar]
- 18.Xie H, Pallero MA, Gupta K, Chang P, Ware MF, Witke K, et al. EGF receptor regulation of cell motility: EGF induces disassembly of focal adhesions independently of the motility-associated PLCγ signaling pathway. J Cell Sci. 1998;111:615–24. doi: 10.1242/jcs.111.5.615. [DOI] [PubMed] [Google Scholar]
- 19.Glading A, Uberall F, Keyse SM, Lauffenburger DA, Wells A. Membrane-proximal ERK signaling is required for M-calpain activation downstream of EGF receptor signaling. J Biol Chem. 2001;276:23341–8. doi: 10.1074/jbc.M008847200. [DOI] [PubMed] [Google Scholar]
- 20.Schneider IC, Hays CK, Waterman CM. EGF-induced contraction regulates paxillin phosphorylation to temporally separate traction generation from deadhesion. Molec Biol Cell. 2009;13:3155–67. doi: 10.1091/mbc.E09-03-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veevers-Lowe J, Ball SG, Shuttleworth A, Kielty CM. Mesenchymal stem cell migration is regulated by fibronectin through α5β1 integrin-mediated activation of PDGFRβ and potentiation of growth factor signals. J Cell Sci. 2011;124:1288–1300. doi: 10.1242/jcs.076935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huveneers S, Danen EH. Adhesion signaling -- crosstalk between integrins, Src, and Rho. J Cell Sci. 2009;122:1059–69. doi: 10.1242/jcs.039446. [DOI] [PubMed] [Google Scholar]
- 23.Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nature Cell Biol. 2004;6:154–61. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 24.Fiedler J, Etzel N, Brenner RE. To go or not to go: migration of human mesenchymal progenitor cells stimulated by isoforms of PDGF. J Cell Biochem. 2004;93:990–8. doi: 10.1002/jcb.20219. [DOI] [PubMed] [Google Scholar]
- 25.Ware MF, Wells A, Lauffenburger DA. Epidermal growth factor alters fibroblast migration speed and directional persistence reciprocally and in a matrix-dependent manner. J Cell Sci. 1998;111:2423–32. doi: 10.1242/jcs.111.16.2423. [DOI] [PubMed] [Google Scholar]
- 26.Hautaniemi S, Kharait S, Iwabu A, Wells A, Lauffenburger DA. Modeling of signal-response cascades using decision tree analysis. Bioinformatics. 2005;21:2027–35. doi: 10.1093/bioinformatics/bti278. [DOI] [PubMed] [Google Scholar]
- 27.Kharait S, Hautaniemi S, Wu S, Iwabu A, Lauffenburger DA, Wells A. Decision tree modeling predicts effects of inhibiting contractility signaling on cell motility. BMC Syst Biol. 2007;1:9. doi: 10.1186/1752-0509-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okamoto T, Aoyama T, Nakayama T, Nakamata T, Hosaka T, Nishijo K, et al. Clonal heterogeneity in differentiation potential of immortalized human mesenchymal stem cells. Biochem Biophys Res Comm. 2002;295:354–61. doi: 10.1016/s0006-291x(02)00661-7. [DOI] [PubMed] [Google Scholar]
- 29.Irvine DJ, Mayes AM, Griffith LG. Nanoscale clustering of RGD peptides at surfaces using Comb polymers. 1. Synthesis and characterization of Comb thin films. Biomacromolecules. 2001;2:85–94. doi: 10.1021/bm005584b. [DOI] [PubMed] [Google Scholar]
- 30.Irvine DJ, Ruzette AV, Mayes AM, Griffith LG. Nanoscale clustering of RGD peptides at surfaces using comb polymers. 2. Surface segregation of comb polymers in polylactide. Biomacromolecules. 2001;2:545–56. doi: 10.1021/bm015510f. [DOI] [PubMed] [Google Scholar]
- 31.Kuhl PR, Griffith-Cima LG. Tethered epidermal growth factor as a paradigm for growth factor induced stimulation from the solid phase. Nature Medicine. 1996;2:1022–7. doi: 10.1038/nm0996-1022. [DOI] [PubMed] [Google Scholar]
- 32.Dickinson RB, Tranquillo RT. Optimal estimation of cell movement indices from the statistical analysis of cell tracking data. AIChE J. 1993;39:1995–2010. [Google Scholar]
- 33.Geurts P, Irrthum A, Wehenkel L. Supervised learning with decision tree-based methods in computational and systems biology. Molec BioSyst. 2009;5:1593–605. doi: 10.1039/b907946g. [DOI] [PubMed] [Google Scholar]
- 34.Platt MO, Roman AJ, Wells A, Lauffenburger DA, Griffith LG. Sustained epidermal growth factor receptor levels and activation by tethered ligand binding enhances osteogenic differentiation of multi-potent marrow stromal cells. J Cell Physiol. 2009;221:306–17. doi: 10.1002/jcp.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kundu AK, Putnam AJ. Vitronectin and collagen I differentially regulate osteogenesis in mesenchymal stem cells. Biochem Biophys Res Comm. 2006;347:347–57. doi: 10.1016/j.bbrc.2006.06.110. [DOI] [PubMed] [Google Scholar]
- 36.Peyton SR, Kalcioglu ZI, Cohen JC, Runkle AP, Van Vliet KJ, Lauffenburger DA, et al. Marrow-derived stem cell motility in 3D synthetic scaffold is governed by geometry along with adhesivity and stiffness. Biotech Bioeng. 2011;108:1181–93. doi: 10.1002/bit.23027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caplan MR, Shah MM. Translating biomaterial properties to intracellular signaling. Cell Biochem Biophys. 2009;54:1–10. doi: 10.1007/s12013-009-9048-5. [DOI] [PubMed] [Google Scholar]
- 38.Zmbuzzi WF, Coelho PG, Alves GG, Granjeiro JM. Intracellular signal transduction as a factor in the development of biomaterials for bone tissue engineering. Biotech Bioeng. 2011;108:1246–50. doi: 10.1002/bit.23117. [DOI] [PubMed] [Google Scholar]
- 39.Platt MO, Wilder CL, Wells A, Griffith LG, Lauffenburger DA. Multipathway kinase signatures of multipotent stromal cells are predictive for osteogenic differentiation. Stem Cells. 2009;27:2804–14. doi: 10.1002/stem.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss MS, PenalverBarnabe B, Bellis AD, Broadbelt LJ, Jeruss JS, Shea LD. Dynamic, large-scale profiling of transcription factor activity from live cells in 3D culture. PLoS ONE. 2010;5:e14026. doi: 10.1371/journal.pone.0014026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito Y. Covalently immobilized biosignal molecule materials for tissue engineering. Soft Matter. 2008;4:46–56. doi: 10.1039/b708359a. [DOI] [PubMed] [Google Scholar]