Abstract
Biological specimens have to be prepared for imaging in the electron microscope in a way that preserves their native structure. Two-dimensional (2D) protein crystals to be analyzed by electron crystallography are best preserved by sugar embedding. One of the sugars often used to embed 2D crystals is trehalose, a disaccharide used by many organisms for protection against stress conditions. Sugars such as trehalose can also be added to negative staining solutions used to prepare proteins and macromolecular complexes for structural studies by single-particle electron microscopy (EM). In this review, we describe trehalose and its characteristics that make it so well suited for preparation of EM specimens and we review specimen preparation methods with a focus on the use of trehalose.
1. Introduction
Electron microscopy (EM) is a versatile technique that can be used to visualize biological specimens ranging from cells and tissues to individual macromolecules. Compared to X-rays, electrons have a higher scattering power and can thus be used to image very thin samples. Furthermore, because electrons carry charge, electromagnetic lenses can be used to focus an electron beam and to form an image that contains phase information. Current transmission electron microscopes equipped with field-emission guns are capable of taking images that contain sub-Angstrom resolution information (O'Keefe et al., 2005), but such very high resolution information can currently only be obtained with inorganic samples. Organic and in particular biological samples have characteristics that make them difficult to study by EM and limit the resolution that can be achieved.
Biological samples consist mostly of light atoms, such as hydrogen, carbon, nitrogen and oxygen. Since electron scattering is proportional to the atomic number, Z, of the scattering atom, their light atoms make biological specimens weakly scattering objects that thus have inherently low contrast. Furthermore, the ratio of elastic to inelastic scattering cross sections is proportional to the atomic number of the scattering atom divided by 20 (σel/σin ~ Z/20) (Egerton, 1976; Reimer and Ross-Messemer, 1990). Thus, for light atoms approximately two damaging inelastic scattering events occur for every useful elastic scattering event, making biological specimens very sensitive to beam damage. Finally, because electrons have a very short mean free path, electron beams have to be generated in a vacuum. Vacuum presents a problem for biological specimens, which consist of up to 70% of water, as dehydration causes a collapse of the structure. To overcome these issues, biological specimens have to be prepared in a way that makes it possible to image them in an electron microscope without destroying their structure.
Early specimen preparation techniques involved the use of heavy metal atoms to boost electron scattering and thus the contrast of biological specimens. In metal shadowing approaches, in which the specimen is decorated with a layer of metal atoms, specimens are often freeze-dried to minimize structural collapse (Cheong et al., 1993; Williams and Wyckoff, 1946). In the negative staining technique, specimens are bathed in a heavy metal salt solution. As the sample is dried, the heavy metal atoms form microcrystals that embed the specimen and help, to some degree, maintain its structure (Ohi et al., 2004). While enhancing image contrast and making the specimen more resistant to beam damage, both metal shadowing and negative staining limit the resolution that can be obtained to about 20 Å and often introduce preparation artifacts.
To prepare specimens for the electron microscope vacuum without introducing artifacts and without limiting the achievable resolution, Nigel Unwin and Richard Henderson introduced sugar embedding for two-dimensional (2D) crystals (Unwin and Henderson, 1975), and Jacques Dubochet and co-workers developed vitrification for the preparation of any kind of biological specimen (Dubochet et al., 1982). 2D crystals are used for structure determination, most commonly of membrane proteins, by electron crystallography (reviewed in Fujiyoshi, 2008; Hite et al., 2007; Raunser and Walz, 2009), a method developed by Unwin and Henderson to study the structure of bacteriorhodopsin (bR) (Unwin and Henderson, 1975). To preserve them in a hydrated state, purple membranes (naturally occurring bR 2D crystals) were dried in the presence of 1% glucose before introducing them into the vacuum of the electron microscope for data collection, making it possible to determine the structure of bR to 7 Å resolution (Henderson, 1975). In vitrification, a specimen in aqueous solution is quick-frozen by plunging the sample into liquid ethane that is cooled to liquid nitrogen temperature (Dobro et al., 2010). Due to the high freezing speed, the water solidifies without having time to form ice crystals, which would cause damage to biological specimens. The resulting water phase, a liquid with very high viscosity, is known as amorphous or vitrified ice, hence the term vitrification. To preserve the vitrified ice layer, vitrified specimens have to be imaged in the electron microscope at cryogenic temperatures, which gave rise to the term cryo-EM. The first vitrified specimens imaged by cryo-EM were viruses (Adrian et al., 1984), but other specimens soon followed (Dubochet et al., 1988). Both sugar embedding and vitrification do not use contrasting agents, and specimens prepared by these techniques thus have low inherent contrast.
Cooling the specimen to low temperature not only preserves the vitrified ice, but it also helps reduce the effects of beam damage. While inelastic scattering events still cause breakage of covalent bonds in polypeptides (Baker and Rubinstein, 2010; Egerton, 2004), due to the lower diffusion coefficients at lower temperatures the fragments formed by radiolysis remain better localized and the effects of beam damage are thus less visible (Taylor and Glaeser, 1974). Cryo-EM is most commonly performed at liquid nitrogen temperature, but electron microscopes have been developed and optimized that can keep the specimen at liquid helium temperature (Fujiyoshi, 1991; Fujiyoshi et al., 1998). While cooling the specimen reduces the effects of beam damage, the only way to minimize beam damage itself is to minimize the electron dose to which the specimen is exposed. EM studies of biological specimens are thus always performed using low-dose procedures (Kuo, 1975). In this technique, all instrumental adjustments such as correction for astigmatism and focusing are performed on a specimen area next to the area of interest, and the area of interest is only exposed for the actual data collection using a very low electron dose, typically in the range of 10 electrons/Å2. As a result, low-dose images are noisy and require substantial computational processing to extract the biologically relevant information.
Trehalose is a sugar that is best known in EM specimen preparation for its use as an embedding medium for 2D crystals, but it has also been used for the preparation of negatively stained specimens and lipid monolayer specimens. In this review, we first provide a brief description of trehalose and its basic properties, and then discuss EM specimen preparation with a focus on the use of trehalose.
2. Trehalose
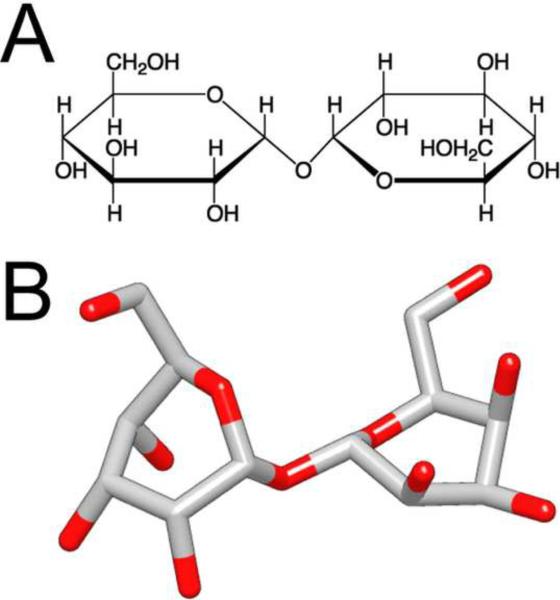
Trehalose was first discovered in 1832 in studies of the ergot of rye (Wiggers, 1832) and was first isolated in 1858 from the cocoon of the parasitic beetle Trehala manna, giving it the name trehalose (Harding, 1923). Trehalose (α,D-glucopyrannosyl-1,1-α,D-glucopyrannoside) is a non-reducing disaccharide formed by two glucose units joined by an α-1,1-glycosidic linkage (Figure 1). This chemical linkage makes trehalose very resistant to acid hydrolysis and thus very stable in aqueous solutions. The 1,1-glucosidic bond also keeps trehalose and other non-reducing sugars in a closed-ring form, which prevents glycation, a non-enzymatic glycosylation reaction in which the aldehyde or ketone end-groups react with lysine or arginine residues of proteins.
Figure 1. The structure of trehalose.
Trehalose is a non-reducing disaccharide composed of two glucose units with a α-1,1-glycosidic linkage. (A) Chemical drawing and (B) three-dimensional structure of trehalose. Red indicates oxygen atoms.
2.1. Role of trehalose in biology
Trehalose serves numerous functions in vivo, which include serving as carbon source (Thevelein, 1984) and structural component of bacterial cell walls (Daffe and Draper, 1998), acting as a regulator of plant growth (Rolland et al., 2002) and osmotic pressure (Kempf and Bremer, 1998), and mediating protection against dehydration (Crowe et al., 1992; Crowe, 2002; Singer and Lindquist, 1998) and freezing (Crowe et al., 1987). Recently, trehalose was also found to be involved in many cell processes associated with other stress responses (Bravim et al., 2010; De Virgilio et al., 1994; Elliott et al., 1996; Fernandez et al., 2010; Reinders et al., 1999; Viner and Clegg, 2001). In yeast, for example, heat shock results not only in the expression of chaperones but also in the production of a large amount of trehalose that helps stabilize proteins (Ellis, 1987; Singer and Lindquist, 1998). Synthesis and accumulation of trehalose in the cytoplasm also plays a crucial role in cold adaptation of Escherichia coli (Kandror et al., 2002). Because of its ability to stabilize proteins, trehalose has been used for a long time in biotechnological and medical applications, serving as a nontoxic cryo-protectant to preserve and store enzymes, membranes, vaccines, animal and plant cells, as well as organs for surgical transplants (Paiva and Panek, 1996).
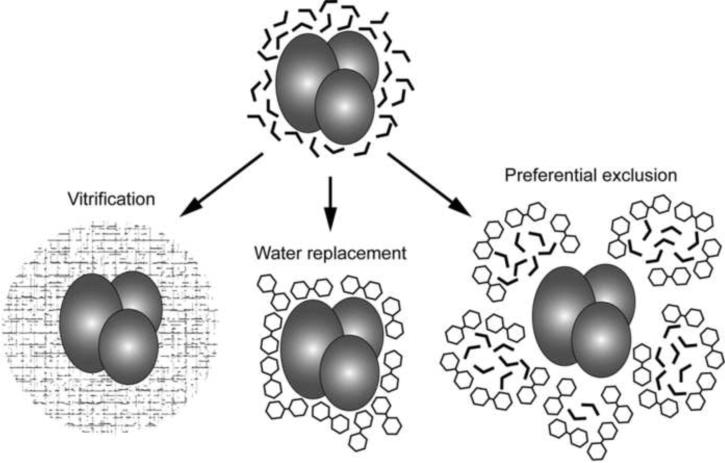
The precise mechanism of how trehalose protects cells from damage remains unclear, but three different theories have been put forward to explain how trehalose may stabilize proteins (Figure 2). In the “vitrification theory”, trehalose forms a viscous glass in the cytoplasm that entraps proteins and preserves them in a native state (Crowe et al., 1984). Glasses are solids with an amorphous (non-crystalline) structure that show a reversible phase transition from a glass-like to a rubber-like state (Zarzycki, 1991). Trehalose has the highest glass-transition temperature among all disaccharides (Chen et al., 2000; Green and Angell, 1989) and can thus transition into a glass state under ambient conditions. Biological molecules are protected and stabilized by the high viscosity and hydrogen bonding interactions provided by the trehalose glass. In the “water replacement theory”, trehalose stabilizes proteins by substituting for their water shell (Crowe et al., 1984; Crowe et al., 1996). Because trehalose has multiple hydroxyl groups, it can interact with the hydrophilic part of proteins and lipid head groups, thus stabilizing proteins and lipid membranes (Crowe et al., 1984; Lee et al., 1986). In the “preferential exclusion theory”, trehalose does not directly interact with proteins but instead sequesters water molecules away from the proteins, thus decreasing their hydration radius and thereby compacting and stabilizing the proteins (Jain and Roy, 2010; Kilburn et al., 2006). Unlike other disaccharides, trehalose does not form internal hydrogen bonds, so that the hydroxyl groups are more likely to interact with solvent molecules. Moreover, the radius of the hydration shell of trehalose is at least 2.5 times larger than that of other sugars, which means that it can sequester more water molecules than any other sugar (Sola-Penna and Meyer-Fernandes, 1998).
Figure 2. Three proposed theories how trehalose may protect proteins from damage.
In addition to protecting proteins from dehydration and preserving their structural integrity, trehalose also functions as an antioxidant, possibly serving as a free radical scavenger (Chen and Haddad, 2004; Elbein et al., 2003; Leekumjorn et al., 2008; Oku et al., 2005), protects cells from damage by oxygen radicals during oxidative stress (Benaroudj et al., 2001), and reduces peroxidation of unsaturated fatty acids by heat and oxygen radicals (Herdeiro et al., 2006; Oku et al., 2005).
2.2. Trehalose for the preparation of EM specimens
Trehalose is used by many organisms for protection against dehydration and freezing, and it is thus an obvious choice to be used as a cryo-protectant in the preparation of EM specimens. However, trehalose has additional advantages. Since trehalose is inert (Colaco et al., 1992; Jain and Roy, 2009), it does not perturb the structure of biological specimens. It can thus be used to embed 2D crystals for electron crystallographic data collection (Hirai et al., 1999) or can be added to negative staining solutions to reduce specimen flattening upon drying of the grid (Harris et al., 1995; Harris, 1996). Most notably, in a comparison with sucrose, trehalose was also found to reduce beam damage and to provide higher image contrast (De Carlo et al., 1999). While remarkable, the reasons for these effects are not clear, although the observed reduction in beam damage may be related to the ability of trehalose to scavenge free radicals (Chen and Haddad, 2004; Elbein et al., 2003; Leekumjorn et al., 2008; Oku et al., 2005).
3. Trehalose in the preparation of membrane protein 2D crystals
Electron crystallography is a method that can be used to determine the structure of proteins that form 2D crystals. While it has been used to analyze the structure of soluble proteins, most notably that of the α,β tubulin dimer (Nogales et al., 1995; Nogales et al., 1998), electron crystallography is particularly well suited for structural analysis of membrane proteins, which can be studied in their native environment, the lipid bilayer (Fujiyoshi, 2008; Hite et al., 2007; Raunser and Walz, 2009). While some membrane proteins form regular arrays naturally, 2D crystals are typically produced by reconstituting a detergent-solubilized membrane proteins into lipid bilayers at a low lipid-to-protein ratio (Abeyrathne et al., 2010; Hasler, 1998; Kühlbrandt, 1992). The 2D crystals can then be used to collect images and diffraction patterns in the electron microscope (Hite et al., 2010b), which provide phase and amplitude information, respectively, that can be used to calculate a density map of the crystallized protein (Schenk et al., 2010). The resolution of the density map that can be obtained is determined by the order of the 2D crystals; the better the order of the crystals, the higher the resolution that can be obtained. However, even the best-ordered 2D crystals only yield high-resolution information if they are correctly prepared for data collection (Hite et al., 2010b; Wang and Downing, 2011).
Unlike the preparation of other biological specimens for EM data collection, the preparation of 2D crystals not only has to protect the crystals from dehydration but also has to produce atomically flat specimens. The reason is that structure determination by electron crystallography requires the recording of data from tilted specimens. Imperfect crystal flatness will result in blurring of the diffraction spots in the direction perpendicular to the tilt axis and thus in the loss of resolution (Wang and Downing, 2011). The preparation of specimens flat enough for collection of near-atomic resolution data requires the use of molybdenum grids, which eliminates cryo-crinkling, and atomically flat carbon support film (reviewed in Hite et al., 2010b). The remaining variables in specimen preparation of 2D crystals are the preparation method and the embedding medium.
3.1. Preparation methods
Vitrification is the most common way specimens are prepared for data collection by cryo-EM (Adrian et al., 1984; Dubochet et al., 1988; Harris and Adrian, 1999). In this technique, the specimen is applied to a grid, which is then blotted and quickly plunged into a cryogen, usually liquid ethane cooled by liquid nitrogen, resulting in the preservation of the specimen in a layer of vitrified ice (Grassucci et al., 2007). While vitrification is very well suited for the preparation of specimens for single-particle EM and electron tomography, it is not ideal for the preparation of 2D crystals. The main problem is presumably that specimens for vitrification cannot be extensively blotted to avoid complete drying of the sample. As a result, vitrified 2D crystals tend to be not very flat, causing significant difficulties for collecting data from tilted specimens (Hite et al., 2010b). This problem may be the reason why only one protein structure was determined at high resolution by electron crystallography using vitrified 2D crystals (Ren et al., 2001).
The best way to prepare 2D crystals for high-resolution data collection is sugar embedding, a method introduced for the structural analysis of purple membranes (Henderson, 1975; Unwin and Henderson, 1975). It is the method with which almost all specimens were prepared that yielded atomic models by electron crystallography (Gonen et al., 2004; Gonen et al., 2005; Grigorieff, 1996; Henderson et al., 1990; Hiroaki et al., 2006; Hite et al., 2010a; Holm et al., 2006; Jegerschöld et al., 2008; Kimura et al., 1997; Kühlbrandt, 1994; Murata, 2000; Nogales et al., 1998; Tani et al., 2009). The back-injection method was the first technique used to prepare sugar-embedded specimens, which was later complemented by the carbon sandwich technique (Gyobu et al., 2004). In both methods, for which detailed protocols are provided in (Hite et al., 2010b), the most crucial parameter that has to be optimized is the thickness of the sugar layer. If the layer is too thick, the electron beam will not penetrate the sample. If the layer is too thin, the crystals may become too dry and sustain damage. The thickness of the sugar layer can be modified by adjusting the concentration of the sugar solution and the blotting time. Sugar embedding is, however, not very reproducible and typically several specimens have to be prepared to produce one that is suitable for high-resolution data collection. The reproducibility can be improved by performing specimen preparation in a controlled environment such as in a cold room.
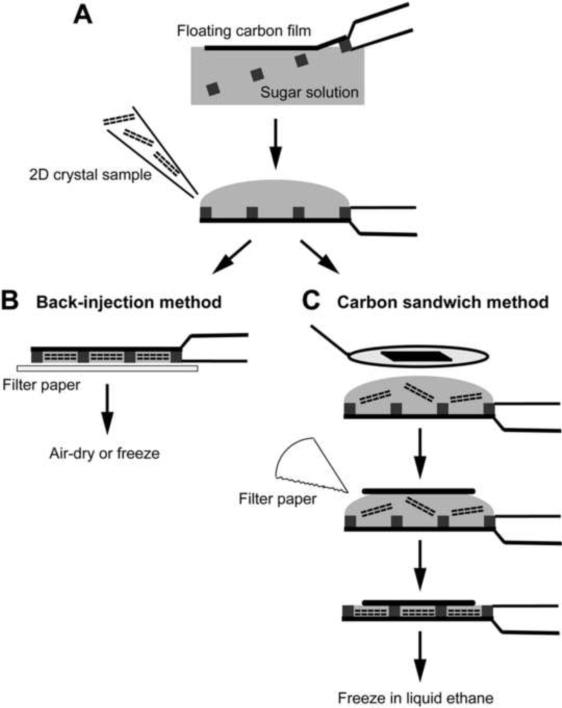
The back-injection method (Figure 3)
Figure 3. Sugar embedding methods used for the preparation of 2D crystals.
(A) A piece of carbon film is floated off mica onto the embedding solution, picked up with an EM grid, and the 2D crystal sample is applied to the opposite side of the grid. (B) In the back-injection method, the grid is simply blotted and then typically allowed to air-dry and cooled down in the electron microscope, but the grid can also be quick-frozen. (C) In the carbon sandwich method, a second, smaller piece of carbon film is placed on the sample using a wire loop. Excess solution is blotted away with a filter paper and the grid quick-frozen in liquid ethane.
The back-injection method is the easiest preparation method for 2D crystals. Briefly, a small piece of thin carbon film on mica is floated off on the surface of the embedding solution and picked up with an EM grid. The crystal solution is applied to the side of the EM grid opposite of the carbon film and mixed with the embedding solution by agitation with a pipetman. Excess solution is blotted away using filter paper, with the blotting time defining the thickness of the sugar layer. The grid is then usually allowed to air-dry and only cooled down once it has been transferred into the electron microscope. Alternatively, the grid can be frozen by plunging it into liquid nitrogen, in which case cryo-transfer is required to introduce the specimen into the electron microscope.
The carbon sandwich method (Figure 3)
Beam-induced movement is a serious problem for high-resolution data collection, which substantially reduces the yield of useful images that can be recorded of highly tilted specimens (Brink et al., 1998; Downing et al., 2004). The carbon sandwich method dramatically reduces beam-induced movement and thus allows for much higher yields of high-resolution images, especially of highly tilted specimens (Gyobu et al., 2004). Furthermore, this method provides better protection against specimen dehydration and therefore improves preparations of 2D crystals that are sensitive to drying. In the carbon sandwich technique, the crystal suspension is applied to a carbon film on a grid, and a wire loop is then used to place a second carbon film on top of the specimen. The grid is blotted and quick-frozen in liquid nitrogen before being cryo-transferred into the electron microscope. The technique is described in (Gyobu et al., 2004), a detailed protocol is provided in (Hite et al., 2010b), and a movie showing the procedure can be found on the web (http://2dx.org/download/movies/Gyobu-Sandwich.mov/view).
3.2. Embedding media
The first sugar used to prepare 2D crystals was glucose, in the preparation of purple membranes (Henderson, 1975). It is usually used to prepare specimens with the back-injection method, and this combination yielded the first atomic structure of a membrane protein, bR, determined by electron crystallography (Henderson et al., 1990). However, glucose is a reducing sugar, and in its open form has a reactive carbonyl group that can promote protein glycation. Even though glycation can occur, this reaction does not appear to have a detrimental effect on the results that can be obtained with 2D crystals prepared in glucose.
Wang and Kühlbrandt prepared their 2D crystals of plant light-harvesting complex II (LHC-II) with tannin (neutralized tannic acid). Tannin has a sugar backbone with polyphenolic groups, which provide many hydroxyl groups that can interact with water molecules. For the LHC-II crystals, which are formed by the detergent-solubilized protein in the absence of lipids, embedding in tannin consistently produced better results than embedding in glucose (Wang and Kühlbrandt, 1991). Adding tannin to glucose was also found to produce the best specimens for data collection of tubulin 2D crystals (Nogales et al., 1995).
Trehalose was first used to prepare 2D crystals of the bacterial outer membrane protein PhoE (Jap et al., 1990). The Fujiyoshi group later used trehalose to prepare 2D crystals of bR (Kimura et al., 1997). A systematic study showed that glucose preserves bR crystals better than trehalose when the specimens are air-dried, but the best results were obtained when bR crystals were embedded in trehalose and then quick-frozen (Hirai et al., 1999). Indeed, the highest-resolution data (3 Å or better) recorded of protein 2D crystals so far were almost all recorded with trehalose-embedded specimens (Gonen et al., 2005; Hite et al., 2010a; Kimura et al., 1997; Tani et al., 2009; Walian and Jap, 1990).
3.3. What method should be chosen to prepare 2D crystals?
Currently there are no clear rules that predict what preparation method will best preserve 2D crystals of a given protein. Due to its simplicity, however, electron crystallographic studies typically begin with air-dried specimens prepared in glucose with the back-injection method. The first parameter to optimize is the sugar concentration, which can range for glucose (and other sugars) from 1% to 20%. If the sample already contains glycerol, a smaller sugar concentration in the embedding medium will be necessary. If the glycerol concentration in the sample is very high, it may be beneficial to remove the glycerol from the sample by dialysis prior to preparing specimens by sugar embedding. Air-dried specimens prepared in glucose with the back-injection method usually allow the collection of data at a resolution of 3.5 Å or better (e.g., Gonen et al., 2004; Henderson et al., 1990). If 2D crystals are sufficiently well ordered to yield data at a resolution of 3.5 Å with glucose embedding, it is worth testing other sugars and the carbon sandwich technique.
Interestingly, glucose-embedded and air-dried 2D crystals of the water channel aquaporin-0 (AQP0) prepared by the back-injection method allowed determination of the AQP0 structure at 3-Å resolution when electron diffraction data were collected at liquid nitrogen temperature (Gonen et al., 2004), but specimens prepared in the same way yielded much poorer diffraction patterns when data were collected at liquid helium temperature (unpublished results). However, when the same crystals were prepared with the carbon sandwich technique in trehalose and then quick-frozen, data collected at liquid helium temperature allowed determination of the AQP0 structure at 1.9-Å resolution (Gonen et al., 2005). In contrast, specimens prepared with the carbon sandwich technique in trehalose produced data of approximately the same quality when electron diffraction patterns were recorded at liquid helium or liquid nitrogen temperature (unpublished results).
The carbon sandwich method was essential for determining the first structure of AQP4 (Hiroaki et al., 2006), was crucial for recording high-resolution data of AQP0 2D crystals (Gonen et al., 2005; Hite et al., 2010a), and yielded superior results for the double-layered 2D crystals formed by the H+,K+-ATPase, which are rather thick and sensitive to dehydration (Abe et al., 2009; Abe et al., 2011). However, the technique is not always applicable. For example, when 2D crystals formed by connexin-26 were prepared with the carbon sandwich technique, the quality of the collected data was very poor (Yoshinori Fujiyoshi, personal communication) and much better when the crystals were prepared with only a single carbon support film (Oshima et al., 2007). The carbon sandwich technique even limited the quality of data collected of AQP4 2D crystals to about 3 Å. The carbon sandwich technique likely exerts mechanical force, in particular shearing forces, on the 2D crystals, which would explain why it yields superior results for stable crystals such as those formed by AQP0 and the H+,K+-ATPase but compromises results obtained with more labile crystals such as those formed by AQP4 and connexin-26.
4. Trehalose in the preparation of 2D crystals formed on lipid monolayers
Uzgiris and Kornberg introduced lipid monolayers as a tool to produce 2D protein arrays (Uzgiris and Kornberg, 1983). In this technique, lipids solubilized in an organic solvent are added to an aqueous buffer solution, causing them to form a monolayer at the air-water interface. The polar head groups of the lipid monolayer, which are in contact with the aqueous solution, then bind and concentrate proteins contained in the aqueous solution. Under the appropriate conditions, protein arrays spontaneously form beneath the lipid monolayer in as little as 30 minutes (Kubalek et al., 1991). Since setting up 2D crystallization trials on lipid monolayers is easy and requires only very small quantities of purified protein, this technique has been used to prepare 2D arrays of a number of soluble and membrane proteins (Ellis, 2001; Lebeau et al., 2001).
4.1. Preparation of 2D arrays on lipid monolayers and transfer to EM grids
For a comprehensive description of the monolayer crystallization technique, the reader is referred to the book “Strategies for Two-Dimensional Crystallization of Proteins Using Lipid Monolayers” (Dietrich, 2005). Briefly, to grow protein arrays on lipid monolayers, ~25 μl aliquots of purified protein in buffers of various composition are placed into wells drilled into a Teflon block. 1 to 2 μl of a lipid mixture dissolved in chloroform is applied to the wells, and the block is placed into a Petri dish lined with wet filter paper and sealed with parafilm. The samples are then incubated at room temperature or at 4°C to allow formation of crystalline protein arrays.
Two methods are commonly used to transfer lipid monolayer samples to EM grids: direct transfer (Kubalek et al., 1991; Uzgiris and Kornberg, 1983) and loop transfer (Asturias and Kornberg, 1995). In the direct transfer method, an EM grid is directly placed on the sample with the carbon film touching the lipid monolayer. After a 30 seconds to 1 minute incubation, to allow the hydrophobic acyl chains to attach to the hydrophobic carbon film, the grid with the adsorbed monolayer sample is lifted off the aqueous solution. In the loop transfer method, a platinum wire loop is used to lift the lipid monolayer from the teflon block and to deposit it onto the carbon film of a glow-discharged EM grid.
4.3. Negative staining, vitrification and sugar embedding of lipid monolayer samples
Once transferred to an EM grid, lipid monolayer samples can be prepared by negative staining, vitrification or sugar embedding. Negative staining is performed as with any other specimen, i.e., the grid is blotted, washed with distilled water or buffer (if desired or needed), stained with a heavy metal salt solution, and typically air-dried (e.g., De Carlo and Harris, 2011; Ohi et al., 2004).
For vitrification, the grid is blotted by applying a piece of filter paper directly against the sample face-on until the liquid interface between the grid and the filter paper wicks away and separates from the grid. The sample is then quickly plunged into liquid ethane (Taylor et al., 2007). As for other samples, the blotting time has to be optimized to avoid generating a vitrified ice layer that is too thick or too thin.
For sugar embedding, the monolayer is transferred to an EM grid, excess aqueous solution is removed with a pipetman, and 2 – 3 μl of sugar solution is immediately added to the grid. The grid is then blotted as described in the previous paragraph. If glucose is used for embedding, the specimen is usually allowed to air-dry, although it can also be quick-frozen. If trehalose is used for embedding, the grid is usually quick-frozen by plunging it into liquid nitrogen or liquid ethane.
4.4. Comparison of vitrification and trehalose embedding of lipid monolayer specimens
Sugar embedding is the method of choice for the preparation of 2D crystals formed by membrane proteins reconstituted into lipid bilayers, but it has not been widely used for the preparation of 2D crystals formed on lipid monolayers. An early study with streptavidin crystals grown on biotinylated lipid monolayers showed that embedding in 1% glucose preserved the crystals better than vitrification (Kubalek et al., 1991). Diffraction spots in electron diffraction patterns of the vitrified crystals were limited to a resolution of 3 Å, whereas glucose-embedded crystals diffracted beyond 3 Å resolution (Kubalek et al., 1991). However, a later study that crystallized streptavidin on a monolayer with a slightly different lipid composition obtained electron diffraction patterns that showed diffraction spots to a resolution better than 2.5 Å with 2D crystals prepared with both glucose embedding and vitrification (Avila-Sakar and Chiu, 1996).
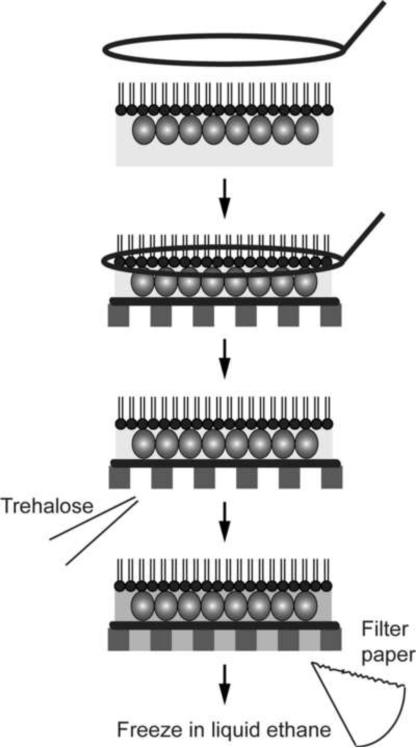
Trehalose embedding has only recently been tested for the preparation of lipid monolayer crystals. The S-layer protein SbpA that forms crystalline arrays on the surface of Bacillus sphaericus has been studied extensively by EM. While its propensity to form 2D arrays make it an ideal specimen, structural information of SbpA obtained from electron crystallographic studies was limited in resolution to about 20 Å (Aebi et al., 1974; Lepault and Pitt, 1984; Lepault et al., 1986; Sleytr et al., 1994). A recent comparison of different transfer methods, embedding media and freezing conditions found that SbpA 2D crystals are best preserved when the loop transfer is used and when crystals are embedded in 5% trehalose and quick-frozen in liquid ethane (Figure 4). This preparation method allowed calculation of a projection map at 7 Å resolution (Norville et al., 2007), while all other tested preparation methods resulted in specimens that were significantly less well preserved (Figure 5).
Figure 4. Trehalose embedding used for 2D crystals grown on lipid monolayers.
The lipid monolayer with the attached 2D crystal is transferred with a wire loop onto the carbon film on an EM grid, and the trehalose solution is applied to the opposite side of the grid. Excess solution is blotted away with a filter paper and the grid quick-frozen in liquid ethane.
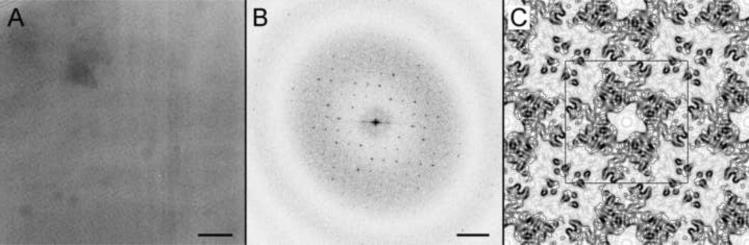
Figure 5. Structure analysis of trehalose-embedded SbpA monolayer 2D crystals.
(A) Image of a trehalose-embedded SbpA 2D crystal grown on a lipid monolayer. Scale bar is 200 nm. (B) Power spectrum of the image shown in (A). Scale bar is (5 nm)−1. (C) Projection map of SbpA at 7 Å resolution. A unit cell with lattice constants a = b = 13.3 nm is outlined in black.
5. Trehalose in the preparation of single-particle EM specimens
The most commonly used techniques to prepare specimens for single-particle EM are negative staining and vitrification, and neither one of these two techniques traditionally employs sugar. Trehalose has been added to single-particle samples before vitrification and was found to reduce beam damage, drying artifacts and water evaporation (De Carlo et al., 1999), but adding sugar to a sample that is being vitrified brings the density of the buffer closer to that of the protein that is being imaged. Such density matching reduces the image contrast and can cause problems, for example, if the biological sample contains a high glycerol concentration. Similarly, drying of proteins in a 1% trehalose solution on holey carbon grids produces thin vitreous trehalose films across the holes that embed and preserve the proteins. Because of the similar density of the trehalose layer and the embedded proteins, this method only produces good results for inherently electron-dense particles, such as ferritin, frataxin, viruses, gold and polymer particles and the likes (Harris and Scheffler, 2002; Harris, 2008).
On the other hand, adding glycerol and sugars to negative staining solution has advantages as their addition helps counteract specimen flattening during drying of the specimen. In particular, with the advent of cryo-negative staining techniques, glycerol and sugars have become more widespread in the preparation of negatively stained specimens (De Carlo and Stark, 2010; De Carlo and Harris, 2011).
5.1. Trehalose in the preparation of negatively stained specimens for single-particle EM
To prepare a negatively stained specimen for single-particle EM, the protein solution is typically applied to a glow-discharged EM grid covered with a continuous carbon film. The grid is then often washed with deionized water, stained with a heavy metal salt solution and dried. In images of negatively stained samples, the protein appears as regions that exclude the stain, hence the name negative staining. While the stain forms microcrystals that embed and somewhat stabilize the structure of the proteins, negative staining introduces artifacts (e.g., Cheng et al., 2006). For example, any part of a molecule that protrudes from the stain layer will be invisible in the image. Also, the drying of the grid often causes the molecules to flatten, an effect that becomes increasingly severe as the thickness of the molecule increases. These issues can be minimized either by adding sugar to the staining solution or by quick-freezing the stained specimen or by a combination of both.
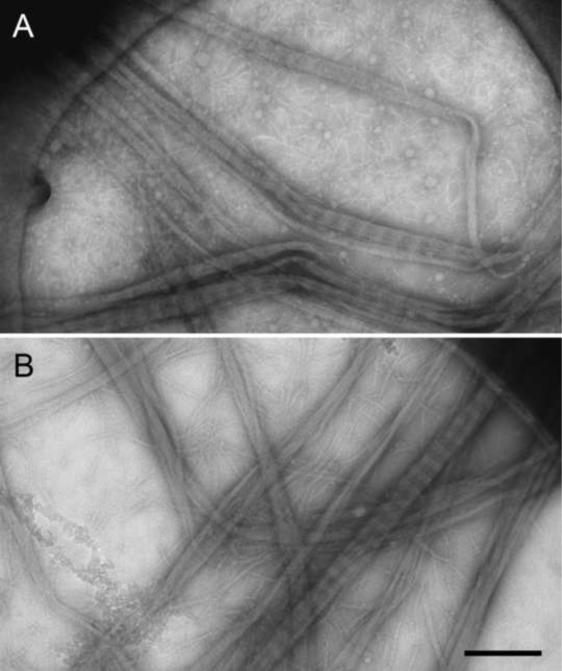
An early implementation of adding sugar to the staining solution to provide for better stain embedding and to reduce specimen flattening upon drying was aurothioglucose, a glucose molecule with a covalently linked gold atom. Aurothioglucose was first used to prepare 2D arrays of eukaryotic ribosomes (Kühlbrandt, 1982) but was later also applied to single particle specimens (e.g., Baumeister et al., 1988). In a related approach, sugar is simply added to the staining solution, which was again first applied to 2D crystalline specimens. For example, sucrose was added to a sodium phosphotungstate solution to prepare the hexagonally packed interlayer (HPI-layer) of Deinococcus radiodurans (Baumeister et al., 1986) and glucose was added to prepare the tetragonal surface layer of Sporosarcina ureae (Engelhardt et al., 1986). Later, sugar was also added to staining solution for the preparation of specimens for single-particle EM imaging. For example, glucose was added to an ammonium molybdate solution to image keyhole limpet hemocyanin (Orlova et al., 1997). In a further development, single-particle and filamentous specimens were prepared using an ammonium molybdate solution containing trehalose on holey carbon films, producing preparations that were superior compared to those on continuous carbon support (Figure 6A) (Harris and Scheffler, 2002; Harris, 2008). While trehalose was first added to an ammonium molybdate stain, it can also be added to other staining solutions, including sodium phosphotungstate stain, which gives particularly good results (Robin Harris, personal communication), and uranium-based stains (see below). Sugars are very beam sensitive and tend to bubble upon exposure to an electron beam. Therefore, specimens prepared with a staining solution containing sugar are best imaged at liquid nitrogen temperature.
Figure 6. Collagen type 1 fibers prepared by different staining methods.
(A) Image of collagen fibers that were spread across a holey carbon film, negatively stained with a trehalose-containing ammonium molybdate solution and air-dried. (B) Image of collagen fibers that were quick-frozen in a saturated ammonium molybdate solution. Scale bar is 300 nm. Figure adapted from Harris, 2008.
Cryo-negative staining is another approach that was developed to improve stain embedding and to minimize specimen flattening. Cryo-negative staining comes in two flavors (De Carlo and Stark, 2010). Adrian and co-workers added a high concentration of ammonium molybdate to the protein solution and then vitrified the specimen by plunging the grid into liquid ethane (Adrian et al., 1998) (see Figure 6B for an example). While ammonium molybdate boosts the image contrast, not every specimen, in particular not every macromolecular complex, can withstand the high ionic strength introduced by the high ammonium molybdate concentration. Stark and co-workers developed an alternative cryo-negative staining procedure (Stark, 2010; see also Ohi et al., 2004 for a slightly modified protocol). In this preparation, the protein is adsorbed to a glow-discharged EM grid covered with a carbon film and embedded and stained with a glycerol-containing uranyl formate solution. The specimen is then covered with a second carbon film and frozen in liquid nitrogen. The preparation is challenging because specimens are often too thick, so that the electron beam cannot penetrate, or too thin, so that the particles are squashed between the two carbon films (Cheng et al., 2006). However, in a perfectly prepared specimen the added glycerol and the two carbon films guarantee that the molecules are completely embedded in stain, and the glycerol in the stain layer combined with quick-freezing ensure minimal flattening of the molecules. Molecules prepared in this way usually show preferred orientations and 3D reconstructions thus have to be calculated using the random conical tilt approach (Radermacher et al., 1987). The resulting density maps are commonly limited in resolution to about 20 to 30 Å, but they can serve as reliable initial models for alignment of particle images from vitrified specimens (e.g., Ohi et al., 2007a; Ohi et al., 2007b; Yip et al., 2010).
5.2. Trehalose in the preparation of Affinity Grid specimens
Monolayer purification and Affinity Grids have recently been developed to combine protein purification and specimen preparation for single-particle EM into a single, fast step (Kelly et al., 2008a; Kelly et al., 2008b). These techniques are based on lipid monolayers, either on an air-water interface (monolayer purification) or pre-adsorbed to the carbon film on an EM grid (Affinity Grid), that contain lipids functionalized with a Nickel-nitrilotriacetic acid (Ni-NTA) group. These monolayers can be used to recruit His-tagged proteins or macromolecular complexes from an impure protein solution or even directly from a cell extract. Using an adaptor system consisting of His-tagged protein A and a specific antibody, Affinity Grids can also be used to prepare non-His-tagged molecules (Kelly et al., 2010a). Once the target molecules have been bound to the Affinity Grid, the specimen can be prepared by negative staining or vitrification. For a review on the use of monolayer purification and Affinity Grid, see (Kelly et al., 2010b).
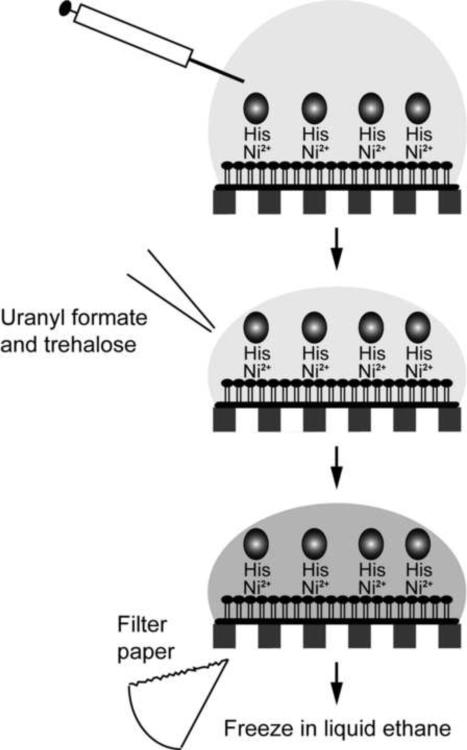
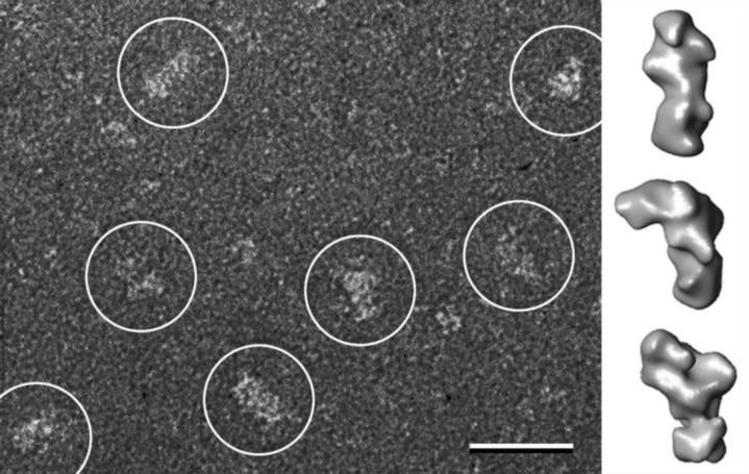
Monolayer purification and Affinity Grids are based on functionalized lipid monolayers. While lipid monolayers are compatible with negative staining and vitrification, they are sensitive to high concentrations of glycerol. Although Affinity Grids, in which the lipid monolayer is stabilized by the interaction with the carbon support film, can tolerate glycerol to some degree (Kelly et al., 2008b), they do not survive the glycerol-based cryo-negative staining procedure introduced by Stark and co-workers. Furthermore, the ammonium molybdate-based cryo-negative staining procedure developed by Adrian and co-workers tends to produce specimens that are too thick. Thus, the only cryo-negative staining procedure that could be used to prepare Affinity Grid specimens was to add trehalose to the uranyl formate staining solution, followed by blotting and freezing in liquid ethane (Figure 7). This procedure was used to prepare cryo-negatively stained Affinity Grid specimens of the Notch extracellular domain, which allowed calculation of 3D density maps at 25 Å resolution by the random conical tilt approach (Figure 8) (Kelly et al., 2010c).
Figure 7. Cryo-negative staining of Affinity Grid preparations.
After incubating the Affinity Grid with sample, excess solution is removed with a Hamilton syringe and trehalose-containing staining solution is added to the grid. The grid is then blotted with filter paper and quick-frozen in liquid ethane.
Figure 8. Structure analysis of the Notch extracellular domain (NECD) using a cryo-negatively stained Affinity Grid preparation.
The left panel shows a raw image with the white circles indicating individual particles. The scale bar is 25 nm. The right panels show 3D reconstructions of the NECD in three different conformations. Figure adapted from Kelly et al., 2010c.
6. Conclusions
Because of its superior characteristics in stabilizing proteins, trehalose is used by many organisms for protection against stress conditions. The same characteristics make trehalose an excellent additive for the preparation of specimens for cryo-EM. In combination with the carbon sandwich technique, it appears to be the optimal embedding medium for 2D crystals for analysis by electron crystallography. In combination with a heavy metal solution, it can also be used to prepare cryo-negatively stained specimens for single-particle EM, and in particular it is currently the only established embedding medium for cryo-negative staining of Affinity Grid specimens.
Acknowledgments
Work in the Walz laboratory is supported by NIH grants P01 GM062580 (to Stephen C. Harrison), R01 GM082927 and R01 EY015107. TW is an investigator in the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe K, Tani K, Nishizawa T, Fujiyoshi Y. Inter-subunit interaction of gastric H+,K+-ATPase prevents reverse reaction of the transport cycle. EMBO J. 2009;28:1637–1643. doi: 10.1038/emboj.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe K, Tani K, Fujiyoshi Y. Conformational rearrangement of gastric H+,K+- ATPase induced by an acid suppressant. Nat. Commun. 2011;2:155. doi: 10.1038/ncomms1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeyrathne PD, Chami M, Pantelic RS, Goldie KN, Stahlberg H. Preparation of 2D crystals of membrane proteins for high-resolution electron crystallography data collection. Methods Enzymol. 2010;481:25–43. doi: 10.1016/S0076-6879(10)81001-8. [DOI] [PubMed] [Google Scholar]
- Adrian M, Dubochet J, Lepault J, McDowall AW. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- Adrian M, Dubochet J, Fuller SD, Harris JR. Cryo-negative staining. Micron. 1998;29:145–160. doi: 10.1016/s0968-4328(97)00068-1. [DOI] [PubMed] [Google Scholar]
- Aebi U, Bijlenga R, v d Broek J, v d Broek H, Eiserling F, Kellenberger C, Kellenberger E, Mesyanzhinov V, Muller L, Showe M, Smith R, Steven A. The transformation of tau particles into T4 heads. II. Transformations of the surface lattice and related observations on form determination. J. Supramol. Struct. 1974;2:253–275. doi: 10.1002/jss.400020218. [DOI] [PubMed] [Google Scholar]
- Asturias FJ, Kornberg RD. A novel method for transfer of two-dimensional crystals from the air/water interface to specimen grids. EM sample preparation/lipid-layer crystallization. J. Struct. Biol. 1995;114:60–66. doi: 10.1006/jsbi.1995.1005. [DOI] [PubMed] [Google Scholar]
- Avila-Sakar AJ, Chiu W. Visualization of β-sheets and side-chain clusters in two-dimensional periodic arrays of streptavidin on phospholipid monolayers by electron crystallography. Biophys. J. 1996;70:57–68. doi: 10.1016/S0006-3495(96)79597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker LA, Rubinstein JL. Radiation damage in electron cryomicroscopy. Methods Enzymol. 2010;481:371–388. doi: 10.1016/S0076-6879(10)81015-8. [DOI] [PubMed] [Google Scholar]
- Baumeister W, Barth M, Hegerl R, Guckenberger R, Hahn M, Saxton WO. Three-dimensional structure of the regular surface layer (HPI layer) of Deinococcus radiodurans. J. Mol. Biol. 1986;187:241–250. doi: 10.1016/0022-2836(86)90231-7. [DOI] [PubMed] [Google Scholar]
- Baumeister W, Dahlmann B, Hegerl R, Kopp F, Kuehn L, Pfeifer G. Electron microscopy and image analysis of the multicatalytic proteinase. FEBS Lett. 1988;241:239–245. doi: 10.1016/0014-5793(88)81069-x. [DOI] [PubMed] [Google Scholar]
- Benaroudj N, Lee DH, Goldberg AL. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J. Biol. Chem. 2001;276:24261–24267. doi: 10.1074/jbc.M101487200. [DOI] [PubMed] [Google Scholar]
- Bravim F, de Freitas JM, Fernandes AA, Fernandes PM. High hydrostatic pressure and the cell membrane: stress response of Saccharomyces cerevisiae. Ann. N. Y. Acad. Sci. 2010;1189:127–132. doi: 10.1111/j.1749-6632.2009.05182.x. [DOI] [PubMed] [Google Scholar]
- Brink J, Sherman MB, Berriman J, Chiu W. Evaluation of charging on macromolecules in electron cryomicroscopy. Ultramicroscopy. 1998;72:41–52. doi: 10.1016/s0304-3991(97)00126-5. [DOI] [PubMed] [Google Scholar]
- Chen T, Fowler A, Toner M. Literature review: supplemented phase diagram of the trehalose-water binary mixture. Cryobiology. 2000;40:277–282. doi: 10.1006/cryo.2000.2244. [DOI] [PubMed] [Google Scholar]
- Chen Q, Haddad GG. Role of trehalose phosphate synthase and trehalose during hypoxia: from flies to mammals. J. Exp. Biol. 2004;207:3125–3129. doi: 10.1242/jeb.01133. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Wolf E, Larvie M, Zak O, Aisen P, Grigorieff N, Harrison SC, Walz T. Single particle reconstructions of the transferrin-transferrin receptor complex obtained with different specimen preparation techniques. J. Mol. Biol. 2006;355:1048–1065. doi: 10.1016/j.jmb.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Cheong GW, Guckenberger R, Fuchs KH, Gross H, Baumeister W. The structure of the surface layer of Methanoplanus limicola obtained by a combined electron microscopy and scanning tunneling microscopy approach. J. Struct. Biol. 1993;111:125–134. [Google Scholar]
- Colaço C, Sen S, Thangavelu M, Pinder S, Roser B. Extraordinary stability of enzymes dried in trehalose: simplified molecular biology. Biotechnology (New York) 1992;10:1007–1011. doi: 10.1038/nbt0992-1007. [DOI] [PubMed] [Google Scholar]
- Crowe JH, Crowe LM, Chapman D. Preservation of membranes in anhydrobiotic organisms: the role of trehalose. Science. 1984;223:701–703. doi: 10.1126/science.223.4637.701. [DOI] [PubMed] [Google Scholar]
- Crowe JH, Crowe LM, Carpenter JF, Aurell Wistrom C. Stabilization of dry phospholipid bilayers and proteins by sugars. Biochem. J. 1987;242:1–10. doi: 10.1042/bj2420001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe JH, Hoekstra FA, Crowe LM. Anhydrobiosis. Annu. Rev. Physiol. 1992;54:579–599. doi: 10.1146/annurev.ph.54.030192.003051. [DOI] [PubMed] [Google Scholar]
- Crowe JH, Hoekstra FA, Nguyen KH, Crowe LM. Is vitrification involved in depression of the phase transition temperature in dry phospholipids? Biochim. Biophys. Acta. 1996;1280:187–196. doi: 10.1016/0005-2736(95)00287-1. [DOI] [PubMed] [Google Scholar]
- Crowe LM. Lessons from nature: the role of sugars in anhydrobiosis. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2002;131:505–513. doi: 10.1016/s1095-6433(01)00503-7. [DOI] [PubMed] [Google Scholar]
- Daffe M, Draper P. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 1998;39:131–203. doi: 10.1016/s0065-2911(08)60016-8. [DOI] [PubMed] [Google Scholar]
- De Carlo S, Adrian M, Kalin P, Mayer JM, Dubochet J. Unexpected property of trehalose as observed by cryo-electron microscopy. J. Microsc. 1999;196:40–45. doi: 10.1046/j.1365-2818.1999.00591.x. [DOI] [PubMed] [Google Scholar]
- De Carlo S, Stark H. Cryonegative staining of macromolecular assemblies. Methods Enzymol. 2010;481:127–145. doi: 10.1016/S0076-6879(10)81006-7. [DOI] [PubMed] [Google Scholar]
- De Carlo S, Harris JR. Negative staining and cryo-negative staining of macromolecules and viruses for TEM. Micron. 2011;42:117–131. doi: 10.1016/j.micron.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C, Hottiger T, Dominguez J, Boller T, Wiemken A. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. I. Genetic evidence that trehalose is a thermoprotectant. Eur. J. Biochem. 1994;219:179–186. doi: 10.1111/j.1432-1033.1994.tb19928.x. [DOI] [PubMed] [Google Scholar]
- Dietrich J, Venien-Bryan C. Strategies for two-dimensional crystallization of proteins using lipid monolayers. Imperial College Press; 2005. [Google Scholar]
- Dobro MJ, Melanson LA, Jensen GJ, McDowall AW. Plunge freezing for electron cryomicroscopy. Methods Enzymol. 2010;481:63–82. doi: 10.1016/S0076-6879(10)81003-1. [DOI] [PubMed] [Google Scholar]
- Downing KH, McCartney MR, Glaeser RM. Experimental characterization and mitigation of specimen charging on thin films with one conducting layer. Microsc. Microanal. 2004;10:783–789. doi: 10.1017/s143192760404067x. [DOI] [PubMed] [Google Scholar]
- Dubochet J, Chang JJ, Freeman R, Lepault J, McDowall AW. Frozen aqueous suspensions. Ultramicroscopy. 1982;10:55–61. [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Egerton RF, Li P, Malac M. Radiation damage in the TEM and SEM. Micron. 2004;35:399–409. doi: 10.1016/j.micron.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Elbein AD, Pan YT, Pastuszak I, Carroll D. New insights on trehalose: a multifunctional molecule. Glycobiology. 2003;13:17r–27r. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- Elliott B, Haltiwanger RS, Futcher B. Synergy between trehalose and Hsp104 for thermotolerance in Saccharomyces cerevisiae. Genetics. 1996;144:923–933. doi: 10.1093/genetics/144.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Hebert H. Structure analysis of soluble proteins using electron crystallography. Micron. 2001;32:541–550. doi: 10.1016/s0968-4328(00)00049-4. [DOI] [PubMed] [Google Scholar]
- Ellis T. Proteins as molecular chaperones. Nature. 1987;328:378–379. doi: 10.1038/328378a0. [DOI] [PubMed] [Google Scholar]
- Engelhardt H, Saxton WO, Baumeister W. Three-dimensional structure of the tetragonal surface layer of Sporosarcina ureae. J. Bacteriol. 1986;168:309–317. doi: 10.1128/jb.168.1.309-317.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez O, Bethencourt L, Quero A, Sangwan RS, Clement C. Trehalose and plant stress responses: friend or foe? Trends Plant. Sci. 2010;15:409–417. doi: 10.1016/j.tplants.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Fujiyoshi Y, Mizusaki T, Morikawa K, Yamagishi H, Aoki Y, Kihara H, Harada Y. Development of a superfluid helium stage for high-resolution electron microscopy. Ultramicroscopy. 1991;38:241–251. [Google Scholar]
- Fujiyoshi Y. The structural study of membrane proteins by electron crystallography. Adv. Biophys. 1998;35:25–80. doi: 10.1016/s0065-227x(98)90004-1. [DOI] [PubMed] [Google Scholar]
- Fujiyoshi Y, Unwin N. Electron crystallography of proteins in membranes. Curr. Opin. Struct. Biol. 2008;18:587–592. doi: 10.1016/j.sbi.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen T, Sliz P, Kistler J, Cheng Y, Walz T. Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature. 2004;429:193–197. doi: 10.1038/nature02503. [DOI] [PubMed] [Google Scholar]
- Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassucci RA, Taylor DJ, Frank J. Preparation of macromolecular complexes for cryo-electron microscopy. Nat. Protoc. 2007;2:3239–3246. doi: 10.1038/nprot.2007.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JL, Angell CA. Phase relations and vitrification in saccharide-water solutions and the trehalose anomaly. J. Phys. Chem. 1989;93:2880–2882. [Google Scholar]
- Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. Electron-crystallographic refinement of the structure bacteriorhodopsin. J. Mol. Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- Gyobu N, Tani K, Hiroaki Y, Kamegawa A, Mitsuoka K, Fujiyoshi Y. Improved specimen preparation for cryo-electron microscopy using a symmetric carbon sandwich technique. J. Struct. Biol. 2004;146:325–333. doi: 10.1016/j.jsb.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Harding TS. History of trehalose, its discovery and methods of preparation. Sugar. 1923;25:476–478. [Google Scholar]
- Harris JR, Gebauer W, Markl J. Keyhole limpet haemocyanin: negative staining in the presence of trehalose. Micron. 1995;26:25–33. [Google Scholar]
- Harris RJ, Gerber M, Gebauer W, Wernicke W, Markl J. Negative stains containing trehalose: application to tubular and filamentous structures. J. Microsc. Soc. Am. 1996;2:42–53. [Google Scholar]
- Harris JR, Adrian M. Preparation of thin-film frozen-hydrated/vitrified biological specimens for cryoelectron microscopy. Methods Mol. Biol. 1999;117:31–48. doi: 10.1385/1-59259-201-5:31. [DOI] [PubMed] [Google Scholar]
- Harris JR, Scheffler D. Routine preparation of air-dried negatively stained and unstained specimens on holey carbon support films: a review of applications. Micron. 2002;33:461–480. doi: 10.1016/s0968-4328(01)00039-7. [DOI] [PubMed] [Google Scholar]
- Harris JR. Negative staining across holes: application to fibril and tubular structures. Micron. 2008;39:168–176. doi: 10.1016/j.micron.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Hasler L, Heymann JB, Engel A, Kistler J, Walz T. 2D crystallization of membrane proteins: rationales and examples. J. Struct. Biol. 1998;121:162–171. doi: 10.1006/jsbi.1998.3960. [DOI] [PubMed] [Google Scholar]
- Henderson R. The structure of the purple membrane from Halobacterium hallobium: analysis of the X-ray diffraction pattern. J. Mol. Biol. 1975;93:123–138. doi: 10.1016/0022-2836(75)90123-0. [DOI] [PubMed] [Google Scholar]
- Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Herdeiro RS, Pereira MD, Panek AD, Eleutherio ECA. Trehalose protects Saccharomyces cerevisiae from lipid peroxidation during oxidative stress. Biochim. Biophys. Acta Gen. Subj. 2006;1760:340–346. doi: 10.1016/j.bbagen.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Hirai T, Murata K, Mitsuoka K, Kimura Y, Fujiyoshi Y. Trehalose embedding technique for high-resolution electron crystallography: application to structural study on bacteriorhodopsin. J. Electron Microsc. (Tokyo) 1999;48:653–658. doi: 10.1093/oxfordjournals.jmicro.a023731. [DOI] [PubMed] [Google Scholar]
- Hiroaki Y, Tani K, Kamegawa A, Gyobu N, Nishikawa K, Suzuki H, Walz T, Sasaki S, Mitsuoka K, Kimura K, Mizoguchi A, Fujiyoshi Y. Implications of the aquaporin-4 structure on array formation and cell adhesion. J. Mol. Biol. 2006;355:628–639. doi: 10.1016/j.jmb.2005.10.081. [DOI] [PubMed] [Google Scholar]
- Hite RK, Raunser S, Walz T. Revival of electron crystallography. Curr. Opin. Struct. Biol. 2007;17:389–395. doi: 10.1016/j.sbi.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hite RK, Li Z, Walz T. Principles of membrane protein interactions with annular lipids deduced from aquaporin-0 2D crystals. EMBO J. 2010a;29:1652–1658. doi: 10.1038/emboj.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hite RK, Schenk AD, Li Z, Cheng Y, Walz T. Collecting electron crystallographic data of two-dimensional protein crystals. Methods Enzymol. 2010b;481:251–282. doi: 10.1016/S0076-6879(10)81011-0. [DOI] [PubMed] [Google Scholar]
- Holm PJ, Bhakat P, Jegerschöld C, Gyobu N, Mitsuoka K, Fujiyoshi Y, Morgenstern R, Hebert H. Structural basis for detoxification and oxidative stress protection in membranes. J. Mol. Biol. 2006;360:934–945. doi: 10.1016/j.jmb.2006.05.056. [DOI] [PubMed] [Google Scholar]
- Jain NK, Roy I. Effect of trehalose on protein structure. Protein Sci. 2009;18:24–36. doi: 10.1002/pro.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain NK, Roy I. Trehalose and protein stability. Curr. Protoc. Protein Sci. 2010;Chapter 4:4.9.1–4.9.12. doi: 10.1002/0471140864.ps0409s59. [DOI] [PubMed] [Google Scholar]
- Jap BK, Downing KH, Walian PJ. Structure of PhoE porin in projection at 3.5 Å resolution. J. Struct. Biol. 1990;103:57–63. doi: 10.1016/1047-8477(90)90086-r. [DOI] [PubMed] [Google Scholar]
- Jegerschöld C, Pawelzik SC, Purhonen P, Bhakat P, Gheorghe KR, Gyobu N, Mitsuoka K, Morgenstern R, Jakobsson PJ, Hebert H. Structural basis for induced formation of the inflammatory mediator prostaglandin E2. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11110–11115. doi: 10.1073/pnas.0802894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandror O, DeLeon A, Goldberg AL. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9727–9732. doi: 10.1073/pnas.142314099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DF, Dukovski D, Walz T. Monolayer purification: a rapid method for isolating protein complexes for single-particle electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 2008a;105:4703–4708. doi: 10.1073/pnas.0800867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DF, Abeyrathne PD, Dukovski D, Walz T. The Affinity Grid: a prefabricated EM grid for monolayer purification. J. Mol. Biol. 2008b;382:423–433. doi: 10.1016/j.jmb.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DF, Dukovski D, Walz T. Strategy for the use of Affinity Grids to prepare non-His-tagged macromolecular complexes for single-particle electron microscopy. J. Mol. Biol. 2010a;400:675–681. doi: 10.1016/j.jmb.2010.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DF, Dukovski D, Walz T. A practical guide to the use of monolayer purification and affinity grids. Methods Enzymol. 2010b;481:83–107. doi: 10.1016/S0076-6879(10)81004-3. [DOI] [PubMed] [Google Scholar]
- Kelly DF, Lake RJ, Middelkoop TC, Fan HY, Artavanis-Tsakonas S, Walz T. Molecular structure and dimeric organization of the Notch extracellular domain as revealed by electron microscopy. PLoS One. 2010c;5:e10532. doi: 10.1371/journal.pone.0010532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf B, Bremer E. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 1998;170:319–330. doi: 10.1007/s002030050649. [DOI] [PubMed] [Google Scholar]
- Kilburn D, Townrow S, Meunier V, Richardson R, Alam A, Ubbink J. Organization and mobility of water in amorphous and crystalline trehalose. Nat. Mater. 2006;5:632–635. doi: 10.1038/nmat1681. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Vassylyev DG, Miyazawa A, Kidera A, Matsushima M, Mitsuoka K, Murata K, Hirai T, Fujiyoshi Y. Surface of bacteriorhodopsin revealed by high-resolution electron crystallography. Nature. 1997;389:206–211. doi: 10.1038/38323. [DOI] [PubMed] [Google Scholar]
- Kubalek EW, Kornberg RD, Darst SA. Improved transfer of two-dimensional crystals from the air/water interface to specimen support grids for high-resolution analysis by electron microscopy. Ultramicroscopy. 1991;35:295–304. doi: 10.1016/0304-3991(91)90082-h. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W. Discrimination of protein and nucleic acids by electron microscopy using contrast variation. Ultramicroscopy. 1982;7:221–232. doi: 10.1016/0304-3991(82)90169-3. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W. Two-dimensional crystallization of membrane proteins. Q. Rev. Biophys. 1992;25:1–49. doi: 10.1017/s0033583500004716. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W, Wang DN, Fujiyoshi Y. Atomic model of plant light-harvesting complex by electron crystallography. Nature. 1994;367:614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- Kuo AM, Glaeser RM. Development of methodology for low exposure, high resolution electron microscopy of biological specimens. Ultramicroscopy. 1975;1:53. doi: 10.1016/s0304-3991(75)80007-6. [DOI] [PubMed] [Google Scholar]
- Lebeau L, Lach F, Venien-Bryan C, Renault A, Dietrich J, Jahn T, Palmgren MG, Kühlbrandt W, Mioskowski C. Two-dimensional crystallization of a membrane protein on a detergent-resistant lipid monolayer. J. Mol. Biol. 2001;308:639–647. doi: 10.1006/jmbi.2001.4629. [DOI] [PubMed] [Google Scholar]
- Lee CW, Waugh JS, Griffin RG. Solid-state NMR study of trehalose/1,2-dipalmitoyl-sn-phosphatidylcholine interactions. Biochemistry. 1986;25:3737–3742. doi: 10.1021/bi00361a001. [DOI] [PubMed] [Google Scholar]
- Leekumjorn S, Wu Y, Sum AK, Chan C. Experimental and computational studies investigating trehalose protection of HepG2 cells from palmitate-induced toxicity. Biophys. J. 2008;94:2869–2883. doi: 10.1529/biophysj.107.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepault J, Pitt T. Projected structure of unstained, frozen-hydrated T-layer of Bacillus brevis. EMBO J. 1984;3:101–105. doi: 10.1002/j.1460-2075.1984.tb01768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepault J, Martin N, Leonard K. Three-dimensional structure of the T-layer of Bacillus sphaericus P-1. J. Bacteriol. 1986;168:303–308. doi: 10.1128/jb.168.1.303-308.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Zhang SX, Downing KH. Preservation of 2-D crystals of tubulin for electron crystallography. J. Struct. Biol. 1995;115:199–208. doi: 10.1006/jsbi.1995.1044. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the α,β tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Norville JE, Kelly DF, Knight TF, Jr., Belcher AM, Walz T. 7 Å projection map of the S-layer protein SbpA obtained with trehalose-embedded monolayer crystals. J. Struct. Biol. 2007;160:313–323. doi: 10.1016/j.jsb.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe MA, Allard LF, Blom DA. HRTEM imaging of atoms at sub-Ångström resolution. J. Elec. Micros. 2005;54:169–180. doi: 10.1093/jmicro/dfi036. [DOI] [PubMed] [Google Scholar]
- Oku K, Kurose M, Kubota M, Fukuda S, Kurimoto M, Tujisaka Y, Okabe A, Sakurai M. Combined NMR and quantum chemical studies on the interaction between trehalose and dienes relevant to the antioxidant function of trehalose. J. Phys. Chem. B. 2005;109:3032–3040. doi: 10.1021/jp045906w. [DOI] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol. Proced. Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Feoktistova A, Ren L, Yip C, Cheng Y, Chen JS, Yoon HJ, Wall JS, Huang Z, Penczek PA, Gould KL, Walz T. Structural organization of the anaphase-promoting complex bound to the mitotic activator Slp1. Mol. Cell. 2007a;28:871–885. doi: 10.1016/j.molcel.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Ren L, Wall JS, Gould KL, Walz T. Structural characterization of the fission yeast U5.U2/U6 spliceosome complex. Proc. Natl. Acad. Sci. U.S.A. 2007b;104:3195–3200. doi: 10.1073/pnas.0611591104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova EV, Dube P, Harris JR, Beckman E, Zemlin F, Markl J, van Heel M. Structure of keyhole limpet hemocyanin type 1 (KLH1) at 15 Å resolution by electron cryomicroscopy and angular reconstitution. J. Mol. Biol. 1997;271:417–437. doi: 10.1006/jmbi.1997.1182. [DOI] [PubMed] [Google Scholar]
- Oshima A, Tani K, Hiroaki Y, Fujiyoshi Y, Sosinsky GE. Three-dimensional structure of a human connexin26 gap junction channel reveals a plug in the vestibule. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10034–10039. doi: 10.1073/pnas.0703704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva CL, Panek AD. Biotechnological applications of the disaccharide trehalose. Biotechnol. Annu. Rev. 1996;2:293–314. doi: 10.1016/s1387-2656(08)70015-2. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J. Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- Raunser S, Walz T. Electron crystallography as a technique to study the structure on membrane proteins in a lipidic environment. Annu. Rev. Biophys. 2009;38:89–105. doi: 10.1146/annurev.biophys.050708.133649. [DOI] [PubMed] [Google Scholar]
- Reimer L, Ross-Messemer M. Contrast in the electron spectroscopic imaging mode of a TEM. II. Z-ratio, structure-sensitive and phase contrast. J. Microsc. 1990;159:143. [Google Scholar]
- Reinders A, Romano I, Wiemken A, De Virgilio C. The thermophilic yeast Hansenula polymorpha does not require trehalose synthesis for growth at high temperatures but does for normal acquisition of thermotolerance. J. Bacteriol. 1999;181:4665–4668. doi: 10.1128/jb.181.15.4665-4668.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Reddy VS, Cheng A, Melnyk P, Mitra AK. Visualization of a water-selective pore by electron crystallography in vitreous ice. Proc. Natl. Acad. Sci. U.S.A. 2001;98:1398–1403. doi: 10.1073/pnas.041489198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J. Sugar sensing and signaling in plants. Plant Cell. 2002;14(Suppl):S185–S205. doi: 10.1105/tpc.010455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk AD, Castano-Diez D, Gipson B, Arheit M, Zeng X, Stahlberg H. 3D reconstruction from 2D crystal image and diffraction data. Methods Enzymol. 2010;482:101–129. doi: 10.1016/S0076-6879(10)82004-X. [DOI] [PubMed] [Google Scholar]
- Singer MA, Lindquist S. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol. Cell. 1998;1:639–648. doi: 10.1016/s1097-2765(00)80064-7. [DOI] [PubMed] [Google Scholar]
- Sleytr UB, Sara M, Messner P, Pum D. Two-dimensional protein crystals (S-layers): fundamentals and applications. J. Cell Biochem. 1994;56:171–176. doi: 10.1002/jcb.240560209. [DOI] [PubMed] [Google Scholar]
- Sola-Penna M, Meyer-Fernandes JR. Stabilization against thermal inactivation promoted by sugars on enzyme structure and function: why is trehalose more effective than other sugars? Arch. Biochem. Biophys. 1998;360:10–14. doi: 10.1006/abbi.1998.0906. [DOI] [PubMed] [Google Scholar]
- Stark H. GraFix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 2010;481:109–126. doi: 10.1016/S0076-6879(10)81005-5. [DOI] [PubMed] [Google Scholar]
- Tani K, Mitsuma T, Hiroaki Y, Kamegawa A, Nishikawa K, Tanimura Y, Fujiyoshi Y. Mechanism of aquaporin-4's fast and highly selective water conduction and proton exclusion. J. Mol. Biol. 2009;389:694–706. doi: 10.1016/j.jmb.2009.04.049. [DOI] [PubMed] [Google Scholar]
- Taylor DW, Kelly DF, Cheng A, Taylor KA. On the freezing and identification of lipid monolayer 2-D arrays for cryoelectron microscopy. J. Struct. Biol. 2007;160:305–312. doi: 10.1016/j.jsb.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, Glaeser RM. Electron diffraction of frozen, hydrated protein crystals. Science. 1974;186:1036–1037. doi: 10.1126/science.186.4168.1036. [DOI] [PubMed] [Google Scholar]
- Thevelein JM. Regulation of trehalose mobilization in fungi. Microbiol. Rev. 1984;48:42–59. doi: 10.1128/mr.48.1.42-59.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin PN, Henderson R. Molecular structure determination by electron microscopy of unstained crystalline specimens. J. Mol. Biol. 1975;94:425–440. doi: 10.1016/0022-2836(75)90212-0. [DOI] [PubMed] [Google Scholar]
- Uzgiris EE, Kornberg RD. Two-dimensional crystallization technique for imaging macromolecules, with application to antigen-antibody-complement complexes. Nature. 1983;301:125–129. doi: 10.1038/301125a0. [DOI] [PubMed] [Google Scholar]
- Viner RI, Clegg JS. Influence of trehalose on the molecular chaperone activity of p26, a small heat shock/α-crystallin protein. Cell Stress Chaperones. 2001;6:126–135. doi: 10.1379/1466-1268(2001)006<0126:iototm>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walian PJ, Jap BK. Three-dimensional electron diffraction of PhoE porin to 2.8 Å resolution. J. Mol. Biol. 1990;215:429–438. doi: 10.1016/s0022-2836(05)80362-6. [DOI] [PubMed] [Google Scholar]
- Wang DN, Kühlbrandt W. High-resolution electron crystallography of light-harvesting chlorophyll a/b-protein complex in three different media. J. Mol. Biol. 1991;217:691–699. doi: 10.1016/0022-2836(91)90526-c. [DOI] [PubMed] [Google Scholar]
- Wang H, Downing KH. Specimen preparation for electron diffraction of thin crystals. Micron. 2011;42:132–140. doi: 10.1016/j.micron.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggers HAL. Untersuchung über das Mutterkorn, Secale cornutum. Annalen der Pharmacie. 1832;1:129–182. [Google Scholar]
- Williams RC, Wyckoff RWG. Applications of metallic shadow-casting to microscopy. J. Appl. Phys. 1946;17:23–33. [Google Scholar]
- Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol. Cell. 2010;38:768–774. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarzycki J. Glasses and the vitreous state. Cambridge Univeristy Press; 1991. [Google Scholar]