Abstract
Several organoselenium compounds including benzyl selenocyanate (BSC), 1,2-phenylenebis(methylene)selenocyanate (o-XSC), 1,3-phenylenebis(methylene)selenocyanate (m-XSC), and 1,4-phenylenebis(methylene)selenocyanate (p-XSC) have been shown to prevent cancers caused by polycyclic aromatic hydrocarbons (PAHs) and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) in experimental animals; these chemical carcinogens are activated by human P450 1 and 2A family enzymes, respectively, to carcinogenic metabolites. In this study, we examined whether these selenium compounds interact with and inhibit human P450 1 and 2A enzymes in vitro. Four organoselenium compounds induced Reverse Type I binding spectra with P450 1A1, 1A2, and 1B1 and Type I binding spectra with P450 2A6 and 2A13. The spectral dissociation constants (Ks) for the interaction of P450 1B1 with these chemicals were 3.6–5.7 µM; the values were lower than those with seen with P450 1A1 (19–30 µM) or 1A2 (6.3–13 µM). The Ks values for Type I binding of P450 2A13 with m-XSC and BSC were both 0.20 µM; the values were very low compared to the interaction of P450 2A6 with m-XSC (5.7 µM) and BSC (2.0 µM). Four selenium compounds directly inhibited 7-ethoxyresorufin O-deethylation activities catalyzed by P450 1A1, 1A2, and 1B1 with IC50 values <1.0 µM, except for the inhibition of P450 1A2 by BSC (1.3 µM). Coumarin 7-hydroxylation activities of P450 2A13 were more inhibited by four selenium compounds than those of P450 2A6, with IC50 values of 0.22–1.4 µM for P450 2A13 and 2.4–6.2 µM for P450 2A6. Molecular docking studies of the interaction of four organoselenium compounds with human P450 enzymes suggest that these chemicals can be docked into the active sites of these human P450 enzymes and that the sites of the selenocyanate functional groups of these chemicals differ between the P450 1 and 2A family enzymes.
INTRODUCTION
Several organoselenium compounds have been shown to prevent lung, mammary, tongue, and colon cancers caused by polycyclic aromatic hydrocarbons (PAHs), e.g. 7,12-demethylbenzanthracene (DMBA) and benzo[a]pyrene, azoxymethane, and tobacco-related carcinogens, e.g. 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), in experimental animal models.1–5 Mechanisms underlying prevention of such carcinogenesis caused by these selenium compounds still remain unclear; however, evidence has accumulated that 1,4-phenylenebis(methylene)selenocyanate (p-XSC) is able to prevent the initiation phase of carcinogenesis caused by DMBA and NNK by inhibiting formation of DNA adducts.6–8 These results suggest that one of the mechanisms of chemoprevention by these selenium compounds may be related to their abilities to inhibit the xenobiotic-metabolizing enzymes that mediate formation of reactive metabolites of chemical carcinogens.9–11
A number of chemical carcinogens exist in our environment, and most of these carcinogens induce cancers only after metabolic activation to carcinogenic metabolites by xenobiotic-metabolizing enzymes, such as P450.11,12 Multiple forms of P450 enzymes have been identified that catalyze the activation of diverse chemical carcinogens with different, but overlapping, substrate specificities.12 The family 1 P450 enzymes—P450 1A1, 1B1, and 1A2—can activate PAHs and heterocyclic amines to carcinogenic metabolites.13 Recent studies have also shown that both P450 2A6 and 2A13 are the enzymes that catalyze the transformation of tobacco related nitrosamines, such as NNK, to carcinogenic metabolites.14–16 Because several selenium compounds have been reported to cause prevention of cancers caused by PAHs and NNK in experimental animals,5, 7–9 it is of interest to examine whether these selenium compounds interact with and inhibit P450 1 and 2A family enzymes in humans. In 1997, we showed that BSC and three XSC isomers induce “Type II” difference spectra with liver microsomes of β-naphthoflavone-treated rats and humans and that these chemicals inhibit the metabolism of xenobiotic and procarcinogen in rat and human liver microsomes.17 However, it has not fully been understood if these selenium compounds interact with and inhibit individual forms of P450 1A1, 1A2, 1B1, 2A6, and 2A13, the major P450s that activate PAHs and tobacco-related nitrosamines to carcinogenic metabolites.13,15,16
In this study, we first used purified recombinant P450 1A1, 1A2, 1B1, 2A6, and 2A13 isolated from membranes of Escherichia coli in examining the spectral interaction with BSC and o-, m-, and p-XSC. We also determined the effects of these four selenium compounds on EROD activities by P450 1A1, 1A2, and 1B1 and coumarin 7-hydroxylation activities by P450 2A6 and 2A13, using bacterial enzyme systems in which cDNAs of NADPH-P450 reductase and individual forms of P450 have been introduced. Molecular docking studies of interaction of selenium compounds with the active sites of P450s are also reported.
MATERIALS AND METHODS
Chemicals
Four organoselenium compounds including benzyl selenocyanate (BSC), 1,2-phenylenebis(methylene)selenocyanate (o-XSC), 1,3-phenylenebis(methylene)selenocyanate (m-XSC), 1,4-phenylenebis(methylene)selenocyanate (p-XSC) were kindly donated by Dr. El-Bayoumy of Pennsylvania State University College of Medicine (Hershey, PA) and their chemical structures are shown (Figure 1). Purities of these four chemicals were >99%. 7-Ethoxyresorufin, resorufin, coumarin, and 7-hydroxycoumarin were purchased from Sigma Chemical Co. (St. Louis, MO). All of the selenium compounds and substrates for P450 assays were dissolved in (CH3)2SO and added directly to the incubation mixtures; the final concentration of organic solvent in the assay was <0.4%. Other chemicals and reagents used in this study were obtained from the sources described previously or were of the highest quality commercially available.18–21
Figure 1.
Chemical structures of selenium compounds used in this study.
Enzymes
Bacterial "bicistronic" P450 1A1, 1A2, 1B1, 2A6, and 2A13 systems were prepared as described.18, 19, 22, 23 The plasmids for the expression of P450s 1A1, 1A2, 1B1, 2A6, or 2A13 plus human NADPH-P450 reductase were introduced into E. coli DH5□ cells and the bacterial membranes were prepared and suspended in 10 mM Tris-HCl buffer (pH 7.4) containing 1.0 mM EDTA and 20% glycerol (v/v) as described.18–21
For purification of P450 1A1, 1A2, and 1B1 enzymes, the bacterial membranes were solubilized in 0.10 M potassium phosphate buffer (pH 7.4) containing 20% glycerol (v/v), 0.5 M NaCl, 10 mM β-mercaptoethanol, 0.5% sodium cholate (w/v), 1% Triton N-101 (w/v), and 30 µM α-naphthoflavone. The solubilized membranes were centrifuged, the supernatant was applied to a nickel-nitrilotriacetic acid column (Qiagen), and the P450 proteins were purified by the method as described previously.24,25 Methods for purification of P450 2A6 and 2A13 from the bacterial membranes have been described elsewhere;25,26 the purified P450 2A13 was kindly donated by Dr. Emily E. Scott and Natasha M. DeVore of University of Kansas (Lawrence, KS). Recombinant P450 2C9, 2E1, and 3A4 were purified from E. coli membranes as described previously.27–29
Methods for isolation and purification of P450 1A1 and 1A2 from liver microsomes of 3-methylcholanthrene-treated rats and rabbits have been described previously.30–33
Enzyme Assays
The 50% inhibition concentration (IC50) of EROD activities of P450 1A1, 1A2, or 1B1 was determined in a standard incubation mixture (0.5 mL) consisting of P450 1A1 (0.03 µM), P450 1A2 (0.05 µM), or P450 1B1 (0.04 µM) in bacterial membranes co-expressing human NADPH-P450 reductase, chemical inhibitors, 100 mM potassium phosphate buffer (pH 7.4), and an NADPH-generating system consisting of 0.5 mM NADP+, 5 mM glucose 6-phosphate, and 0.5 unit of yeast glucose 6-phosphate dehydrogenase/mL.18,19 7-Ethoxyresorufin (4.0 µM) was added to start the reaction and the formation of resorufin was determined in a Hitachi F-4500 spectrofluorometer using an excitation wavelength of 571 nm and an emission wavelength of 585 nm. Time course studies of inhibition of P450-dependent EROD activities by selenium compounds were determined as follows. P450 1A1, 1A2, or 1B1 was mixed with 0.10 M potassium phosphate buffer (pH 7.4) containing chemical inhibitors and 7-ethoxyresorufin, and the reaction was started by the addition of NADPH; the formation of resorufin was directly monitored for 0–6 min. Metabolism-dependent changes in inhibition of P450 by chemicals was determined using the pseudo-first-order time-dependent losses of EROD activity, essentially according to the methods described previously.19,20 (19,20). Briefly, semi-logarithmic plots of the percent relative activity (activities with versus without inhibitors) were determined and the losses of activities were calculated from initial linear decreases in activities per min. The IC50 values were estimated using GraphPad Prism software (GraphPad Software, San Diego, CA).
Coumarin 7-hydroxylation was determined using P450 2A6 and 2A13 in membranes of E. coli co-expressing human NADPH-P450 reductase as described previously.22,34
Spectral Binding Titrations
Purified P450 enzymes were diluted to 1.0 µM in 0.10 M potassium phosphate buffer (pH 7.4) containing 20% glycerol (v/v) and the binding spectra were recorded with subsequent additions of chemical inhibitors in a Jasco V-550 spectrophotometer as described previously.21 Briefly, the chemical inhibitors were added to the buffer with or without P450 and the spectra were recorded between 350 nm and 500 or 700 nm. The substrate binding spectra were obtained by subtracting the blank spectra (in the absence of P450) from the P450 spectra (in the presence of P450). Spectral dissociation constants (Ks) were estimated using GraphPad Prism software (GraphPad Software, San Diego, CA).
Other Assays
P450 and protein concentrations were estimated by the methods described previously.35,36
Docking Simulation of m-XSC into Human P450 Enzymes
The crystal structures of P450 1A2, 1B1, 2A6, and 2A13 have recently been reported.26,37–40 The human P450 1A1 primary sequence was aligned with human P450 1A2 (Protein Data Bank code 2HI4) in the MOE software (ver. 2009.10, Computing Group, Montreal, Canada) for modeling of a three-dimensional structure.37,41 Prior to docking, the energy of the P450 structures was minimized using the CHARMM22 force field. Docking simulation was carried out for m-XSC binding to P450 enzymes using the MMFF94x force field distributed in the MOE Dock software. Twenty solutions were generated for each docking experiment and ranked according to total interaction energy (U value).
RESULTS
Reverse Type I Binding Spectra of Human P450 1A1, 1A2, and 1B1 with Four Organoselenium Compounds
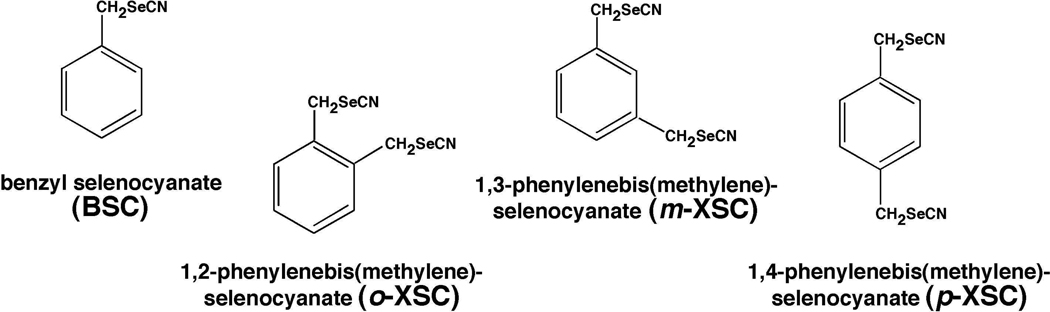
Our purified P450 1A1 preparation contained mostly low-spin P450 heme but had small amounts of the high-spin form (Figure 2A). BSC (at 1.25–80 µM) changed the P450 spectrum, in terms of its Soret maximum, thus forming Reverse Type I binding spectra with a peak wavelength at 414 nm and a trough peak at 387 nm in the difference spectrum (Figure 2D). Purified P450 1A2 was mostly high-spin P450 and was slightly shifted to low-spin upon binding with 1.25–80 µM p-XSC (Figure 2B and 2E). The Reverse Type I binding spectra of P450 1B1 with different concentrations of 1.25–80 µM m-XSC was clearly observed, with a peak wavelength of 418 nm and a trough wavelength at 389 nm in the difference spectra (Figure 2C and 2F).
Figure 2.
Spectral interaction of BSC with P450 1A1 (A and D), p-XSC with P450 1A2 (B and E), and m-XSC with P450 1B1 (C and F). Absolute spectra (A, B, and C) and difference spectra (D, E, and F) were recorded between 350 and 500 nm.
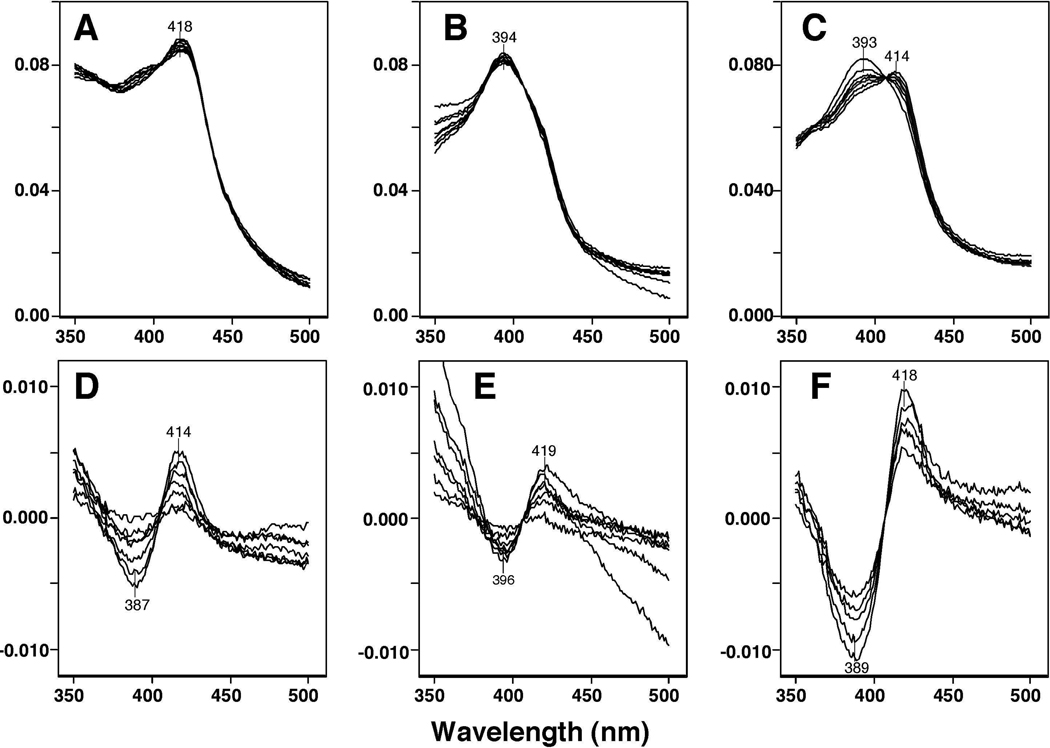
Spectral analysis was also done with P450 1A1 and 1A2 purified from liver microsomes of 3-methylcholanthrene-treated rats and rabbits to determine if these P450s interact with BSC (Figure 3). Rat P450 1A1 did not show any spectral changes with 1.25~80 µM BSC; the absolute spectra showed peak wavelengths of the α-, β-, and Soret bands at 568, 530, and 417 nm, respectively (Figure 3A). Rat and rabbit liver P450 1A2 had peak wavelengths at 393 and 392 nm, respectively, in the absolute spectra (Figure 3B and 3C) and showed spectral changes with BSC with peak wavelengths of 419 and 423 nm, respectively, and trough wavelengths of 391 and 389 nm, respectively (Figure 3D and 3E). The spectral dissociation constants (Ks) for the interaction of BSC with rat and rabbit P450 1A2 were estimated to be 16 ± 2.4 µM and 90 ± 29 µM, respectively.
Figure 3.
Spectral interaction of BSC with rat liver P450 1A1 (A), rat liver 1A2 (B and D), and rabbit liver P450 1A2 (C and E). BSC (at concentrations of 1.25, 2.5 5.0, 10, 20, 40, and 80 µM) were added to the buffer with 1.5 µM rat P450 1A1 or 1A2 or 1.0 µM rabbit P450 1A2 and the spectra were recorded between 350 and 700 nm for rat P450 1A1 (A) and 350 and 500 nm for rat and rabbit P450 1A2 (B and C). The difference spectra of interaction of P450s with selenium compounds are shown in Figures 3D and 3E.
The Ks and ΔAmax values for the interaction of four selenium compounds with human P450 1A1, 1A2, and 1B1 were determined (Table 1). The Ks values with P450 1A1 were 23, 26, 30, and 19 µM for the complexes containing BSC, o-XSC, m-XSC, and p-XSC, respectively, and the ΔAmax/Ks values were estimated to be 0.45–0.79 (×10−3 M−1) with these chemicals. The Ks values with human P450 1A2 were found to be 6.3–13 µM and ΔAmax/Ks values were between 0.38–0.88 (×10−3 M−1). Among these three P450 enzymes examined, P450 1B1 showed clear interactions with these compounds: the Ks values were found to be 3.6–5.1 µM with these chemicals and the ΔAmax/Ks values were the highest among these P450 enzymes.
Table 1.
Reverse Type I Binding Spectra of P450 1A1, 1A2, and 1B1 Induced by Organoselenium Compounds
| P450 | chemical | Ks (µM) | ΔAmax (× 10−3) | ΔAmax/Ks (× 10−3) |
|---|---|---|---|---|
| P450 1A1 | BSC | 23 ± 4.1 | 14 ± 1.0 | 0.61 |
| o-XSC | 26 ± 4.6 | 20 ± 1.2 | 0.77 | |
| m-XSC | 30 ± 4.5 | 18 ± 1.2 | 0.60 | |
| p-XSC | 19 ± 1.6 | 8.6 ± 0.24 | 0.45 | |
| P450 1A2 | BSC | 11 ± 2.3 | 9.5 ± 0.57 | 0.88 |
| o-XSC | 6.3 ± 1,6 | 2.4 ± 0.20 | 0.38 | |
| m-XSC | 13 ± 1.9 | 5.8 ± 0.25 | 0.46 | |
| p-XSC | 12 ± 1.2 | 9.7 ± 0.40 | 0.81 | |
| P450 1B1 | BSC | 5.3 ± 0.96 | 19 ± 0.11 | 3.60 |
| o-XSC | 4.6 ± 0.75 | 15 ± 0.51 | 3.30 | |
| m-XSC | 5.7 ± 1.0 | 21 ± 1.0 | 3.70 | |
| p-XSC | 3.6 ± 0.33 | 21 ± 0.48 | 5.83 |
Spectral interaction was determined in a system containing 1 µM P450 and 6–10 concentrations of selenium compounds in 0.10 M potassium phosphate buffer (pH 7.4) containing 20% glycerol (v/v). Results are presented as means ± SE.
Type I Binding Spectra of P450 2A6 and 2A13 with Four Organoselenium Compounds
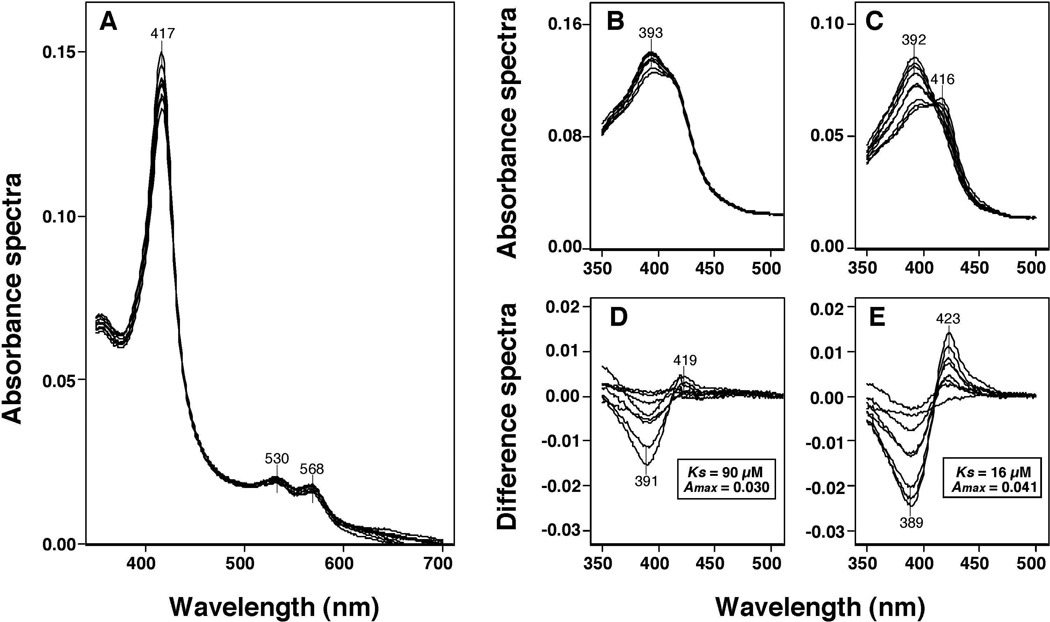
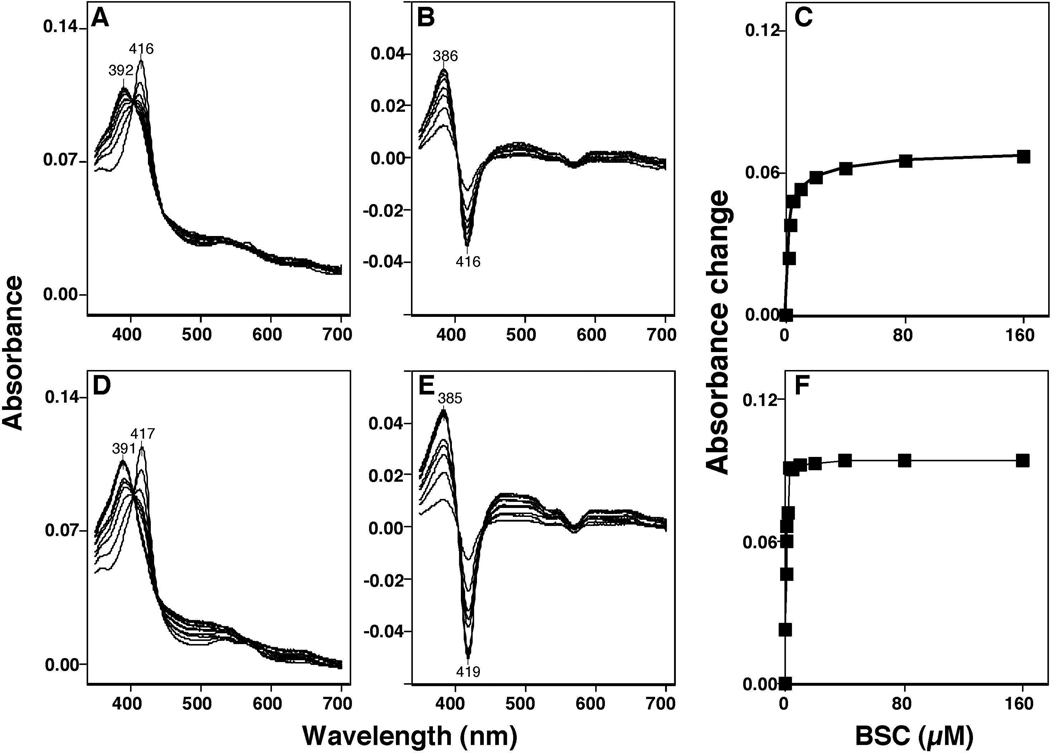
Human P450 2A6 and 2A13 had Soret peak wavelengths at 416 and 417 nm, respectively, and these peak wavelengths were changed to 392 and 391 nm, respectively, when concentrations of 0.125–160 µM BSC were added to the P450 samples (Figures 4A and 4D). The difference spectra of P450 2A6 and 2A13 for the interaction with BSC showed typical Type I binding with peak wavelengths at 386 and 385 nm, respectively, and trough wavelengths at 416 and 419 nm, respectively (Figures 4B and 4E). The concentration dependent interaction of BSC with P450 2A6 and 2A13 showed that the Ks values were 2.0 µM and 0.20 µM, respectively and the ΔAmax/Ks values were 0.066 and 0.094, respectively (Figures 4C and 4F).
Figure 4.
Spectral interaction of BSC with P450 2A6 (A, and B) and P450 2A13 (D and E). Chemicals (at concentration of 0.078–160 µM) were added to the buffer with or without 1 µM each P450 and the spectra were recorded between 350 and 700 nm (A and D). The difference spectra of interaction of P450s with selenium compounds are shown in Figures 4B and 4E. The concentration dependent interaction of BSC with P450 2A6 and 2A13 are shown in Figures 4C and 4F, respectively.
Ks, ΔAmax, and ΔAmax/Ks values for the interaction of P450 2A6 and 2A13 with four selenium compounds were determined (Table 2). In case of P450 2A6, BSC had the lowest Ks value of four selenium compounds tested (2.0 µM), thus resulting in the highest ΔAmax/Ks value of 0.032. Interestingly, the Ks values of P450 2A13 with BSC and m-XSC were found to be very low (0.20 µM); the values were 10- and 28-fold lower than those obtained with P450 2A6. As a result, the ΔAmax/Ks values of 0.47 and 0.40 were very high for the interaction of P450 2A13 with BSC and m-XSC, respectively. The spectral interactions of o-XSC and p-XSC with P450 2A13 were somewhat less pronounced than those with BSC and m-XSC but were stronger than those of interaction with P450 2A6.
Table 2.
Type I Binding Spectra of P450 2A6 and 2A13 with Organoselenium Compounds
| P450 | chemical | Ks (µM) | ΔAmax (× 10−3) | ΔAmax/Ks (× 10−3) |
|---|---|---|---|---|
| P450 2A6 | BSC | 2.0 ± 0.14 | 66 ± 0.93 | 32.00 |
| o-XSC | 5.1 ± 0.45 | 40 ± 0.82 | 7.80 | |
| m-XSC | 5.7 ± 0.52 | 37 ± 0.88 | 6.50 | |
| p-XSC | 14 ± 1.9 | 31 ± 1.2 | 2.20 | |
| P450 2A13 | BSC | 0.20 ± 0.021 | 94 ± 1.5 | 470.00 |
| o-XSC | 0.75 ± 0.044 | 69 ± 0.49 | 92.00 | |
| m-XSC | 0.20 ± 0.015 | 79 ± 0.91 | 395.00 | |
| p-XSC | 1.5 ± 0.28 | 69 ± 2.6 | 46.00 |
Spectral interaction was determined in a system containing 1 µM P450 and 6–10 concentrations of selenium compounds in 0.10 M potassium phosphate buffer (pH 7.4) containing 20% glycerol (v/v). Results are presented as means ± SE.
We also determined the spectral changes of P450 2C9, 2E1, and 3A4 with these four selenium compounds and found no apparent spectral interaction observed with these P450 enzymes (results not shown).
Inhibition of P450 1A1-, 1A2, and 1B1-Dependent EROD Activities by Four Organoselenium Compounds
Four organoselenium compounds were examined for their abilities to inhibit EROD activities catalyzed by P450 1A1, 1A2, and 1B1 (Table 3). These four selenium compounds significantly inhibited EROD activities catalyzed by P450 1A1 with IC50 values between 0.1 and 0.6 µM, P450 1A2 with IC50 values between 0.2 and 0.6 µM, and P450 1B1 with IC50 values between 0.1 and 0.3 µM.
Table 3.
Inhibition of P450 1A1-, 1A2, and 1B1-Dependent EROD Activities by Organoselenium Compounds
| IC50 for inhibition of EROD activity (µM) |
||||
|---|---|---|---|---|
| P450 | BSC | o-XSC | m-XSC | p-XSC |
| P450 1A1 | 0.45 ± 0.038 | 0.11 ± 0.021 | 0.10 ± 0.013 | 0.26 ± 0.031 |
| P450 1A2 | 1.3 ± 0.22 | 0.39 ± 0.042 | 0.20 ± 0.021 | 0.63 ± 0.059 |
| P450 1B1 | 0.27 ± 0.031 | 0.14 ± 0.027 | 0.13 ± 0.011 | 0.16 ± 0.009 |
IC50 values were obtained by measuring EROD activities catalyzed by P450 1A1, 1A2, and 1B1. IC50 values represent means ± SE.
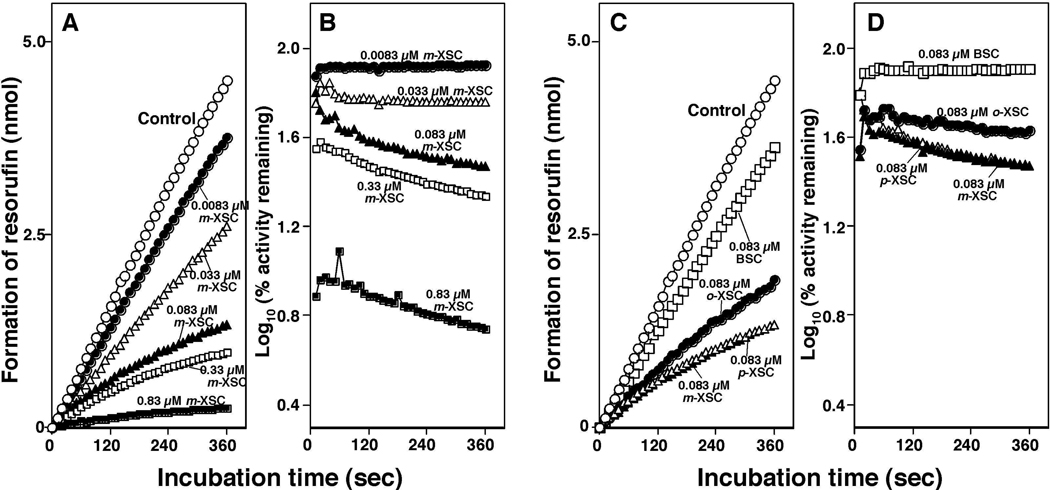
Time course studies of inhibition of P450 1B1-dependent EROD activities by different concentrations of m-XSC showed that resorufin was formed linearly (with respect to time) on incubation of 7-ethoxyresorufin with P450 1B1 system, and its formation was found to be inhibited by 0.0083, 0.033, 0.083, 0.33, and 0.83 µM m-XSC (Figure 5A). The semilogarithmic plots of percent relative activity of the inhibition of EROD activities by m-XSC showed that there were slight decreases in the formation of resorufin with increasing incubation time, indicating the possibility of metabolism-dependent inhibition of EROD activities by m-XSC (Figure 5B). Similar results were also obtained for the inhibition of P450 1B1-dependent EROD activities by 0.083 µM BSC, o-XSC, and p-XSC (Figure 5C and 5D). Studies involving the time course of inhibition of EROD activities by m-XSC in bacterial membranes containing P450 1A1 or 1A2 (and NADPH-P450 reductase) showed very similar results to those obtained with the P450 1B1 enzyme system described above (results not shown).
Figure 5.
Time-dependent changes in the formation of resorufin by incubating 7-ethoxyresorufin with P450 1B1 in the absence (Control) and in the presence of 0.0083 µM, 0.033 µM, 0.083 µM, 0.33 µM, and 0.83 µM m-XSC in Figure 5A and in the presence of 0.083 µM o-XSC, 0.083 µM m-XSC, 0.083 µM p-XSC, and 0.083 µM BSC in Figure 5C. E. coli membranes expressing P450 1B1 and NADPH-P450 reductase were mixed with 7-ethoxyresorufin and chemical inhibitors, and the reactions were started by adding NADPH. The semi-logarithmic plots of the percent relative activity (activities with vs without inhibitors) are shown in different concentrations of m-XSC (B) and 0.083 µM each of o-, m-, and p-XSC and BSC (D).
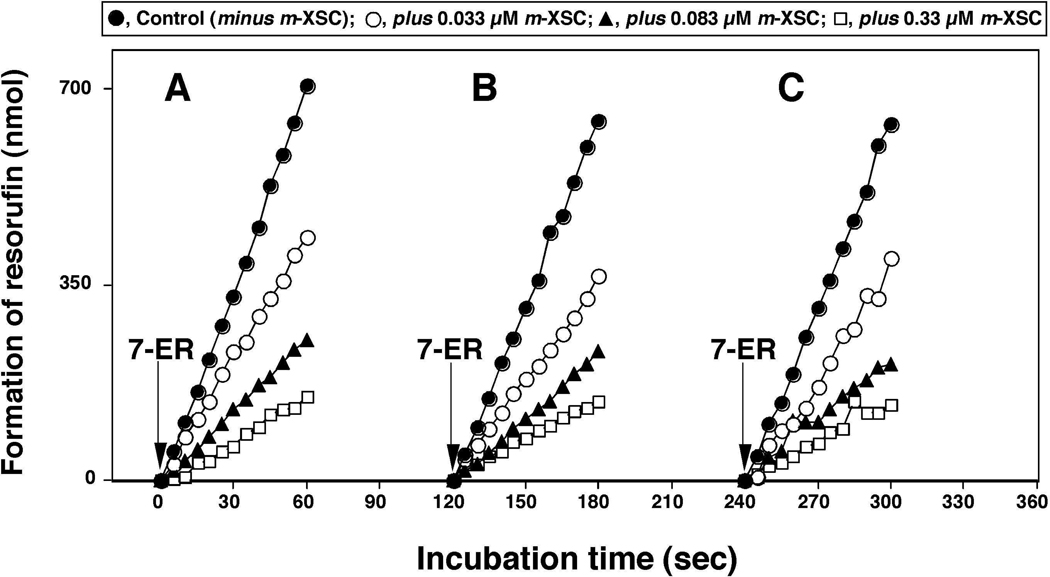
In order to confirm if these selenium compounds are oxidized by P450 1B1 to active products that inhibit P450 1B1 itself, we examined the effects of oxidation of m-XSC with the P450 1B1 enzyme system on the inhibition of EROD activities (Figure 6). When m-XSC (at 0.033, 0.083, and 0.33 µM) was added together with 7-ethoxyresorufin in the P450 1B1 enzyme system, the formation of resorufin was decreased as compared with the control incubation in which m-XSC was not added (Figure 6A). In another set of experiments, we first incubated different concentrations of m-XSC with P450 1B1 enzyme system for 2 min (Figure 6B) or 4 min (Figure 6C) and then added 7-ethoxyresorufin (measuring EROD activity). The results showed that preincubation of m-XSC for 2 and 4 min did not change the inhibitory potencies as compared without preincubation (Figure 6A), indicating that there were no metabolism-dependent inhibition of P450 1B1 by m-XSC.
Figure 6.
Effects of preincubation time on the inhibition of P450 1B1-dependnet EROD activities by m-XSC. E. coli membranes expressing P450 1B1 and NADPH-P450 reductase were first incubated without (Control) or with 0.033 µM, 0.083 µM, and 0.33 µM m-XSC in the presence of NADPH. 7-Ethoxyresorufin was added at incubation time of 0 min (A, without preincubation), 2 min (B, with preincubation for 2 min), and 4 min (C, with preincubation for 4 min) and the formation of resorufin was determined as a function of time.
Inhibition of P450 2A6 and 2A13-Dependent Coumarin 7-Hydroxylation Activities by Four Organoselenium Compounds
Four organoselenium compounds were examined to inhibit coumarin 7-hydroxylation activities catalyzed by P450 2A6 and 2A13 (Table 4). IC50 values for the interaction of P450 2A6-dependent coumarin 7-hydroxylation activities by o- and m-XSC were 2.7 and 2.4 µM, respectively, and with BSC and p-XSC were 4.3 and 6.2 µM, respectively. IC50 values of coumarin 7-hydroxylation activities catalyzed by P450 2A13 were much lower than those seen with P450 2A6, and we found that m-XSC inhibited P450 2A13 most potently with an IC50 value of 0.22 µM, followed by BSC, o-XSC, and p-XSC with IC50 values of 1.2, 1.2, and 1.4 µM, respectively.
Table 4.
Inhibition of P450 2A6- and 2A13-Dependent Coumarin 7-Hydroxylation Activities by Organoselenium Compounds
| IC50 for inhibition of coumarin 7-hydroxylation activity (µM) |
||||
|---|---|---|---|---|
| P450 | BSC | o-XSC | m-XSC | p-XSC |
| P450 2A6 | 4.3 ± 0.36 | 2.7 ± 0.34 | 2.4 ± 0.19 | 6.2 ± 0.55 |
| P450 2A13 | 1.2 ± 0.19 | 1.2 ± 0.13 | 0.22 ± 0.031 | 1.4 ± 0.21 |
IC50 values were obtained by measuring coumarin 7-hydroxylation activities catalyzed by P450 2A6 and 2A13. IC50 values represent means ± SE.
Preincubation for 15 min of BSC and m-XSC with baculosome P450 2A6 and 2A13 systems fortified with NADPH did not change the inhibition potencies of coumarin 7-hydroxylation activities as compared with the systems without preincubation (Table 5).
Table 5.
Effects of Preincubation Time on the Inhibition of P450 2A6- and 2A13-Dependent Coumarin 7-Hydroxylation Activities by BSC and m-XSC
| coumarin 7-hydroxylation (nmol/min/nmol P450) |
||
|---|---|---|
| without preincubation | with preincubation | |
| P450 2A6 | ||
| control | 3.26 (100) | 3.06 (100) |
| + BSC (4.3 µM) | 1.70 (52) | 1.52 (50) |
| + m-XSC (0.68 µM) | 1.56 (48) | 1.46 (48) |
| P450 2A13 | ||
| control | 1.04 (100) | 0.98 (100) |
| + BSC (1.2 µM) | 0.49 (47) | 0.48 (49) |
| + m-XSC (0.22 µM) | 0.59 (56) | 0.53 (54) |
BSC and m-XSC were preincubated with recombinant P450 2A6 and 2A13 systems fortified with NADPH for 0 or 15 min and the catalytic activities were determined after adding 1.0 µM coumarin. Values in parentheses indicate % of control. Results are presented as means of triplicate determinations. There were no significant differences in the inhibition of coumarin 7-hydroxylation activities by BSC and m-XSC with and without preincubation.
Molecular Docking of Interaction of m-XSC with P450 1A1, 1A2, 1B1, 2A6, and 2A13
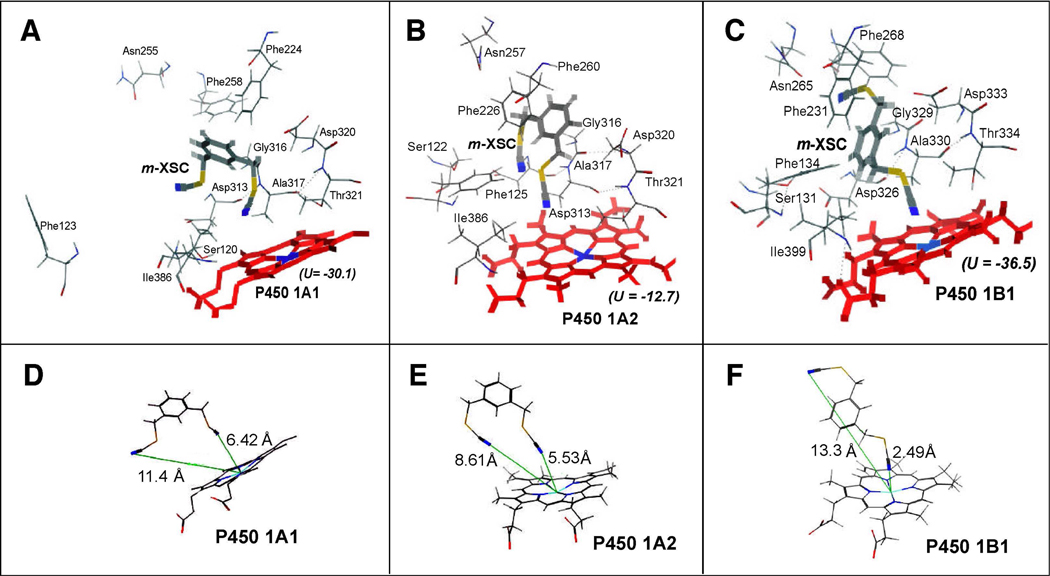
m-XSC was used for the analysis of molecular docking into the active sites of P450s 1A1, 1A2, and 1B1 according to the methods as described in MATERIALS AND METHODS (Figure 7). In the figures, the key amino acid residues in the substrate recognition sites (SRS) 1, 2, 3, 4, and 5 are shown.42 In these three cases, one of the selenium moieties of m-XSC was positioned towards the active sites of P450 1A1, 1A2, and 1B1 and the ligand-P450 interaction energies (U values) were found to be −30.1, −12.7, and −36.5, respectively. We also found that m-XSC was surrounded by several amino acids, including Asp313, Gly316, Ala317, Asp320, and Thr 321 (in SRS4) of P450 1A1 and 1A2 and Asp326, Gly329, Ala330, Asp333, and Thr334 (in SRS4) of P450 1B1 (Figure 7A–7C). The interaction between m-XSC and P450 1A1 or 1A2 seemed to be not so tight as compared with the amino acids in P450 1B1. The distances between the N-atom in one of the −CH2SeCN moieties of m-XSC and the Fe- atom in P450 1A1, 1A2, and 1B1 were calculated using in silico analysis (Figure 7D–7F). By comparing theses distances in P450 1A1, 1A2, and 1B1, it was found that one of the selenium moieties was closely situated in the active sites of P450 1B1 (2.49 Å); these distances were 5.53 Å and 6.42 Å in P450 1A1 and 1A2, respectively.
Figure 7.
Docking simulation of interaction of m-XSC with P450 1A1 (A), 1A2 (B), and 1B1 (C). The heme group of the P450 is shown at the lower part of each of the figures and the amino acid residues that may interact with m-XSC are presented. In the figure, oxygen, nitrogen, sulfur, selenium, and iron are colored with red, blue, yellow, dark yellow, and light blue, respectively. Heme and m-XSC are shown in thick lines and colored with red and gray, respectively. The distances (shown in lines with green) between the N-atom in the −CH2SeCN moieties of m-XSC and the Fe- atom in P450 1A1, 1A2, and 1B1 are also shown in part 7D, 7E, or 7F, respectively.
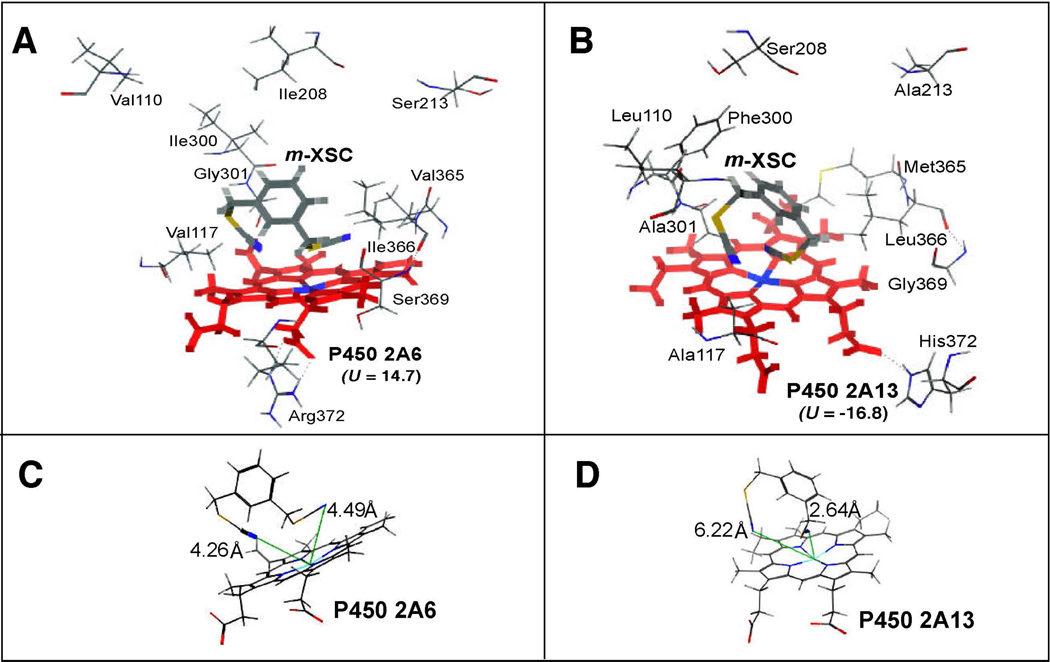
m-XSC was also docked into the active sites of P450 2A6 and 2A13 (Figure 8). In contrast to the cases in P450 family 1 enzymes, both selenium moieties at 1- and 3-positions of m-XSC were docked near the heme of P450 2A6 and 2A13. The ligand-P450 interaction energies were smaller in P450 2A13 (U = −16.8) than in P450 2A6 (U = 14.7). In P450 2A13, Phe300 and Ala301 have been identified as critical in interaction of P450 2A13 with the substrates phenacetin, phenthyl isothiocyanate, and 2’-methoxyacetophenone,43,44 but these residues were not observed to be so different in the interactions of P450 2A6 and 2A13 with m-XSC. However, the positions of Leu110 and Ala117 in P450 2A13 were slightly different with the positions of Val110 and Val117 in P450 2A6. The distance between the N-atom of m-XSC and the Fe- atom of P450 2A13 (2.64 Å) (Figure 8C) was also, close as compared with P450 2A6 (4.26 or 4.49 Å) (Figure 8D).
Figure 8.
Docking simulation of m-XSC with P450 2A6 (A) and 2A13 (B). The distance between the N-atom in the −CH2SeCN moieties of m-XSC and the Fe-atom in P450 2A6 or 2A13 is also shown in part 8C or 8D, respectively. Other details are the same as in the legend to Figure 7.
DISCUSSION
It has been reported that p-XSC and BSC can inhibit tumorigenesis induced by PAHs, e.g. DMBA and benzo[a]pyrene, in laboratory animals4,7–9 and that o- and m-XSC appear to be equally effective to BSC and p-XSC in inhibiting DMBA-DNA adduct formation in the rat mammary gland.8,45 Our present studies show that all of the four selenium compounds are able to interact with and inhibit P450 family 1 enzymes that have been reported to activate a variety of PAHs.11,13 The affinities of four selenium compounds in interactions with P450 family 1 enzymes were not very different; the Ks values obtained in the spectral interactions of four selenium compounds with P450 1A1, 1A2, and 1B1 were 19–30 µM, 6.3–13 µM, and 3.6–5.7 µM, respectively, and the IC50 values for these chemicals were 0.10–0.45 µM for P450 1A1, 0.20–1.3 µM for P450 1A2, and 0.13–0.27 µM for P450 1B1. However, these organoselenium compounds were found to have relatively higher affinities for P450 1B1 than P450 1A1 and 1A2, because the Ks values in P450 1B1 were lower and the ΔAmax/Ks values in P450 1B1 were higher than those in the cases of the latter two enzymes. Molecular docking studies support the view that one of the selenium moieties of m-XSC is more closely situated to the heme of P450 1B1 than in the cases of P450 1A1 and 1A2; the distance between Fe-atom in the P450 heme and the N-atom in the −CH2SeCN moiety was calculated to be 2.49 Å in P450 1B1 as compared with 6.42 and 5.53 Å in P450s 1A1 and 1A2, respectively.
Both P450 2A6 and 2A13 have been shown to activate tobacco-related nitrosamines, e.g. NNK, to reactive products that attack DNA to cause cell transformation.14–16 It has been reported that P450 2A13 is mainly expressed in the respiratory tracts, e.g. nasal mucosa, trachea, and lung while 2A6 is more abundant in the liver than in the lung.46–48 Recent studies have identified that P450 2A13 is much more active than P450 2A6 in metabolizing several of these tobacco carcinogens and may be more important in understanding the basis of carcinogenesis in respiratory systems in humans.16,47,48 Our current studies showed that i) four selenium compounds bind to P450 2A6 and 2A13 to show typical Type I binding spectra, ii) both P450 2A6 and 2A13-dependent coumarin 7-hydroxylation activities are significantly inhibited by these selenium compounds, and iii) the spectral changes and catalytic inhibition by these chemicals are more profoundly observed with P450 2A13 than P4502A6. These results suggest the possibility that potent inhibition of P450 2A13—as well as P450 2A6—seems to be important to understand the basis of mechanisms of prevention of cancers caused by tobacco-related carcinogens. It is interesting in this regard to note the results of Sohn et al.,49 who showed that total selenium levels in blood, liver, kidney, and mammary gland increase in rats fed three XSC isomers (5 or 15 ppm as Se) for 3 days and that the XSC levels in blood, liver, kidney, and mammary gland were roughly estimated to be 1.5–1.8 µM, 7–15 µM, 12–25 µM, and 6–9 µM, respectively. The levels in these tissues seem to be very high enough to inhibit P450 1 and 2A activities when these XSC isomers are dosed in laboratory animals in vivo.
Our molecular docking studies show that m-XSC is surrounded by Val117, Ile300, Gly301, and Val365 of P450 2A6 and Ala117, Phe300, Ala301, and Met365 of P450 2A13; the differences in amino acid residues in the two P450 enzymes may be one of the factors controlling different affinities of these selenium compounds with P450 2A6 and 2A13.26,38,39,43,44 Several amino acid residues of P450 2A13 (that are different from those of P450 2A6) may provide a wider pocket for substrate binding over the heme and drive the more stable U energy in the docking of m-XSC into P450 2A13 than P450 2A6 (Figure 8). One of the selenium moieties of m-XSC was more closely situated to the heme of P450 2A13 than that of P450 2A6,as calculated to be 2.64 Å in the former case and 4.26 Å in the latter case. It is interesting to note the results of Devore et al.43,44 in that P450 2A13 residues, Phe300 and Ala301 play very important roles in phenacetin O-deethylation, followed by Ala117, Ser208, Met365, and Gly369. Additionally, each of these residues is key to the affinity of P450 2A13 for 2’-methoxyacetophenone and phenethyl isothiocyanate.
Our results show that both BSC and m-XSC directly inhibit EROD activity catalyzed by P450 1A1, 1A2, and 1B1 and coumarin 7-hydroxylation activity catalyzed by P450 2A6 and 2A13. Although these results suggest that these P450 enzymes may not convert these selenium compounds to active metabolites that inhibit catalytic activities, it is not known at present whether these chemicals can be oxidized by these human P450 enzymes; further work is necessary to solve these questions.9 Other human P450 enzymes, e.g. P450 2C9, 2E1, and 3A4, did not show any apparent spectral changes with these selenium compounds, supporting the view that the interaction of these selenium compounds with P450 family 1 and 2A enzymes may be rather specific in humans.
Little is known about the metabolism of organoselenium compounds by P450 enzymes in humans, as well as in laboratory animals. El-Bayoumy et al.50 have reported that the cumulative percentages of radioactivity excreted in rats dosed with p-[14C]XSC were 24% in urine and 75% in feces. A tetraselenocyclophane was identified as one of the metabolites in feces, and several sulfate and glucuronic acid conjugates were found in urine; the structures of these conjugate metabolites have not yet been identified.50 The same group has also reported that p-XSeSG (a GSH conjugate of p-XSC) shows similar effects to p-XSC in preventing the formation of DNA adducts in rats dosed with DMBA.51
We have previously shown that P450 1B1 displays Reverse Type I binding spectra with various compounds such as flavonoids and their hydroxylated and methoxylated derivatives, pyrene and its ethynyl, propyl, and butynyl derivatives, naphthalene and its propargyl ether derivatives, phenanthrene and its propargyl ether derivatives, and biphenyl and its propargyl ether derivatives and that these spectral potencies are correlated with their inhibition potencies to inhibit EROD activity catalyzed by P450 1B1.21,41 Our current studies showed that four selenium compounds also induced Reverse Type I binding spectra on interaction with P450 1A1 and 1A2 as well as P450 1B1 and inhibit EROD activity catalyzed by these P450 enzymes. The spectral interaction of these selenium compounds was rather specific for the high-spin, but not the low-spin form, of human P450 1A1 and 1B1. We did not find any spectral interaction of these selenium compounds with the low-spin form of rat liver P450 1A1, supporting the above conclusion. Our results also showed that rabbit liver P450 1A2 was much more reactive in interacting with BSC to form spectral changes than rat and human P450 1A2 enzymes.
Previously we showed that these selenium compounds induce Type II difference spectra with human and rat liver microsomes.17 The wavelength maximum was obtained at 393–395 nm and a trough at 430 nm in the difference spectra of interaction of these selenium compounds with the liver microsomes.17 In our current studies with purified human P450 family 1 enzymes and rat and rabbit P450 1A2 enzymes, the peak and trough wavelengths in the difference spectra were at 386–391 nm and at 430 nm respectively, indicating that there are differences in peak wavelengths between microsomes and purified P450 family 1 enzymes. It is not known whether these selenium compounds can interact with other P450s as well as P450 1A2 and 2A6 in human liver microsomes; however, our current results showed that P450 2C9, 2E1, and 3A4—three major P450s in human liver52,53—did not show any binding spectra with these compounds.
Mechanisms underlying chemoprevention of chemical carcinogenesis by organoselenium compounds are still unclear, although several possible mechanisms have been proposed.5,8–10,45,54 The inhibition of DNA adduct levels formed by interaction of reactive metabolites of PAHs and NNK with DNA in vivo has been reported and these mechanisms are of interest since these reactive metabolites have been identified to be formed by P450 family 1 and 2A enzymes.11,12,14,48 Recent studies have also established that there are another mechanisms in association of chemoprevention by selenium compounds, following analysis of gene responses in cancer cells. El-Bayoumy et al.55 have shown that p-XSC inhibit cell growth in a dose-dependent manner and induce apoptosis in non-small cell lung cancer cell lines. These authors observed the reduction of expression of cyclooxygenase-2 and phospholipase A2; p-XSC upregulates 22 genes and downregulates 13 genes on cDNA microarray analysis of the cancer cells. The same laboratory has also reported that both BSC and p-XSC interact with and bind to the Cys62 residue in the active site of the NF-kB (p50) protein in the lung cancer cells.56
In conclusion, the present results showed that four organoselenium compounds interact with human P450 1A1, 1A2, 1B1, 2A6, and 2A13, thus causing Reverse Type I binding spectra in the former three enzymes and Type I binding spectra in the latter two P450 2A enzymes. All of these P450 enzymes were inhibited by these selenium compounds as judged by analysis of EROD activities catalyzed by family 1 P450 enzymes and coumarin 7-hydroxylation activity catalyzed by P450 2A6 and 2A13. These selenium compounds have higher affinities for P450 1B1 than P450 1A1 and 1A2 as judged by analysis of spectral interaction and molecular docking studies, although the IC50 values of the inhibition of EROD activities by these selenium compounds were not very different in these three P450 enzymes examined. In addition, P450 2A13 was found to be more susceptible to these selenium compounds than P450 2A6 in forming Type I binding spectra and in inhibition of catalytic activity. These results suggest that one of the mechanisms underlying prevention of cancers caused by PAHs and tobacco-related carcinogens with these selenium compounds may be the result of inhibition of P450 family 1 and 2A enzymes, particularly P450 1B1 and 2A13, at the initial step of carcinogen activation.
ACKNOWLEDGEMENTS
We thank Dr. E. E. Scott and N. M. DeVore (University of Kansas) for the purified P450 2A13 and Dr. K. El-Bayoumy (Pennsylvania State University) for the organoselenium compounds used in this study.
Funding Sources
This work was supported in part by Grants from the Ministry of Education, Science, and Culture of Japan, the Ministry of Health and Welfare of Japan (T.S., N.M., K.T., S.T., H.Y., M.K.) and NIH grants R37 CA090426 and P30 ES000267 (F.P.G.).
ABBREVIATIONS
- EROD
7-ethoxyresorufin O-deethylation
- BSC
benzyl selenocyanate
- o-XSC
1,2-phenylenebis(methylene)selenocyanate
- m-XSC
1,3-phenylenebis(methylene)selenocyanate
- p-XSC
1,4-phenylenebis(methylene)selenocyanate
- PAHs
polycyclic aromatic hydrocarbons
- DMBA
7,12-dimethylbenz[a]anthracene
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
REFERENCES
- 1.Reddy BS, Upadhyaya P, Simi B, Rao CV. Evaluation of organoselenium compounds for potential chemopreventive properties in colon carcinogenesis. Anticancer Res. 1994;14:2509–2514. [PubMed] [Google Scholar]
- 2.Ip C, El-Bayoumy K, Upadhyaya P, Ganther H, Vadhanavikit S, Thompson H. Comparative effect of inorganic and organic selenocyanate derivatives in mammary cancer chemoprevention. Carcinogenesis. 1994;15:187–192. doi: 10.1093/carcin/15.2.187. [DOI] [PubMed] [Google Scholar]
- 3.Fiala ES, Joseph C, Sohn OS, El-Bayoumy K, Reddy BS. Mechanism of benzylselenocyanate inhibition of azoxymethane-induced colon carcinogenesis in F344 rats. Cancer Res. 1991;51:2826–2830. [PubMed] [Google Scholar]
- 4.Nayini J, El-Bayoumy K, Sugie S, Cohen LA, Reddy BS. Chemoprevention of experimental mammary carcinogenesis by the synthetic organoselenium compound, benzylselenocyanate, in rats. Carcinogenesis. 1989;10:509–512. doi: 10.1093/carcin/10.3.509. [DOI] [PubMed] [Google Scholar]
- 5.Richie JP, Jr, Kleinman W, Desai DH, Da A, Amin SG, Pinto JT, El-Bayoumy K. The organoselenium compound 1,4-phenylenebis(methylene)selenocyanate inhibits 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced tumorgenesis and enhances glutathione-related antioxidant levels in A/J mouse lung. Chem.-Biol. Interact. 2006;161:93–103. doi: 10.1016/j.cbi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Prokopczyk B, Cox JE, Upadhyaya P, Amin S, Desai D, Hoffmann D, El-Bayoumy K. Effects of dietary 1,4-phenylenebis(methylene)selenocyanate on 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced DNA adduct formation in lung and liver of A/J mice and F344 rats. Carcinogenesis. 1996;17:749–753. doi: 10.1093/carcin/17.4.749. [DOI] [PubMed] [Google Scholar]
- 7.El-Bayoumy K, Upadhyaya P, Desai DH, Amin S, Hecht SS. Inhibition of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone tumorigenicity in mouse lung by the synthetic organoselenium compound, 1,4-phenylenebis(methylene)selenocyanate. Carcinogenesis. 1993;14:1111–1113. doi: 10.1093/carcin/14.6.1111. [DOI] [PubMed] [Google Scholar]
- 8.El-Bayoumy K, Chae YH, Upadhyaya P, Meschter C, Cohen LA, Reddy BS. Inhibition of 7,12-dimethylbenz[a]anthracene-induced tumors and DNA adduct formation in the mammary glands of female Sprague-Dawley rats by the synthetic organoselenium compound, 1,4-phenylenebis(methylene)selenocyanate. Cancer Res. 1992;52:2402–2407. [PubMed] [Google Scholar]
- 9.El-Bayoumy K, Sinha R. Mechanisms of mammary cancer chemoprevention by organoselenium compounds. Mutat. Res. 2004;551:181–197. doi: 10.1016/j.mrfmmm.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 10.El-Bayoumy K, Das A, Boyiri T, Desai D, Sinha R, Pittman B, Amin S. Comparative action of 1,4-phenylenebis(methylene)selenocyanate and its metabolites against 7,12-dimethylbenz[a]anthracene-DNA adduct formation in the rat and cell proliferation in rat mammary tumor cells. Chem.-Biol. Interact. 2003;146:179–190. doi: 10.1016/j.cbi.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Shimada T. Xenobiotic-metabolizing enzymes involved in activation and inactivation of carcinogenic polycyclic aromatic hydrocarbons. Drug Metab. Pharmacokinet. 2006;21:257–276. doi: 10.2133/dmpk.21.257. [DOI] [PubMed] [Google Scholar]
- 12.Guengerich FP, Shimada T. Oxidation of toxic and carcinogenic chemicals by human cytochrome P-450 enzymes. Chem. Res. Toxicol. 1991;4:391–407. doi: 10.1021/tx00022a001. [DOI] [PubMed] [Google Scholar]
- 13.Shimada T, Hayes CL, Yamazaki H, Amin S, Hecht SS, Guengerich FP, Sutter TR. Activation of chemically diverse procarcinogens by human cytochrome P450 1B1. Cancer Res. 1996;56:2979–2984. [PubMed] [Google Scholar]
- 14.Hecht SS. Metabolic activation and detoxification of tobacco-specific nitrosamines: a model for cancer prevention strategies. Drug Metab. Rev. 1997;26:373–390. doi: 10.3109/03602539409029803. [DOI] [PubMed] [Google Scholar]
- 15.Yamazaki H, Inui Y, Yun C-H, Guengerich FP, Shimada T. Cytochrome P450 2E1 and 2A6 enzymes as major catalysts for metabolic activation of N-nitrosodialkylamines and tobacco-related nitrosamines in human liver microsomes. Carcinogenesis. 1992;13:1789–1794. doi: 10.1093/carcin/13.10.1789. [DOI] [PubMed] [Google Scholar]
- 16.Chiang HC, Wang CY, Lee HL, Tsou TC. Metabolic effects of CYP2A6 and CYP2A13 on 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK)-induced gene mutation-A mammalian cell-based mutagenesis approach. Toxicol. Appl. Pharmacol. 2011;253:145–152. doi: 10.1016/j.taap.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 17.Shimada T, El-Bayoumy K, Upadhyaya P, Sutter TR, Guengerich FP, Yamazaki H. Inhibition of human cytochrome P450-catalyzed oxidations of xenobiotics and procarcinogens by synthetic organoselenium compounds. Cancer Res. 1997;57:4757–4764. [PubMed] [Google Scholar]
- 18.Shimada T, Guengerich FP. Inhibition of human cytochrome P450 1A1-, 1A2-, and 1B1-mediated activation of procarcinogens to genotoxic metabolites by polycyclic aromatic hydrocarbons. Chem. Res. Toxicol. 2006;19:288–294. doi: 10.1021/tx050291v. [DOI] [PubMed] [Google Scholar]
- 19.Shimada T, Murayama N, Okada K, Funae Y, Yamazaki H, Guengerich FP. Different mechanisms of inhibition for human cytochrome P450 1A1, 1A2, and 1B1 by polycyclic aromatic inhibitors. Chem. Res. Toxicol. 2007;20:489–496. doi: 10.1021/tx600299p. [DOI] [PubMed] [Google Scholar]
- 20.Shimada T, Murayama N, Tanaka K, Takenaka S, Imai Y, Hopkins NE, Foroozesh MK, Alworth WL, Yamazaki H, Guengerich FP, Komori M. Interaction of polycyclic aromatic hydrocarbons with human cytochrome P450 1B1 in inhibiting catalytic activity. Chem. Res. Toxicol. 2008;21:2313–2323. doi: 10.1021/tx8002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimada T, Tanaka K, Takenaka S, Foroozesh MK, Murayama N, Yamazaki H, Guengerich FP, Komori M. Reverse type I binding spectra of human cytochrome P450 1B1 induced by flavonoid, stilbene, pyrene, naphthalene, phenanthrene, and biphenyl derivatives that inhibit catalytic activity: a structure-function relationship study. Chem. Res. Toxicol. 2009;22:1325–1333. doi: 10.1021/tx900127s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamazaki T, Nakamura M, Komatsu T, Ohyama K, Hatanaka N, Asahi S, Shimada N, Guengerich FP, Shimada T, Nakajima T, Yokoi T. Roles of NADPH-P450 reductase and apo- and holo-cytochrome b5 on xenobiotic oxidations catalyzed by 12 recombinant human cytochrome P450s expressed in membranes of Escherichia coli. Protein Express. Purif. 2002;24:329–337. doi: 10.1006/prep.2001.1578. [DOI] [PubMed] [Google Scholar]
- 23.Fukami T, Katoh M, Yamazaki H, Yokoi T, Nakajima T. Human cytochrome P450 2A13 efficiently metabolizes chemicals in air pollutants: naphthalene, styrene, and toluene. Chem. Res. Toxicol. 2008;21:720–725. doi: 10.1021/tx700325f. [DOI] [PubMed] [Google Scholar]
- 24.Guengerich FP, Hosea NA, Martin MV. Purification of cytochromes P450: products of bacterial recombinant expression systems. Methods Mol. Biol. 1998;107:77–83. doi: 10.1385/0-89603-519-0:77. [DOI] [PubMed] [Google Scholar]
- 25.Chowdhury G, Calcutt MW, Guengerich FP. Oxidation of N-Nitrosoalkylamines by human cytochrome P450 2A6: sequential oxidation to aldehydes and carboxylic acids and analysis of reaction steps. J. Biol. Chem. 2010;285:8031–8044. doi: 10.1074/jbc.M109.088039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith BD, Sanders JL, Porubsky PR, Lushington GH, Stout CD, Scott EE. Structure of the human lung cytochrome P450 2A13. J. Biol. Chem. 2007;282:17306–17313. doi: 10.1074/jbc.M702361200. [DOI] [PubMed] [Google Scholar]
- 27.Gillam EM, Guo Z, Guengerich FP. Expression of modified human cytochrome P450 2E1 in Escherichia coli, purification, and spectral and catalytic properties. Arch. Biochem. Biophys. 1994;312:59–66. doi: 10.1006/abbi.1994.1280. [DOI] [PubMed] [Google Scholar]
- 28.Sandhu P, Baba T, Guengerich FP. Expression of modified cytochrome P450 2C10 (2C9) in Escherichia coli, purification, and reconstitution of catalytic activity. Arch. Biochem. Biophys. 1993;306:43–50. doi: 10.1006/abbi.1993.1536. [DOI] [PubMed] [Google Scholar]
- 29.Krauser JA, Guengerich FP. Cytochrome P450 3A4-catalyzed testosterone 6β-hydroxylation stereochemistry, kinetic deuterium isotope effects, and rate-limiting steps. J. Biol. Chem. 2005;280:19496–19506. doi: 10.1074/jbc.M501854200. [DOI] [PubMed] [Google Scholar]
- 30.Harada N, Omura T. Selective induction of two different molecular species of cytochrome P-450 by phenobarbital and 3-methylcholanthrene. J. Biochem. (Tokyo) 1981;89:237–248. doi: 10.1093/oxfordjournals.jbchem.a133187. [DOI] [PubMed] [Google Scholar]
- 31.Shimada T, Sawabe Y. Activation of 3,4,3',4'-tetrachlorobiphenyl to protein-bound metabolites by rat liver microsomal cytochrome P-448-containing monooxygenase system. Toxicol. Appl. Pharmacol. 1983;70:486–493. doi: 10.1016/0041-008x(83)90166-7. [DOI] [PubMed] [Google Scholar]
- 32.Imai Y, Hashimoto-Yutsudo C, Satake H, Girardin A, Sato R. Multiple forms of cytochrome P-450 purified from liver microsomes of phenobarbital- and 3-methylcholanthrene-pretreated rabbits. I. Resolution, purificaton, and molecular properties. J. Biochem. (Tokyo) 1980;88:489–503. doi: 10.1093/oxfordjournals.jbchem.a132996. [DOI] [PubMed] [Google Scholar]
- 33.Shimada T, Imai Y, Sato R. Covalent binding of polychlorinated biphenyls to proteins by reconstituted monooxygenase system containing cytochrome P-450. Chem. Biol. Interact. 1981;38:29–44. doi: 10.1016/0009-2797(81)90151-4. [DOI] [PubMed] [Google Scholar]
- 34.Yamazaki H, Tanaka M, Shimada T. Highly sensitive high-performance liquid chromatographic assay for coumarin 7-hydroxylation and 7-ethoxycoumarin O-deethylation by human liver cytochrome P450 enzymes. J. Chromatogr. B. 1999;721:13–19. doi: 10.1016/s0378-4347(98)00472-1. [DOI] [PubMed] [Google Scholar]
- 35.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J. Biol. Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 36.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 37.Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, Johnson EF. Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J. Biol. Chem. 2007;282:14348–14355. doi: 10.1074/jbc.M611692200. [DOI] [PubMed] [Google Scholar]
- 38.Sansen S, Hsu MH, Stout CD, Johnson EF. Structural insight into the altered substrate specificity of human cytochrome P450 2A6 mutants. Arch. Biochem. Biophys. 2007;464:197–206. doi: 10.1016/j.abb.2007.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yano JK, Hsu MH, Griffin KJ, Stout CD, Johnson EF. Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat. Struct. Mol. Biol. 2005;12:822–823. doi: 10.1038/nsmb971. [DOI] [PubMed] [Google Scholar]
- 40.Wang A, Savas U, Stout CD, Johnson EF. Structural characterization of the complex between alpha-naphthoflavone and human cytochrome P450 1B1. J. Biol. Chem. 2011;286:5736–5743. doi: 10.1074/jbc.M110.204420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimada T, Tanaka K, Takenaka S, Murayama N, Martin MV, Foroozesh MK, Yamazaki H, Guengerich FP, Komori M. Structure-function relationships of inhibition of human cytochromes P450 1A1, 1A2, 1B1, 2C9, and 3A4 by 33 flavonoid derivatives. Chem. Res. Toxicol. 2010;23:1921–1935. doi: 10.1021/tx100286d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 43.DeVore NM, Smith BD, Urban MJ, Scott EE. Key residues controlling phenacetin metabolism by human cytochrome P450 2A enzymes. Drug Metab. Dispos. 2008;36:2582–2590. doi: 10.1124/dmd.108.023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeVore NM, Smith BD, Wang JL, Lushington GH, Scott EE. Key residues controlling binding of diverse ligands to human cytochrome P450 2A enzymes. Drug Metab. Dispos. 2009;37:1319–1327. doi: 10.1124/dmd.109.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El-Bayoumy K, Sinha R. Molecular chemoprevention by selenium: a genomic approach. Mutat. Res. 2005;591:224–236. doi: 10.1016/j.mrfmmm.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 46.Ding X, Kaminsky LS. Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu. Rev. Pharmacol. Toxicol. 2003;43:149–173. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- 47.Wang H, Tan W, Hao B, Miao X, Zhou G, He F, Lin D. Substantial reduction in risk of lung adenocarcinoma associated with genetic polymorphism in CYP2A13, the most active cytochrome P450 for the metabolic activation of tobacco-specific carcinogen NNK. Cancer Res. 2003;63:8057–8061. [PubMed] [Google Scholar]
- 48.Wong HL, Zhang X, Zhang QY, Gu J, Ding X, Hecht SS, Murphy SE. Metabolic activation of the tobacco carcinogen 4-(methylnitrosamino)-(3-pyridyl)-1-butanone by cytochrome P450 2A13 in human fetal nasal microsomes. Chem. Res. Toxicol. 2005;18:913–918. doi: 10.1021/tx0500777. [DOI] [PubMed] [Google Scholar]
- 49.Sohn OS, Fiala FS, Upadhyaya P, Chae Y-H, El-Bayoumy K. Comparative effects of phenylenebis(methylene)selenocyanate isomers on xenobiotic metabolizing enzymes in organs of female CD rats. Carcinogenesis. 1999;20:615–621. doi: 10.1093/carcin/20.4.615. [DOI] [PubMed] [Google Scholar]
- 50.El-Bayoumy K, Upadhyaya P, Sohn O-C, Rosa J, Fiala ES. Synthesis and excretion profile of 1,4-[14C]phenylenebis(methylene)selenocyanate in the rat. Carcinogenesis. 1998;19:1603–1607. doi: 10.1093/carcin/19.9.1603. [DOI] [PubMed] [Google Scholar]
- 51.El-Bayoumy K, Das A, Boyiri T, Desai D, Sinha R, Pittman B, Amin S. Comparative action of 1,4-phenylenebis(methylene)selenocyanate and its metabolites against 7,12-dimethylbenz[a]anthracene-DNA adduct formation in the rats and cell proliferation in rat mammary tumor cells. Chem.-Biol. Interact. 2003;146:179–190. doi: 10.1016/j.cbi.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 52.Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P450 enzymes involved in the oxidation of drugs, carcinogens, and toxic chemicals. Studies with liver microsomes of 30 Japanese and 30 Caucasians. J. Pharmacol. Exp. Ther. 1994;270:414–423. [PubMed] [Google Scholar]
- 53.Guengerich FP. Human cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. 3rd ed. New York: Kluwer Academic/Plenum Press; 2005. pp. 377–530. [Google Scholar]
- 54.El-Bayoumy K, Narayanan BA, Desai DH, Narayanan NK, Pittman B, Amin SG, Schwartz J, Nixon DW. Elucidation of molecular targets of mammary cancer chemoprevention in the rat by organoselenium compounds using cDNA microarray. Carcinogenesis. 2003;24:1505–1514. doi: 10.1093/carcin/bgg103. [DOI] [PubMed] [Google Scholar]
- 55.El-Bayoumy K, Das A, Narayanan B, Narayanan N, Fiala ES, Desai D, Rao CV, Amin S, Sinha R. Molecular targets of the chemopreventive agent 1,4-phenylenebis (methylene)-selenocyanate in human non-small cell lung cancer. Carcinogenesis. 2006;27:1369–1376. doi: 10.1093/carcin/bgi328. [DOI] [PubMed] [Google Scholar]
- 56.Chen KM, Spratt TE, Stanley BA, De Cotiis DA, Bewley MC, Flanagan JM, Desai D, Das A, Fiala ES, Amin S, El-Bayoumy K. Inhibition of nuclear factor-kappaB DNA binding by organoselenocyanates through covalent modification of the p50 subunit. Cancer Res. 2007;67:10475–10483. doi: 10.1158/0008-5472.CAN-07-2510. [DOI] [PubMed] [Google Scholar]