Abstract
It has been posited that animal development evolved from pre-existing mechanisms for regulating cell differentiation in the single celled and colonial ancestors of animals. Although the progenitors of animals cannot be studied directly, insights into their cell biology may be gleaned from comparisons between animals and their closest living relatives, the choanoflagellates. We report here on the life history, cell differentiation and intercellular interactions in the colony-forming choanoflagellate Salpingoeca rosetta. In response to diverse environmental cues, S. rosetta differentiates into at least five distinct cell types, including three solitary cell types (slow swimmers, fast swimmers, and thecate cells) and two colonial forms (rosettes and chains). Electron microscopy reveals that cells within colonies are held together by a combination of fine intercellular bridges, a shared extracellular matrix, and filopodia. In addition, we have discovered that the carbohydrate-binding protein wheat germ agglutinin specifically stains colonies and the slow swimmers from which they form, showing that molecular differentiation precedes multicellular development. Together, these results help establish S. rosetta as a model system for studying simple multicellularity in choanoflagellates and provide an experimental framework for investigating the origin of animal multicellularity and development.
Keywords: origin of animal multicellularity, choanoflagellate, development, intercellular bridges, colony, Salpingoeca rosetta, Proterospongia
Introduction
Choanoflagellates are a group of single-celled and colony-forming microeukaryotes found in diverse marine and freshwater environments. By characterizing the life history and cell biology of choanoflagellates, the closest living relatives of animals, it may be possible to reconstruct the ancestry of animal cell differentiation (Carr et al., 2008; King, 2004; Ruiz-Trillo et al., 2008; Steenkamp et al., 2006). Choanoflagellate cells typically bear a single apical flagellum surrounded by a collar of microvilli (Fig. 1). Flagellar movement generates water currents that draw prey bacteria onto the outer surface of the collar, where the bacteria are phagocytosed (Lapage, 1925; Pettitt et al., 2002). This cell morphology and feeding behavior is conserved in all choanoflagellate species and, within animals, is structurally and functionally conserved in the form of choanocytes, a group of specialized feeding cells found in sponges. The resemblance of choanoflagellates to sponge choanocytes has long been interpreted as evidence of a close relationship between choanoflagellates and animals (James-Clark, 1867; Maldonado, 2004; Nielsen, 2008) and modern phylogenetic analyses now demonstrate that choanoflagellates are the closest known sister group of animals (Carr et al., 2008; King et al., 2008; Ruiz-Trillo et al., 2008; Steenkamp et al., 2006). Furthermore, ancestral character-state reconstruction based on the phylogenetic relationships among choanoflagellates, sponges, and eumetazoans suggests that the last common ancestor of animals and choanoflagellates resembled a modern choanoflagellate (Carr et al., 2008; Nielsen, 2008; Steenkamp et al., 2006).
Fig. 1. Five distinct cell morphologies observed in S. rosetta cultures.
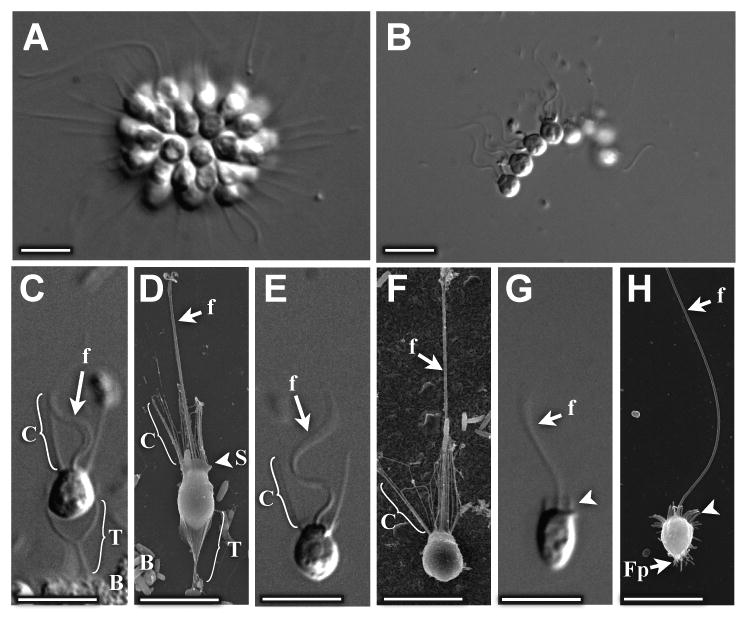
(A) Cells in rosette colonies orient in a sphere around a central focus, with their apical flagella and collars oriented radially outward. (B) Cells in chain colonies attach to one another laterally to form linear arrays of cells. (C,D) Thecate cells have long (∼4μm) collars surrounding apical flagella and attach to substrates via a goblet-shaped theca. (E,F) Slow swimmers have similar morphology to thecate cells, but lack thecae. (G,H) Fast swimmers have no theca and either no collar or a truncated collar (arrowheads), and are often covered in small filopodia. Key: f: flagellum, C: collar, T: theca, S: skirt, Fp: filopodia, B: bacteria. Scale bars = 5μm. (A,B,C,E,G: DIC microscopy, D,F,H: Scanning Electron Microscopy)
Intriguingly, some choanoflagellates are able to form multicelled colonies as part of their life cycle. For example, cells within colonies of Choanoeca perplexa (previously known as Proterospongia choanojuncta) attach to one another via the pairing of collar microvilli (Leadbeater, 1983a). In other species, such as Codosiga botrytis (Hibberd, 1975) and Desmarella Kent (Karpov and Coupe, 1998), neighboring cells in colonies are connected by fine intercellular bridges that, at least superficially, resemble the ring canals that link developing spermatogonia or oogonia in animals (Carlson and Handel, 1988; Greenbaum et al., 2007; Kojima, 1992; Ong and Tan, 2010; Schindelmeiser et al., 1983). Given that colony formation is found in diverse choanoflagellate lineages, it is possible that colony formation was present in the last common ancestor of animals and choanoflagellates (Carr et al., 2008). Therefore, understanding modern choanoflagellate cell biology and colony formation may provide insight into to the earliest forms of animal development.
Salpingoeca rosetta (previously known as Proterospongia sp. ATCC 50818, see Taxonomic Description below) is a recently isolated choanoflagellate species that forms colonies in the laboratory (Fairclough et al., 2010). We have shown previously that this organism expresses members of key cell signaling and adhesion protein families that were previously thought to be exclusively found in animals (King et al., 2003). In addition, a genome project currently in progress should provide genomic resources for rapidly gaining insight into the biology of S. rosetta (Ruiz-Trillo et al., 2007). By studying cell differentiation and development in S. rosetta, it may be possible to characterize the ancestral functions of proteins that regulate animal development.
In the current study, we address two fundamental aspects of the S. rosetta life history: cell differentiation and morphogenesis. We find that S. rosetta undergoes cell differentiation in response to diverse environmental cues. S. rosetta cells in culture can differentiate into at least three solitary forms and two distinct colonial forms: rosette colonies and chain colonies. The development of colonies is preceded by molecular differentiation; only those solitary cells that are competent to develop into colonies stain with wheat germ agglutinin (WGA), as do all of the cells within colonies. Ultrastructural analyses of cell morphology reveal that cells in rosette and chain colonies are connected by a combination of intercellular bridges, extracellular matrix (ECM), and filopodia. These findings expand our understanding of cell differentiation in S. rosetta and provide a foundation for molecular studies probing the origin of animal multicellularity.
Material and methods
Initial isolation of choanoflagellate S. rosetta
A single choanoflagellate rosette colony was isolated by Tom Nerad from mud core samples collected near Hog Island, Virginia, USA (37.45278° N 75.67521° W) in February 2000. After propagation in growth media (King et al., 2009), the choanoflagellate culture (containing associated environmental bacteria) was deposited under strain designation ATCC 50818 (American Tissue Culture Collection, Manassas, VA). Subsequent analyses demonstrated that ATCC 50818 is not contaminated with any other species of eukaryotes (Fairclough and Richter, unpublished).
Growth propagation, and enrichment of S. rosetta cell types
Growth media were prepared in artificial sea water (King et al., 2009) and S. rosetta cultures were maintained by passaging 2 mL of culture into 15 mL fresh medium every 3 days. Cell type enriched cultures (described below) were derived from a rosette colony-free culture (see supplement to (Fairclough et al., 2010)).
Fast-swimmer cell cultures
The supernatant was removed from a rosette colony-free culture and attached cells were washed twice with fresh medium to remove swimming cells. Recovery for one day led the attached thecate cells to produce fast swimmers in the water column. The majority of cells in the supernatant of this culture were fast-swimmers, however the proportion of slow-swimmers increased over time.
Thecate cell cultures
The supernatant from a fast-swimmer culture was diluted into fresh medium and grown overnight to allow cells to attach and differentiate into thecate cells. The attached thecate cells were washed twice with fresh medium, resulting in a population of thecate cells that was relatively free of bacterial biofilm. Over time, fast-swimmers were produced again and accumulated in the water column.
Slow swimmer cell cultures
The supernatant from a fast-swimmer culture was diluted into fresh medium and allowed to recover overnight, generating slow swimmers and thecate cells. Although the resulting supernatant was enriched for slow swimmers, sometimes it also contained significant numbers of fast swimmers.
Chain cultures
Cultures consisting primarily of chain colonies were generated by diluting 2 mL of cells from the supernatant of a rosette colony-deficient culture (see supplement to (Fairclough et al., 2010)) into 15 mL fresh medium every day for 1–2 weeks.
Rosette cultures
Rosette colonies were produced using two different strategies. In the first approach, a chain culture containing mixed environmental bacteria was inoculated with live Algoriphagus machipongonensis bacteria (previously known as Algoriphagus sp. PR1; Alegado et al., 2011, submitted). Addition of A. machipongonensis induces the development of rosette colonies, which became the dominant form in the culture within 2 days.
In the second approach, a monoxenic strain of S. rosetta was generated in which the sole source of bacteria was A. machipongonensis. The undefined population of environmental bacteria in the ATCC 50818 culture was replaced through the following: the culture was treated with a combination of multiple antibiotics (ofloxacin 10 μg ml-1, kanamycin 50 μg ml-1, streptomycin 50 μg ml-1), serially diluted to further reduce the diversity of bacteria associated with the choanoflagellate culture, sorted by choanoflagellate cell size on a DAKOCytomation MoFlo High Speed Cell Sorter (Carpenteria, CA), and finally supplemented with the colony-inducing bacterium A. machipongonensis. The resultant choanoflagellate culture line was propagated over several weeks in antibiotic-free growth media. Bacterial monoxenicity was assessed by plating on modified Zobell medium (Carlucci and Pramer, 1957) and restriction fragment length polymorphism analysis. When split daily for 1–2 weeks, this monoxenic culture produces rosette-only cultures.
Microscopy, Fixation, & WGA staining
Cells were imaged with a Leica DMI6000B Microscope equipped with a DFC350 FX camera. For wheat germ agglutinin (WGA) labeling experiments, we added 0.2 ng/ml Alexa Fluor 488-labeled wheat germ agglutinin (#W11261 Invitrogen, CA) to live cells and imaged immediately. Plasma membrane staining occurred within seconds, and over time (>10 minutes) fluorescent spots also accumulated in the food vacuoles of some cells. Samples for Fig. 4 were fixed in 2% glutaraldehyde + 50mM PIPES pH 8.0 prior to imaging to prevent cell movement.
Fig. 4. WGA+ slow swimmers are competent to differentiate into colonies.
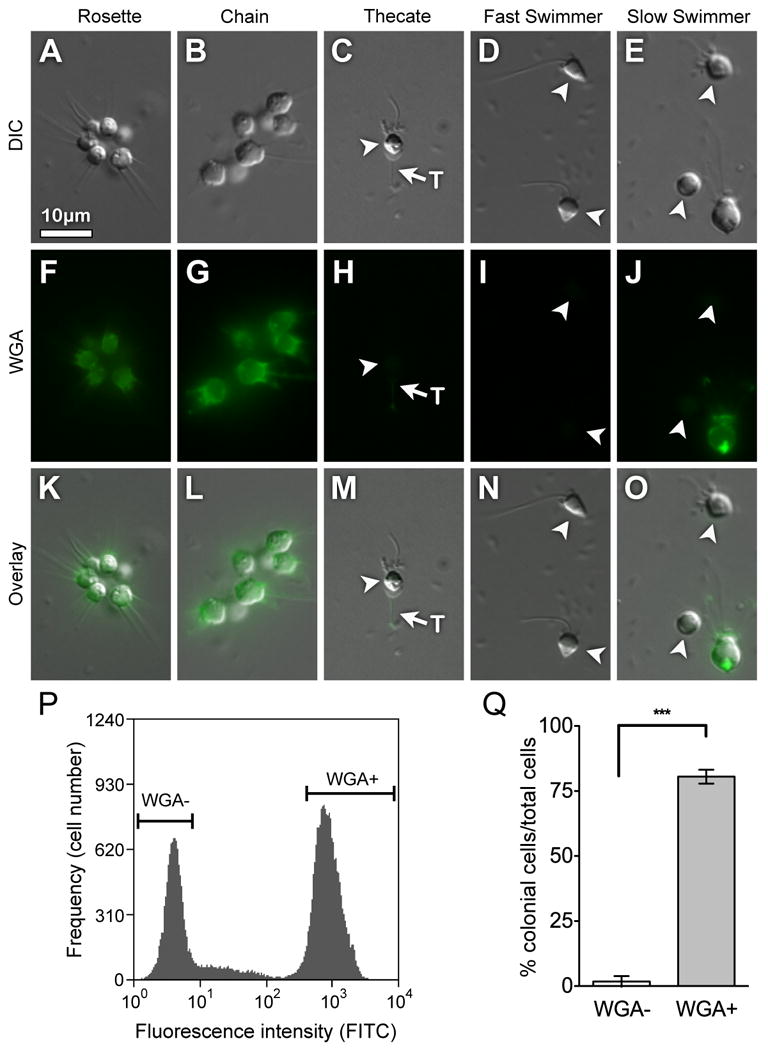
WGA labels the plasma membrane of all cells within rosette and chain colonies (A, B, F, G, K, L). Although WGA does not label thecate cells, it does label the theca weakly (C, H, M). WGA does not label fast swimmers (D, I, N) and labels only a subset of slow swimmer cells (E, J, O). Slow swimmer cells that were either positive or negative for staining with WGA were sorted based on relative fluorescence intensity per cell following incubation with Alexa-488 labeled WGA (representative sort, P). Following induction with A. machipongonensis, only WGA+ cells were competent to develop into colonies (Q). The percentage of cells in rosette colonies (± SE) was measured 48 h after WGA sorting and exposure to A. machipongonensis. Shown is the average of four independent experiments performed in triplicate. (***P < 0.0001). Key: T:theca
Rosette colony development in WGA stained cells
Cells from a colony-free culture were concentrated by centrifugation at 1,000 × g for 5 minutes, incubated with 0.2ng/ml Alexa Fluor 488-labeled WGA, and sorted on a Cytopeia INFLUX sorter (BD Biosciences, Seattle, Washington) into WGA+ and WGA- populations (Fig. 5A). Cells were sorted into artificial seawater, diluted to a final concentration of 106 cells/mL in 11 mL of growth media, inoculated with 10 μl A. machipongonensis culture, and plated (100 μl per well) in 96-well Greiner Micro Clear plates (BioOne, Gloucester UK) in triplicate (i.e. three wells per condition per timepoint). The percentage of WGA+ cells and the percentage of colonial cells in each culture were assessed at 0 hr, 24 hr, and 48 hr after treatment with A. machipongonensis. To monitor the proportion of WGA+ cells at each time point, cells were incubated with WGA as above, fixed with 2% glutaraldehyde + 50 mM PIPES pH 8.0 and imaged. WGA+ staining was measured by fluorescence microscopy using a semi-automated system for image acquisition (ImagePro 6.2.1). Digital images were acquired by collecting three fields of view per well with a 10× objective to obtain images of 100-1000 cells per well in a total imaged area of 0.6-2.4 mm2. These images were quantified manually using the Object Counter function in ImageJ software (NIH) and analyzed for statistical significance using unpaired t-tests with Welch's correction.
Fig. 5. Cells within rosette colonies are attached to one another with ECM, filopodia, and intercellular bridges.
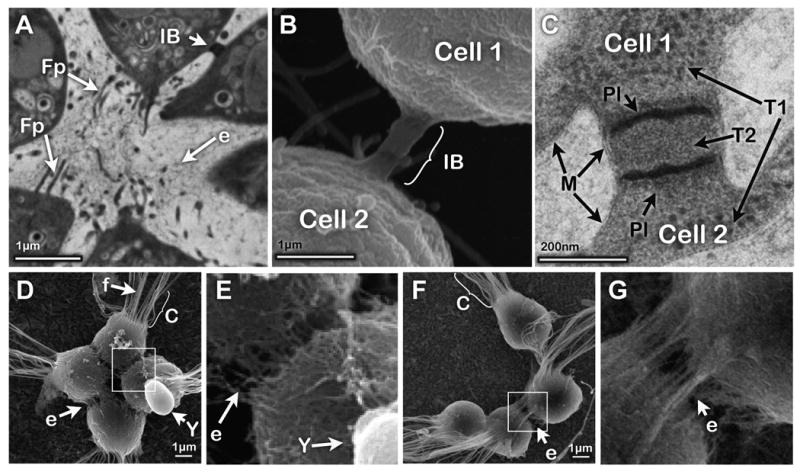
(A) TEM of a thin section through a rosette shows filopodia extending into the central ECM and an intercellular bridge connecting neighboring cells. (B) SEM of an intercellular bridge. (C) TEM of a thin section through an intercellular bridge shows two electron-dense plates trisecting the bridge (see also Movie 5). The texture of the cytoplasm within cells (T1) differs from that within the bridge (T2). (D,E) SEM of a rosette colony shows a shared, filamentous ECM linking S. rosetta cell bodies. ECM is absent from co-cultured yeast cells (included as a negative control). (F,G) Cells in chain colonies share ECM filaments (e), but lack filopodia. Key: f: flagellum, C: collar, IB: intercellular bridge, Fp: filopodia, E: ECM, Y: yeast, Pl: bridge plate, M: cell membrane. Scale bars as marked.
Transmission Electron Microscopy (TEM)
Colonies from the monoxenic culture of S. rosetta-were concentrated by centrifugation (500×g for 20 mins), resuspended in a small volume of artificial sea water, loaded into capillary tubes (Hohenberg et al., 1994), high-pressure frozen (HPF) in a Leica EM PACT2, and fixed by freeze substitution in 0.01% OsO4 + 0.2% uranyl acetate in acetone. Samples were infiltrated with Epon-Araldite (Embed-812), cut into 100nm sections and imaged on a FEI Tecnai 12 Transmission electron microscope. Tilted images of the junction were taken on an automated stage (Fischione instruments 2040 Dual axis tomography holder) then registered using an image registration routine in Matlab (modified from Periaswamy and Farid, 2003).
Scanning Electron Microscopy (SEM)
For SEM of thecate cells (Fig. 1D), cells from a colony-deficient culture were allowed to attach to Leica gold-coated HPF planchettes overnight. These planchettes were rinsed in artificial seawater before the high pressure freezing and freeze-substitution procedures described above. The sample was exchanged into 100% ethanol, critical point dried on a Tousimis AutoSamdri 815 Critical Point Dryer and sputter coated (24 s on Tousimis sputter coater) before imaging on a Hitachi S-5000 Scanning electron microscope.
For SEM of HPF colonies (Fig. 1H, Fig. 3A, B), we first created covalently crosslinked WGA-coated gold HPF planchettes by incubating gold-coated HPF planchettes in 10mg/mL DSP (Di(N-succinimidyl) 3,3′-dithiodipropionate; Sigma #D3669) in DMSO for 30 minutes, rinsing in ddH2O, then incubating with 2mg/mL WGA at 4°C overnight. WGA-planchettes were rinsed before use. Colonies from the monoxenic culture of S. rosetta-were concentrated by centrifugation at 500 × g for 20 mins, added onto the planchettes, and centrifuged at 50 × g for 20 minutes to adhere the live colonies to the WGA. The planchettes were subjected to high pressure freezing and were freeze substituted in acetone with 0.1% glutaraldehyde, 0.25% uranyl acetate and 0.01% OsO4, and processed as above.
Fig. 3. Transitions between thecate cells and fast swimmers.
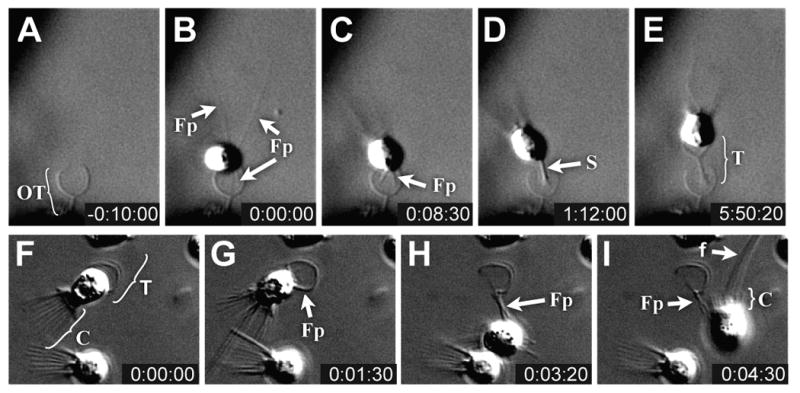
(A-E) Timelapse microscopy of a fast swimmer building a new theca (Movie 1). Although fast swimmers normally attach to environmental substrates, an unusual case of attachment to an empty theca is presented here because the added elevation from the substrate affords a better view of the attachment process. (A) An empty theca (abandoned by another cell). (B) A fast swimmer uses long filopodia to attach to the empty theca. (C) Those filopodia in contact with the empty theca become more refractile and coalesce to form the base of a new stalk projecting from the base of the cell. (D) The coalesced filopodia form a highly refractile stalk which extends from the cell base. (E) The refractile material is replaced by a stable stalk, after which the cell becomes more spherical and secretes the theca cup from its sides, leaving a ∼1μm gap between the theca and cell base. (F– H) Timecourse of a cell releasing from its theca (Movie 3). (F) Prior to release, the thecate cell is attached to the theca sub-equatorially and has a long collar (indicated by bracket). (G, H) As the cell begins to leave its theca, filopodia extend from the sides of the cell thereby lifting the cell out of the theca cup. (I) Prior to complete detachment, the collar retracts, as indicated by the short bracket. Key: OT: Original Theca, Fp: Filopodia, T: Theca, S: Stalk, C: collar, f: flagellum. Times indicated in hours:minutes:seconds.
For SEM of glutaraldehyde-fixed colonies and single cells (Fig. S3B), covalently crosslinked amino-coated silica wafers were first created by incubating silica wafers with 2% 3-Aminopropyltriethoxysilane (#15108 Acros Organics, NJ) in chloroform for 30 minutes at room temperature, rinsing wafers in ddH2O, then baking at 80°C for 30 minutes. Colonies were concentrated by centrifugation as above, fixed by mixing 1:1 with 4% glutaraldehyde/100mM HEPES pH 8.0 in artificial sea water, then centrifuged onto amino-coated silica wafers at 50×g for 20 minutes to fix colonies to wafers. After 1h, glutaraldehyde was quenched with 1/10 volume of 1M glycine pH 8, wafers were rinsed 2× in 1M Cacodylate buffer pH 7.4, incubated in 1% OsO4 for 1h, rinsed again and dehydrated through an ethanol series before critical point drying.
Results
Morphology of cell types
S. rosetta cultures grown under laboratory conditions contain at least five different forms (Table 1, Fig. 1 and 2), including two types of multicelled colonies, spherical “rosettes” and linear “chains.” Cells in rosette colonies are tightly packed, with the base of each cell directed toward a central focus (Fig. 1A, S1). Cells in chain colonies, in contrast, are attached to one another in linear or branched arrangements (Fig. 1B, S2A). Rosette and chain colonies are free-swimming and were not observed to attach to substrates.
Table 1. Characteristics of S. rosetta cell types.
| Form | Theca | Long Collar | WGA staining | Intercellular bridges | Filopodia | Central Focus |
|---|---|---|---|---|---|---|
| Rosette colony | - | + | + | + | +/- | + |
| Chain colony | - | + | + | + | - | - |
| Thecate cell | + | + | - | - | - | - |
| Slow Swimmer | - | + | +/- | - | - | - |
| Fast Swimmer | - | - | - | - | + | - |
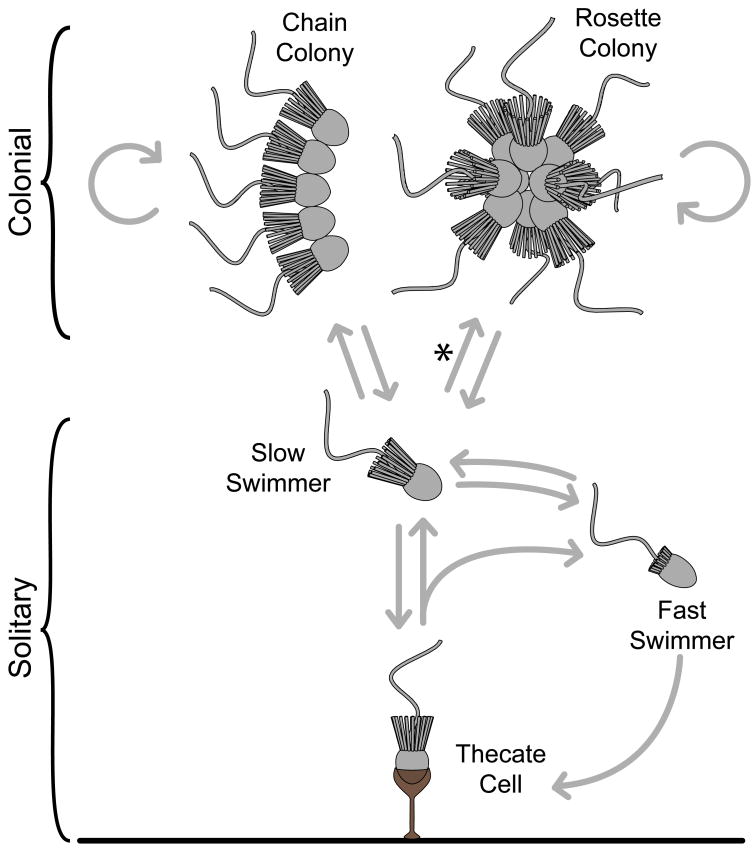
Fig. 2. A model of S. rosetta life history.
S. rosetta cells can differentiate between at least five different forms. Arrows depict observed and inferred transitions that are described in the main text and in Fig. S9. Fast swimmers can settle to produce thecate cells that then produce swimming cells either through cell division or theca abandonment. Under rapid growth conditions, slow swimmer cells proliferate but remain attached via intercellular bridges and ECM to produce chain colonies, or, in the presence of A. machipongonensis bacteria (denoted by ‘*’), rosette colonies that have intercellular bridges, ECM and filopodia.
The three solitary cell types -- thecate, slow swimmer, and fast swimmer -- can be distinguished by their cellular morphology and behavior (Fig. 1C-H). Thecate cells attach to environmental substrates through the construction of a goblet-shaped structure called a theca, which contains a cup atop a slender ∼3 μm long stalk (Fig. 1C, D, S3). Thecate cells display the distinctive morphology of the genus Salpingoeca (James-Clark, 1868), chains display that of Desmarella (Karpov and Coupe, 1998), and rosettes display the Proterospongia morphology (Leadbeater, 1983a), revealing that the cell types historically used to define these genera are life history stages and not valid as taxonomic characters (see also (Karpov and Coupe, 1998) and (Leadbeater, 1983a).
Except for the presence of the theca, the cell bodies of thecate cells resemble those of slow swimmers (Fig. 1E, F), with both cell types having a long (∼5 μm) feeding collar and rounded cell body. Fast swimmers, in contrast, have a reduced or absent feeding collar. With their long flagella and tapered cell bodies, which are often covered with short, ∼1 μm filopodia, they resemble animal sperm (Fig. 1G,H). The existence of multiple colonial and solitary forms of S. rosetta reveals that this species has as many differentiated cell types as some early branching animals (e.g. five cell types in sponges; (Valentine, 2004)).
Transitions among solitary cell types
The existence of multiple cell types reveals the ability of S. rosetta cells to differentiate. To establish a foundation for future studies of cell differentiation in S. rosetta, we have used a combination of video microscopy and modulation of growth conditions to reconstruct dynamic and flexible life history. We find that solitary cells can produce either other types of solitary cells, or colonies (Figs. 2, S9). Fast swimmer cells can directly produce thecate cells (Fig. 3A-E; Movies 1 and 2). Upon contact with a substrate (in the case of Fig. 3B, an empty theca), fast swimmers extend multiple long filopodia (∼10-15 μm) from the basal pole of the cell. Those filopodia that do not contact the substrate soon retract (Fig. 3C), and the remaining filopodia coalesce to form a nascent stalk that lifts the base of the cell several microns away from the surface (Fig. 3D). The stalk can vary in length considerably, ranging from ∼3μm to essentially absent (Fig. S3B). SEM imaging of mature theca stalks shows them to be tubular and attached to the substrate via a splayed base (Fig. S3B), suggesting they result from the secretion of material from the coalesced filopodia and nascent stalk. Upon reaching the cell body, the basal pole of the stalk converts into the rounded shape of the typical mature thecate cell (Movie 1). Finally, a cup is constructed atop the stalk, presumably by secretion from the surface of the cell; the cup and stalk together comprise the theca. The entire process of cell differentiation, from the first contact of fast swimmer cells with an environmental substrate to the differentiation and maturation of thecate cells, takes approximately 6 hours.
Thecate cells, in turn, can produce swimming cells either through theca abandonment or cell division. During theca abandonment, the cell extends long projections (probably filopodia) between the cell equator and the theca cup (Fig. 3G, H, Movie 3). Simultaneously, the collar appears to shorten (Fig. 3I, Movie 3) and eventually the cell releases from the theca and swims away. Swimming cells are also produced when thecate cells divide (Movie 4). During thecate cell division, the flagellum retracts, the cell cleaves and one of the daughter cells swims away.
It is not clear whether the cells produced through thecate cell division or theca abandonment are fast swimmers or slow swimmers. Direct observation of the fate of thecate cells is nearly impossible because of the small size and rapid swimming speed of fast and slow swimmers (e.g. Movie 4 shows cell division, but does not reveal the morphology of the motile daughter). Based upon the observation that fast swimmers become enriched in the water column after the addition of fresh media to pure populations of thecate cells, we hypothesize that thecate cells can give rise to fast swimmers. Nonetheless, it is also possible that thecate cells can produce slow swimmers, which are abundant in cultures grown under standard conditions (King et al., 2009).
Molecular differentiation precedes the onset of colony formation
In addition to characterizing the morphology of S. rosetta cell types, we sought a biochemical marker of cell differentiation. Plant-based lectins, which bind specifically and with high affinity to distinct sugar groups, have been used as cell surface probes for cell differentiation in animals and unicellular eukaryotes (Allen et al., 1988; Falk et al., 1994; Lueken et al., 1981; Ramoino, 1997; Ramoino et al., 2001). We found that one lectin, wheat germ agglutinin (WGA), selectively labels the plasma membrane of all cells in rosette and chain colonies at equal levels (Fig. 4 A, B, F, G, K, L), but does not stain thecate cells or fast swimmers (Fig. 4 C, D, H, I, M, N). Interestingly, WGA also stains a subpopulation of slow swimmers (Fig. 4 E, J, O).
We have previously observed that rosette colonies develop from slow swimmers by serial cell division (Fairclough et al., 2010), although it was not known whether all slow swimmers are competent to form colonies. The observation that WGA stains rosettes, chains, and a subset of slow swimmers raises the possibility that WGA+ slow swimmers may be the cells from which colonies develop. To determine the ability of WGA- and WGA+ slow simmer cells to form rosette colonies, we stained slow swimmer cultures, isolated populations of WGA- and WGA+ cells by FACS (Fig. 4P), and added Algoriphagus machipongonensis bacteria to induce rosette colony development (Methods). At the 0h time point, cells sorted into the high fluorescence-intensity population were 91.6 ± 2.5% (mean ± SD) WGA+ and cells sorted into the low fluorescence-intensity population were 0% WGA+. After 48 hours, 80.5 ± 2.7% (mean ± SE) of cells derived from WGA+ slow swimmers were in rosette colonies, compared to only 1.8 ± 0.06% (mean ± SE) of cells from WGA- populations (Fig. 4Q). Thus, although all slow swimmers appear to be morphologically identical, these data suggest the existence of at least two subpopulations that are differentiated by both their WGA staining patterns and their capacity to form rosette colonies.
Ultrastructural features of S. rosetta colonies
Understanding the cellular features that define S. rosetta colonies may provide insight into the mechanisms of colony development and the evolution of colony formation in choanoflagellates. We find that adjacent cells in rosette and chain colonies are connected laterally by intercellular bridges positioned slightly basal to the cell equators (Fig. 5A-B, S2C-D, S4). No more than one bridge was observed between any given pair of cells. The intercellular bridges are cylindrical structures 198 ± 38 nm wide and 149 ± 76 nm long (mean ± SD, n=10) and usually contain two densely-stained plates positioned 80 ± 21 nm apart (Fig. 5C). The cytoplasm between the two plates differs in granularity from that on either side of the intercellular bridges (Fig. 5C), suggesting that the plates may provide a permeability barrier that prevents unregulated sharing of cytoplasmic contents. For example, ∼17nm puncta, possibly ribosomes, are restricted to the main cytoplasm and apparently occluded from the internal bridge material.
To determine whether the cell membrane is continuous across the intercellular bridges, as in the intercellular bridges (ring canals) of animal spermatogonia and oogonia (Carlson and Handel, 1988; Greenbaum et al., 2007; Kojima, 1992; Ong and Tan, 2010; Schindelmeiser et al., 1983), we performed a TEM-rotational analysis of an ultra-thin section of an S. rosetta intercellular bridge. We found that the cell membrane extends continuously between neighboring cells and across the bridge (Fig. 5C, Movie 5). The continuity of the cell membrane and position of the bridges, coupled with the observation that colonies form through cell division, supports the hypothesis that intercellular bridges in S. rosetta colonies result from incomplete cytokinesis during colony development (Fairclough et al., 2010).
Cells in colonies are also connected by shared ECM; the ECM covers the cell bodies but not their microvilli (Fig. 5D-G). (The apparent absence of ECM in Fig. 5B can be attributed to the fixation of colonies in room temperature glutaraldehyde, which preserves cell structure and membranes, while eliminating most of the obscuring ECM.) The absence of ECM material on yeast cells (co-processed with rosette colonies as a control, Fig. 5D, E) demonstrates that this is not a condensation of material from the medium. Unlike the theca, which is composed of a thin, tightly woven sheet (Fig. S3D), the ECM of colonies is looser and fills the space between cells (Fig. 5E, G).
Finally, we have observed filopodia extending from the basal pole of cells in most, but not all, rosette colonies (Fig. 5A, S4). The filopodia in rosette colonies do not contact neighboring cells directly, but instead appear to terminate in the ECM found in the rosette core. While filopodia were not observed in chain colonies, further work will be necessary to determine if filopodia are specifically required for the formation of rosette colonies.
Cryptic features of the S. rosetta life history
On occasion, we observe additional cell types that hint at the existence of additional phases to the life history of S. rosetta that are not yet easily accessed in the laboratory. For example, in rapidly growing cultures, cells may have up to four flagella and collars (Fig. S2D, see also (Leadbeater, 1985)). In addition, we sometimes see cells that are reminiscent of minute cells from P. choanojuncta (Leadbeater, 1983b). S. rosetta minute cells are ejected from colonies and attach to substrates transiently without building theca (Fig. S5, S6, Movie 6). Clusters of minute cells were sometimes observed in rosette colony cultures (Fig. S6A) and could be distinguished from rosette colonies by their smaller cell size, short or absent collars, and lack of intercellular bridges (Fig. S6B). These minute colonies appeared to be held together by filopodia that extend from the base of the cells into the core.
Even among common cell types, transitions can be hard to follow because of the motility of cells in the water column. Although the fate of colonies is difficult to monitor, time-lapse video microscopy reveals that rosette colonies can reproduce by colony fission (Fig. S7, Movie 7). Daughter rosettes form when two clusters of cells pull away from each other and subsequently close in around new focal points. Chain colonies can also reproduce, but unlike rosettes, these colonies appear to divide by fragmentation as growing chains become increasingly susceptible to shear stress. Further insights into the roles of multiflagellated cells, minute cells, and colony fission in the life history of S. rosetta will require the discovery of new growth conditions that favor their study.
Discussion
Developmental biology generally concerns itself with the processes of differentiation, morphogenesis, growth, and reproduction. The study of animal evolution and development has revealed that these processes are regulated by a conserved set of molecular mechanisms in animals as diverse as sea anemones, fruit flies, and humans (Carroll et al., 2005; De Robertis, 2008; Gerhart, 1999; Pires da Silva and Sommer, 2003). A surprising number of the proteins involved in animal development evolved first in the unicellular ancestors of animals, where they may have functioned to sense and respond to variable environments (Abedin and King, 2008; King et al., 2003; King et al., 2008; Sebe-Pedros et al., 2010). Choanoflagellates, the closest living relatives of animals, provide the opportunity to investigate how cell differentiation in unicellular eukaryotes laid the foundations for animal cell biology and development (Carr et al., 2008; King et al., 2008; Ruiz-Trillo et al., 2008; Steenkamp et al., 2006). The life history of S. rosetta, which alternates between single-celled and colonial states, may therefore serve as a simple model for understanding a transition to multicellularity.
S. rosetta as a simple model for animal multicellularity
In the present study we have charted cell differentiation and morphogenesis during the life history of S. rosetta (Fig. 2). S. rosetta can dynamically interconvert among at least three solitary forms and two colonial forms in response to environmental cues (Table 1, Fig. 1, S9). Cells of S. rosetta propagate by cell division, both in colonial and solitary states. In addition, S. rosetta undergoes morphogenesis during the development of colonies and during rosette colony fission (Fig S8). Our observations in the laboratory reveal that S. rosetta exhibits an unexpected degree of developmental flexibility in response to changing environmental conditions (Methods). While there were five different cell types commonly observed in the laboratory, additional cell types (Fig. S5, S6) were observed on occasion, suggesting that our characterization of the life history of S. rosetta is incomplete. Whereas the behaviors of the fast swimmer cell type (fast swimming, lack of feeding and subsequent differentiation into a thecate cell) seem to indicate a dispersal form, the ecological significance of the other cell types remains obscure. To more fully investigate the S. rosetta life history and understand the ecological significance of each cell type, it will be necessary to document natural S. rosetta microhabitats and thereby refine growth conditions in the laboratory.
Features of S. rosetta cell and colony morphology are evocative of multicellular organisms
Fundamental features of cellular interactions within multicellular organisms -- namely direct cell-cell contacts, ECM, and filopodia -- are also found in S. rosetta. For example, intercellular bridges physically link adjacent cells in rosette and chain colonies (Fig. 5, S3, S4). Similar intercellular connections are widespread in many multicellular organisms including animals, plants, Volvox, and fungi; each of these structures derives from incomplete cytokinesis (Haglund et al., 2011; Hoops et al., 2006; Leys, 2003). While these structures may be homologous (Glotzer, 2005; Otegui et al., 2005), it is also possible that disparate lineages converged on the maintenance of intercellular bridges following incomplete cytokinesis as a simple pathway to multicellularity.
In animals, intercellular connections can appear transiently during cytokinesis in the form of the midbody (Byers and Abramson, 1968; Glotzer, 2001; Glotzer, 2005; Mullins and McIntosh, 1982). Alternatively, stable intercellular bridges are found in syncytia and as ring canals that form by incomplete cytokinesis during animal oogenesis and spermatogenesis (Carlson and Handel, 1988; Fiil, 1978; Greenbaum et al., 2007; Kojima, 1992; Ong and Tan, 2010; Schindelmeiser et al., 1983). In S. rosetta, the placement and continuous membrane of intercellular bridges (Fig. 5) suggest that these structures are also a product of incomplete cytokinesis. However, unlike animal ring canals that enable bulk flow of cytoplasm between daughter cells (Theurkauf and Hazelrigg, 1998), intercellular bridges in S. rosetta have electron-dense plates which are inconsistent with this function. Nevertheless, diffusion of small signaling molecules and membrane-anchored proteins may be possible across the intercellular bridges. Intercellular bridges lacking the electron dense plates are also observed on occasion and indicate that the bridges may be open for free sharing of cytoplasm between cells under certain conditions (e.g. Fig. S8). Determining the composition and function of S. rosetta intercellular bridges in molecular detail will help reveal the extent to which these structures are related to intercellular bridges from other multicellular lineages.
Like cells from diverse multicellular and unicellular lineages, S. rosetta cells also produce ECM. In animals, ECM regulates a number of developmental and physiological processes by coordinating signaling between cells and organizing the orientation of cells relative to one another within a tissue (Hynes, 2009). Three types of S. rosetta cells produce ECM: thecate cells, rosette colonies, and chain colonies (Fig. 5). Consistent with previous descriptions of choanoflagellate thecae, we find the theca of S. rosetta to be comprised of dense goblet-shaped ECM (Fig. S3C) (Carr et al., 2008; Leadbeater, 1977). In contrast, the ECM surrounding colonial cells is seemingly amorphous, loose and space-filling (Fig 5). Because the ECM of colonies is absent from unicellular swimming stages of the S. rosetta life history, we surmise that its function is specific to the development or maintenance of S. rosetta cell and substrate attachment – potentially by contributing to the structural integrity of colonies or by mediating signaling events involved in cell adhesion.
In addition to structures classically associated with cell adhesion in multicellular organisms, S. rosetta fast swimmers and rosette colony cells also produce filopodia (Fig 1H, 3, 6). We hypothesize that extension of filopodia may provide a means to establish stable contacts with substrates or ECM. Consistent with this hypothesis, we find rosette colonies to be more resistant to shear than are chain colonies, which lack filopodia. Alternatively, S. rosetta filopodia may be involved in intercellular signaling through secretion of diffusible molecules into the ECM of rosette colonies, akin to the function of cytonemes, cytoplasmic extensions hypothesized to propagate specific intercellular signals through segregation of signaling components (Affolter and Basler, 2011; Ramirez-Weber and Kornberg, 1999; Roy et al., 2011).
Evolutionary implications of choanoflagellate colony development
The evolution of animals from their single-celled ancestors represents one of the major events in life's history. It has been hypothesized that the origin of animals required at least two innovations: the evolution of multicellularity and the division of labor amongst cells within the multicellular individual (Buss, 1987; King, 2004; Michod, 2007; Pfeiffer et al., 2001). Thus, colony development in the animal stem lineage is thought to represent a transition state in the evolution of multicellularity (Haeckel, 1874; Mikhailov et al., 2009; Nielsen, 2008) and the ability of choanoflagellates to form simple colonies holds special relevance for discussions of animal origins (King, 2004; Leadbeater and Kelly, 2001). Recent phylogenetic studies of choanoflagellates coupled with efforts to reconstruct ancestral character states suggest that colony formation may have evolved before the diversification of two of the three major choanoflagellate clades, if not earlier (Carr et al., 2008). An alternative and less parsimonious possibility is that colony formation evolved multiple times independently within choanoflagellates. The phylogenetic distribution of intercellular bridges in choanoflagellates should shed important light on this issue. Intercellular bridges containing densely-staining plates similar to those we have observed here in S. rosetta have been previously observed connecting cells in colonies from choanoflagellates representing two of the three major choanoflagellate clades (Carr et al., 2008; Hibberd, 1975; Karpov and Coupe, 1998). If the molecular mechanisms for intercellular bridge formation are shared among diverse colonial choanoflagellates, it should be possible to pinpoint the evolutionary origin of these bridges and of colony formation generally-- either within the choanoflagellates or before the divergence of choanoflagellates and animals.
The close evolutionary relationship between choanoflagellates and animals raises the possibility that the mechanisms controlling cell differentiation and multicellular development in S. rosetta are evolutionarily related to those regulating development in the animal lineage. The ability of most choanoflagellates to adhere to surfaces and to their secreted ECM (in the form of the theca) suggests the presence of adhesion mechanisms that could have been co-opted to support intercellular adhesion in animals. The genome of a closely related choanoflagellate, Monosiga brevicollis, encodes homologs of genes required for cell-cell and cell-substrate adhesion, raising the question of how such genes functioned in the last common ancestor of animals (Abedin and King, 2008; King et al., 2008; Sebe-Pedros et al., 2010).
Future questions and challenges
The life history of S. rosetta, in which cell differentiation and morphogenesis can be easily regulated in the laboratory, provides a unique experimental system with which to investigate animal origins and the ecological implications of colony formation. First, by comparing the cell biology, physiology, and genome content of S. rosetta and other choanoflagellates with animals, we can reconstruct the probable features of their last common ancestor and potentially gain insight into the ancestral functions of genes required for the regulation of animal development. Second, by determining the molecular and cellular mechanisms underlying colony development in S. rosetta and other choanoflagellate species, we can investigate whether the last common ancestor of modern choanoflagellates and animals might have been capable of forming simple colonies. Finally, the ability to regulate the switch between single cells and colonies in laboratory cultures of S. rosetta offers a unique experimental system in which to investigate the potential selective advantages and disadvantages of coloniality under controlled conditions.
Our observation that S. rosetta can produce both solitary cells and colonies implies either that there is no significant advantage or disadvantage to being colonial, or that colony formation represents a trade-off, with multicellularity being advantageous in some environmental conditions and a handicap in others. Therefore testing the relative fitness of colonies and single cells from S. rosetta under defined laboratory conditions will help reveal the environmental contexts under which colony formation might have provided a selective advantage. Although the fossil record remains silent about the origin of animal multicellularity, meaningful insights about how molecular and cellular innovations intersected with a changing environment may emerge from the study of S. rosetta and other choanoflagellates.
Proposal of Salpingoeca rosetta sp. nov. as a replacement for the generic name Proterospongia sp. ATCC 50818
ATCC50818 was previously described as a member of the genus Proterospongia (Carr et al., 2008; King et al., 2003). However, “Proterospongia” is now used to refer to the colonial stage in the life histories of certain choanoflagellates, rather than being considered the name of a valid genus (Carr et al., 2008), and the current convention in choanoflagellate nomenclature is to base species names on the characteristics of their unicellular life history stages (B. Leadbeater, personal communication). The molecular phylogeny of ATCC50818 was investigated based on a combined 4-gene alignment ((Carr et al., 2008); the 18S rRNA sequence is available at Genbank under accession number EU011924). Based on this information, plus character trait analysis, we propose that ATCC50818 should be placed in the genus Salpingoeca.
Description of Salpingoeca rosetta sp. nov
Salpingoeca rosetta (ros.et.ta N.L. fem. adj. rosetta meaning rose, after the orientation of cells in the colonial life history stage)
Cells are 2.0–4.5 μm in diameter (usually ∼2.5 μm) and slightly ovoid, with a length:width ratio of ∼5:4. One type of solitary cell that is sedentary, two types of solitary cells that are free-swimming, and two type of colonies--rosette and chain colonies-may be seen within a single culture. The flagellum is 200–250 nm in diameter and ∼15 μm long and surrounded by collar of 25–36 microvilli 75–100 nm in diameter and ∼5 μm long in all cell types but the dispersal type (fast swimmer), which has a short collar <0.5 μm long. Each of the sedentary solitary cells (thecate cells) attaches to an environmental substrate via a secreted goblet-shaped theca that is connected to the cell body, but often with a gap of ∼1 μm between the rim of the cup and the base of the cell body. The theca stalk can vary in length from 0 to 4 μm, but is usually ∼3 μm long. The remaining two solitary cell types are two types of free-swimmers: slow swimmers and fast swimmers. The slow swimmers resemble thecate cells morphologically, except for the absence of a theca. The fast swimmers (the dispersal cell type) resemble the slow swimmers, except for having a shorter (∼0.5 μm) collar. Colonial cells are sparsely coated with filamentous extracellular material that surrounds their cell bodies. Adjacent cells connect via intercellular bridges, surrounded by shared plasma membrane and positioned slightly basal to cell equators. Intercellular bridges are cylindrical structures, 200 nm wide and 150 nm long, partitioned by two parallel densely osmophilic plates 80 nm apart. Chain colonies consist of linear or branched arrangements of cells. Rosette colonies consist of spheroidal arrangements of cells around a central focus. In rosette colonies, but not chains, filopodia often extend from the base of the cells into the ECM at the colony core. Fast swimmers give rise to thecate cells by substrate attachment. Slow swimmers give rise to colonial forms by remaining attached following cell division.
Supplementary Material
Movie 1: Fast-swimmer attaching to substrate and building new theca. Attachment is normally directly to substrate, but attachment to an empty theca is presented as elevation from substrate affords a better view of the attachment process. Fast-swimmer attaches to empty theca (abandoned by another cell) with long filopodia. Those filopodia in contact with empty theca become more refractile and coalesce to form base of new stalk at the base of the cell. Coalesced filopodia form highly refractile stalk which elongates from cell base. As soon as refractile material reaches base, cell becomes more spherical and secretes the theca cup from cell sides, leaving gap (∼1μm) between theca and cell base.
Movie 2: Top view of two fast swimmers attaching to substrate. Cells attach via long filopodia, and move several microns across substrates before building thecae.
Movie 3: Timecourse of three cells releasing from their thecae. As cells begin to leave thecae, multiple filopodia extend from sides of cell maintaining contact with edge of theca cup (clearest in middle cell at 1:02:10--1:30:00, and left cell at 1:01:30). Change in angle of filopodia as it releases from theca in left cell (from 01:01:20 to 01:01:30) shows that these are filopodia and not retraction fibers. As cells release, collar retracts (clearest in right cell at 0:12:30). Times shown in Hours:Minutes:Seconds
Movie 4: Thecate cell division showing that one daughter cell leaves while the other remains in the theca.
Movie 5: Tilt series through an intercellular bridge shows that the cell membrane is continuous across the bridge.
Movie 6: Rosette colony ejects minute cells that adhere to the coverslip.
Movie 7: S. rosetta rosette colonies reproduce by fission.
Highlights.
Salpingoeca rosetta can differentiate into at least 3 solitary and 2 colonial cell types in response to diverse environmental conditions.
Wheat germ agglutinin staining in solitary cells signals competence to develop into multicellular colonies.
S. rosetta colonies are characterized by the presence of intercellular bridges, filopodia, and extracellular matrix.
Acknowledgments
This work would not have been possible without the efforts of Tom Nerad, who established the original S. rosetta culture in 2000 while at the ATCC, and Richard Zuzow, who first determined how to regulate S. rosetta colony development in the lab. We are grateful to Zac Cande, Inaki Ruiz-Trillo, Emina Begovic, Jody Westbrook, Pawel Burkardt, Daniel Richter, and two anonymous reviewers for thoughtful comments on the manuscript and Hector Nolla for technical assistance with FACS analysis. This work was supported by funding from the NIGMS/NIH (grant number GM089977), the Gordon and Betty Moore Foundation Marine Microbiology Initiative, a post-doctoral fellowship to M.J.D. from the Miller Institute for Basic Research in Science, an NIH National Research Service Award and Fellowship grant to R.A.A. (5F32GM086054), NIH Training grant T32 HG00047 support for S.R.F. and an NSF Graduate Research Fellowship award to T.C.L. N.K. is a Scholar in the Integrated Microbial Biodiversity Program of the Canadian Institute for Advanced Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abedin M, King N. The premetazoan ancestry of cadherins. Science. 2008;319:946–8. doi: 10.1126/science.1151084. [DOI] [PubMed] [Google Scholar]
- Affolter M, Basler K. Cell biology. Cytonemes show their colors. Science. 2011;332:312–3. doi: 10.1126/science.1205971. [DOI] [PubMed] [Google Scholar]
- Allen RD, Ueno MS, Fok AK. A survey of lectin binding in Paramecium. J Protozool. 1988;35:400–7. doi: 10.1111/j.1550-7408.1988.tb04117.x. [DOI] [PubMed] [Google Scholar]
- Buss LW. The evolution of individuality. Princeton University Press; Princeton, N.J.: 1987. [Google Scholar]
- Byers B, Abramson DH. Cytokinesis in HeLa: post-telophase delay and microtubule-associated motility. Protoplasma. 1968;66:413–35. doi: 10.1007/BF01255868. [DOI] [PubMed] [Google Scholar]
- Carlson JG, Handel MA. Intercellular bridges and factors determining their patterns in the grasshopper testis. J Morphol. 1988;196:173–85. doi: 10.1002/jmor.1051960206. [DOI] [PubMed] [Google Scholar]
- Carlucci AF, Pramer D. Factors influencing the plate method for determining abundance of bacteria in sea water. Proc Soc Exp Biol Med. 1957;96:392–4. doi: 10.3181/00379727-96-23487. [DOI] [PubMed] [Google Scholar]
- Carr M, Leadbeater BS, Hassan R, Nelson M, Baldauf SL. Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proc Natl Acad Sci U S A. 2008;105:16641–6. doi: 10.1073/pnas.0801667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to diversity: molecular genetics and the evolution of animal design. Blackwell Publishing; Malden, MA: 2005. [Google Scholar]
- De Robertis EM. Evo-devo: variations on ancestral themes. Cell. 2008;132:185–95. doi: 10.1016/j.cell.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairclough SR, Dayel MJ, King N. Multicellular development in a choanoflagellate. Curr Biol. 2010;20:R875–6. doi: 10.1016/j.cub.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk P, Roth KA, Gordon JI. Lectins are sensitive tools for defining the differentiation programs of mouse gut epithelial cell lineages. Am J Physiol. 1994;266:G987–1003. doi: 10.1152/ajpgi.1994.266.6.G987. [DOI] [PubMed] [Google Scholar]
- Fiil A. Follicle cell bridges in the mosquito ovary: syncytia formation and bridge morphology. J Cell Sci. 1978;31:137–43. doi: 10.1242/jcs.31.1.137. [DOI] [PubMed] [Google Scholar]
- Gerhart J. 1998 Warkany lecture: signaling pathways in development. Teratology. 1999;60:226–39. doi: 10.1002/(SICI)1096-9926(199910)60:4<226::AID-TERA7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Glotzer M. Animal cell cytokinesis. Annu Rev Cell Dev Biol. 2001;17:351–86. doi: 10.1146/annurev.cellbio.17.1.351. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–9. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- Greenbaum MP, Ma L, Matzuk MM. Conversion of midbodies into germ cell intercellular bridges. Dev Biol. 2007;305:389–96. doi: 10.1016/j.ydbio.2007.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeckel E. The gastraea-theory, the phylogenetic classification of the animal kingdom and the homology of the germ-lamelle. Quarterly Journal of Microscopical Science. 1874;14:142–165. [Google Scholar]
- Haglund K, Nezis IP, Stenmark H. Structure and functions of stable intercellular bridges formed by incomplete cytokinesis during development. Commun Integr Biol. 2011;4:1–9. doi: 10.4161/cib.4.1.13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd DJ. Observations on the ultrastructure of the choanoflagellate Codosiga botrytis (Ehr.) Saville-Kent with special reference to the flagellar apparatus. Journal of Cell Science. 1975;17:191–219. doi: 10.1242/jcs.17.1.191. [DOI] [PubMed] [Google Scholar]
- Hohenberg H, Mannweiler K, Muller M. High-pressure freezing of cell suspensions in cellulose capillary tubes. J Microsc. 1994;175:34–43. doi: 10.1111/j.1365-2818.1994.tb04785.x. [DOI] [PubMed] [Google Scholar]
- Hoops HJ, Nishii I, Kirk DL. Cytoplasmic Bridges in Volvox and Its Relatives. In: Frantisek Baluska DV, Barlow PW, editors. Cell-Cell Channels. Springer; 2006. p. 321. [Google Scholar]
- Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–9. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James-Clark H. Conclusive proofs on the animality of the ciliate sponges, and their affinities with the Infusoria Flagellata. Memoirs of the Boston Society of Natural History. 1867;1:305–340. [Google Scholar]
- James-Clark H. On the spongiae ciliatae as infusoria flagellata; or observations on the structure, animality, and relationship of Leucosolenia botryoides. Annals and Magazine of Natural History. 1868;1:133–142. 188–215, 250–264. [Google Scholar]
- Karpov SA, Coupe SJ. A revision of choanoflagellate genera Kentrosiga, Schiller, 1953 and Desmarella, Kent, 1880. Acta Protozoologica. 1998;37:23–27. [Google Scholar]
- King N. The unicellular ancestry of animal development. Dev Cell. 2004;7:313–25. doi: 10.1016/j.devcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–3. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–8. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Young SL, Abedin M, Carr M, Leadbeater BS. Starting and maintaining Monosiga brevicollis cultures. Cold Spring Harb Protoc. 2009 doi: 10.1101/pdb.prot5148. 2009, pdb prot5148. [DOI] [PubMed] [Google Scholar]
- Kojima Y. Ultrastructure of goat testes: intercellular bridge between germ cells. J Vet Med Sci. 1992;54:213–9. doi: 10.1292/jvms.54.213. [DOI] [PubMed] [Google Scholar]
- Lapage G. Notes on the Choanoflagellate, Codosiga botrytis, Ehrbg. Quarterly Journal of Microscopical Science. 1925;69:471–508. [Google Scholar]
- Leadbeater BS. Distribution and chemistry of microfilaments in choanoflagellates, with special reference to the collar and other tentacle systems. Protistologica. 1983a;19:157–166. [Google Scholar]
- Leadbeater BSC. Life-history and ultrastructure of a new marine species of Proterospongia (Choanoflagellida) Journal of the Marine Biological Association UK. 1983b;63:135–160. [Google Scholar]
- Leadbeater BS, Kelly M. Evolution of animals - choanoflagellates and sponges. Water and Atmosphere. 2001;9:9–11. [Google Scholar]
- Leadbeater BSC. Observations on Life-History and Ultrastructure of Marine Choanoflagellate Choanoeca-Perplexa-Ellis. Journal of the Marine Biological Association of the United Kingdom. 1977;57:285–&. [Google Scholar]
- Leadbeater BSC. Developmental studies on the loricate choanoflagellate Stephanoeca diplocostata Ellis. IV. Effects of silica deprivation on growth and lorica production. Protoplasma. 1985;127:171–179. [Google Scholar]
- Leys S. The significance of syncytial tissues for the position of the hexactinellida in the metazoa. Integrative and Comparative Biology. 2003;43:19–27. doi: 10.1093/icb/43.1.19. [DOI] [PubMed] [Google Scholar]
- Lueken WW, Breer H, Hartkemeyer M. Local and Temporal Pattern of Con A-Binding Site Aggregation During Conjugation in Euplotes vannus (Ciliophora, Hypotrichida) Journal of Protozoology. 1981;28:414–417. [Google Scholar]
- Maldonado M. Choanoflagellates, choanocytes, and animal multicellularity. Invertebrate Biology. 2004;123:1–22. [Google Scholar]
- Michod RE. Evolution of individuality during the transition from unicellular to multicellular life. Proc Natl Acad Sci U S A. 2007;104 1:8613–8. doi: 10.1073/pnas.0701489104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov KV, Konstantinova AV, Nikitin MA, Troshin PV, Rusin LY, Lyubetsky VA, Panchin YV, Mylnikov AP, Moroz LL, Kumar S, Aleoshin VV. The origin of Metazoa: a transition from temporal to spatial cell differentiation. Bioessays. 2009;31:758–68. doi: 10.1002/bies.200800214. [DOI] [PubMed] [Google Scholar]
- Mullins JM, McIntosh JR. Isolation and initial characterization of the mammalian midbody. J Cell Biol. 1982;94:654–61. doi: 10.1083/jcb.94.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C. Six major steps in animal evolution: are we derived sponge larvae? Evolution & Development. 2008;10:241–257. doi: 10.1111/j.1525-142X.2008.00231.x. [DOI] [PubMed] [Google Scholar]
- Ong S, Tan C. Germline cyst formation and incomplete cytokinesis during Drosophila melanogaster oogenesis. Dev Biol. 2010;337:84–98. doi: 10.1016/j.ydbio.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Otegui MS, Verbrugghe KJ, Skop AR. Midbodies and phragmoplasts: analogous structures involved in cytokinesis. Trends Cell Biol. 2005;15:404–13. doi: 10.1016/j.tcb.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periaswamy S, Farid H. Elastic registration with partial data. Biomedical Image Registration. 2003;2717:102–111. [Google Scholar]
- Pettitt ME, Orme BAA, Blake JR, Leadbeater BSC. The hydrodynamics of filter feeding in choanoflagellates. European Journal of Protistology. 2002;38:313–332. [Google Scholar]
- Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292:504–507. doi: 10.1126/science.1058079. [DOI] [PubMed] [Google Scholar]
- Pires da Silva A, Sommer RJ. The evolution of signalling pathways in animal development. Nat Rev Genet. 2003;4:39–49. doi: 10.1038/nrg977. [DOI] [PubMed] [Google Scholar]
- Ramirez-Weber FA, Kornberg TB. Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell. 1999;97:599–607. doi: 10.1016/s0092-8674(00)80771-0. [DOI] [PubMed] [Google Scholar]
- Ramoino P. Lectin-binding glycoconjugates in Paramecium primaurelia: changes with cellular age and starvation. Histochem Cell Biol. 1997;107:321–9. doi: 10.1007/s004180050117. [DOI] [PubMed] [Google Scholar]
- Ramoino P, Fronte P, Fato M, Beltrame F, Robello M, Diaspro A. Fluid phase and receptor-mediated endocytosis in Paramecium primaurelia by fluorescence confocal laser scanning microscopy. Eur Biophys J. 2001;30:305–12. doi: 10.1007/s002490100166. [DOI] [PubMed] [Google Scholar]
- Roy S, Hsiung F, Kornberg TB. Specificity of Drosophila cytonemes for distinct signaling pathways. Science. 2011;332:354–8. doi: 10.1126/science.1198949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Burger G, Holland PW, King N, Lang BF, Roger AJ, Gray MW. The origins of multicellularity: a multi-taxon genome initiative. Trends Genet. 2007;23:113–8. doi: 10.1016/j.tig.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. A phylogenomic investigation into the origin of metazoa. Mol Biol Evol. 2008;25:664–72. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- Schindelmeiser J, Greven H, Bergmann M. The immature part of the testis in Salamandra salamandra (L.) (Amphibia, Urodela) Arch Histol Jpn. 1983;46:159–72. doi: 10.1679/aohc.46.159. [DOI] [PubMed] [Google Scholar]
- Sebe-Pedros A, Roger AJ, Lang FB, King N, Ruiz-Trillo I. Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc Natl Acad Sci U S A. 2010;107:10142–7. doi: 10.1073/pnas.1002257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenkamp ET, Wright J, Baldauf SL. The protistan origins of animals and fungi. Mol Biol Evol. 2006;23:93–106. doi: 10.1093/molbev/msj011. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE, Hazelrigg TI. In vivo analyses of cytoplasmic transport and cytoskeletal organization during Drosophila oogenesis: characterization of a multistep anterior localization pathway. Development. 1998;125:3655–66. doi: 10.1242/dev.125.18.3655. [DOI] [PubMed] [Google Scholar]
- Valentine JW. On the origin of phyla. University of Chicago Press; Chicago ; London: 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1: Fast-swimmer attaching to substrate and building new theca. Attachment is normally directly to substrate, but attachment to an empty theca is presented as elevation from substrate affords a better view of the attachment process. Fast-swimmer attaches to empty theca (abandoned by another cell) with long filopodia. Those filopodia in contact with empty theca become more refractile and coalesce to form base of new stalk at the base of the cell. Coalesced filopodia form highly refractile stalk which elongates from cell base. As soon as refractile material reaches base, cell becomes more spherical and secretes the theca cup from cell sides, leaving gap (∼1μm) between theca and cell base.
Movie 2: Top view of two fast swimmers attaching to substrate. Cells attach via long filopodia, and move several microns across substrates before building thecae.
Movie 3: Timecourse of three cells releasing from their thecae. As cells begin to leave thecae, multiple filopodia extend from sides of cell maintaining contact with edge of theca cup (clearest in middle cell at 1:02:10--1:30:00, and left cell at 1:01:30). Change in angle of filopodia as it releases from theca in left cell (from 01:01:20 to 01:01:30) shows that these are filopodia and not retraction fibers. As cells release, collar retracts (clearest in right cell at 0:12:30). Times shown in Hours:Minutes:Seconds
Movie 4: Thecate cell division showing that one daughter cell leaves while the other remains in the theca.
Movie 5: Tilt series through an intercellular bridge shows that the cell membrane is continuous across the bridge.
Movie 6: Rosette colony ejects minute cells that adhere to the coverslip.
Movie 7: S. rosetta rosette colonies reproduce by fission.