Abstract
Importance of the field
Miniaturization is key to advancing the state-of-the-art in high content screening (HCS), in order to enable dramatic cost savings through reduced usage of expensive biochemical reagents and to enable large-scale screening on primary cells. Microfluidic technology offers the potential to enable HCS to be performed with an unprecedented degree of miniaturization.
Areas covered in this review
This perspective highlights a real-world example from the authors’ work of HCS assays implemented in a highly miniaturized microfluidic format. Advantages of this technology are discussed, including cost savings, high throughput screening on primary cells, improved accuracy, the ability to study complex time-varying stimuli, and ease of automation, integration, and scaling.
What the reader will gain
The reader will understand the capabilities of a new microfluidics-based platform for HCS, and the advantages it provides over conventional plate-based HCS.
Take home message
Microfluidics technology will drive significant advancements and broader usage and applicability of HCS in drug discovery.
1. Introduction
High content cell screening (HCS) is a biological research tool that uses living cells to assay the effects of drugs, RNAi, or other biological agents and perturbations. HCS is increasingly being used by the pharmaceutical industry for drug discovery 1–5, due to its ability to detect potential alterations of a variety of cellular phenotypes, yielding a much richer understanding of the effects of each compound screened. These phenotypes include but are not limited to subcellular localization and expression of key signaling proteins, cytoskeletal structures, cell shape and size, etc.
However, the development of targeted drug therapies using HCS assays performed in the traditional multi-well format is greatly hindered by highly inefficient usage of consumable materials, including costly biochemical reagents and valuable cells obtained from biopsy or surrogate tissue samples. Improved assays are needed both for preclinical evaluation of candidate therapeutic agents (the results of which correlate with results obtained in animal models or human clinical trials) and for prediction of drug responses using clinical samples (to stratify patients for clinical trial selection).
In the case of high throughput drug screening, e.g. to identify lead compounds, tremendous cost savings may be obtained if the consumption of valuable and expensive drug libraries and antibodies can be markedly decreased. Improved efficiency that results in savings even as low as cents to dollars per datapoint is significant enough to accumulate to large overall savings (potentially tens or hundreds of thousands of dollars), thereby enabling the usage of HCS in large scale drug screens involving as many as a million compounds.
In the case of predicting drug responses, tissue samples used for prediction may be exceedingly small or valuable, e.g. in the case of fine needle biopsy, thereby requiring subculturing just to obtain sufficient cell numbers to test molecular-level responses of a few candidate drugs, doses, or exposure durations. In turn, the subculturing process may result in altered biomolecular responses and yield misleading predictions of overall tumor or organismal responses. Accordingly, assays with markedly reduced tissue sample usage are needed to enable direct measurement of pharmacodynamic responses to many drugs using primary cell samples.
The rapidly advancing field of microfluidics offers the opportunity to address the challenge of materials usage efficiency. Microfluidics enables the manipulation of fluids and particles that are geometrically constrained at the micron scale 6, 7, and offers the promise of a “lab on a chip”, e.g. miniaturized biochemical assays implemented in microfluidic devices 8–10. Over the past decade, microfluidic technology has advanced rapidly from the development of basic components to the emergence of large scale, fully integrated devices. Such devices, especially those used for PCR-based nucleic acid detection and enzymatic reaction-based in vitro assays, have become increasingly common in the biopharma research environment 11–13.
Recently, the concept of “lab on a chip” was extended to the patterning, culture, and stimulation of cells coupled with high-throughput cellular measurements, all performed in a single device14. In principle, microfluidic experimental platforms not only offer the critical advantages of miniaturization and high throughput as compared to traditional cell biological assays, but also provide very well-defined control over the cellular microenvironment due to precise fluid handling15. One microfluidics-based approach that has been proposed involves creation of picoliter-scale droplets within a device, which can be used to perform HCS by encapsulating cells and candidate drugs in the droplet or perform high throughput drug screening via enzymatic assays implemented in such droplets 16–18. While this approach provides the advantages of miniaturization, there are several unresolved issues associated with this approach for HCS applications, e.g. keeping adherent cells viable within droplets, exchange of multiple fluids needed for immunocytochemical staining, imaging of cells with subcellular resolution, etc.
In comparison, we have recently developed novel microfluidic-based technology that fully implements high content screening, including cell culture and stimulation, staining, imaging and image analysis (summarized in Section 2). We anticipate that microfluidic devices such as ours will be very useful for highly multiplexed interrogation of high-priority molecular pharmacodynamic targets, especially in assays utilizing valuable primary cells. In particular, the microfluidic devices represent a general platform for probing multiple targets within and across key signaling pathways in response to multiple drugs or drug combinations. As such, we believe that microfluidic devices will be highly useful in drug discovery and response prediction applications.
2. Case example of microfluidics-based high content screening
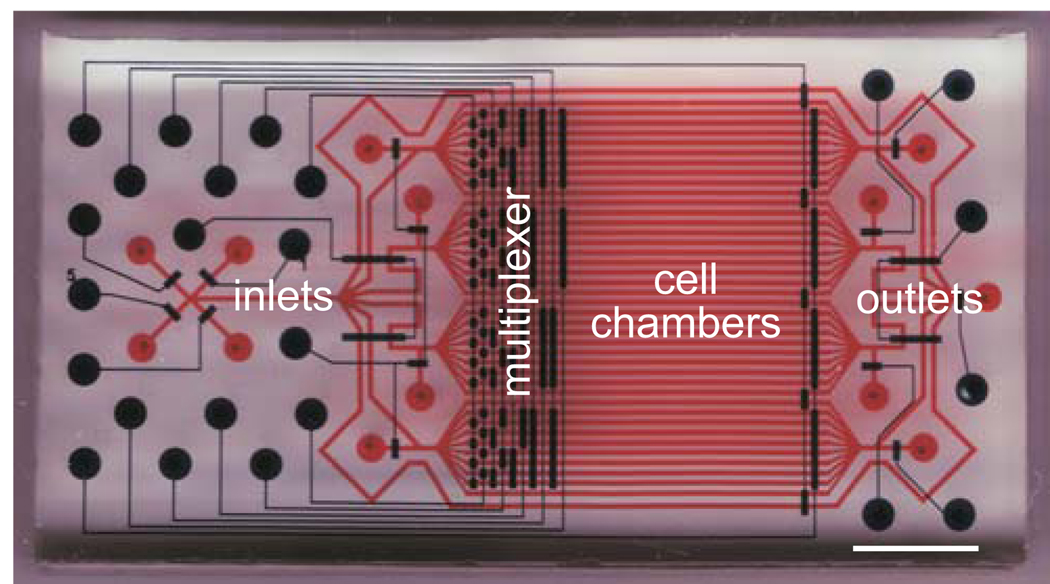
We have designed a microfluidic device for high-throughput, time-resolved high content screening 19, 20 (Fig. 1). The microfluidic device is molded of polymer materials, such as polydimethylsiloxane (PDMS), an inexpensive, bio-compatible, and optically transparent silicon elastomer. In our device design, 32 separate compartments are linked to multiple inlets and outlets, which enable the compartments to be filled with a solution of cells, cell medium containing a drug of interest, fixative and staining reagents, etc. Fluid flow to the compartments is controlled by actuating a manifold of integrated membrane valves21 (Fig. 1). A more detailed description of the design of the fluidic network can be found in our previous work 19, 20.
Figure 1. Design of a microfluidic device for high content screening.
Cell chambers and fluidic conduits are shown in red and integrated membrane valves are shown in blue. Device inlets, outlets, cell chamber, and multiplexed valves are indicated. Scale bar, 5 mm. This figure was originally published in Molecular Cellular Proteomics 19, and is used with permission.
In a typical experiment, about 300 cells are loaded into each compartment, for a total of nearly 10,000 individual cell experiments. The device is coupled to a completely automated system which actuates the membrane valves to precisely expose each compartment to a different combination or concentration of exogenously added factors in the cell medium, for different periods of time. Then, cells in the device are fixed and immunochemically stained, and the cells are imaged, for example, by a motorized microscope or fluorescence scanner (Fig. 2).
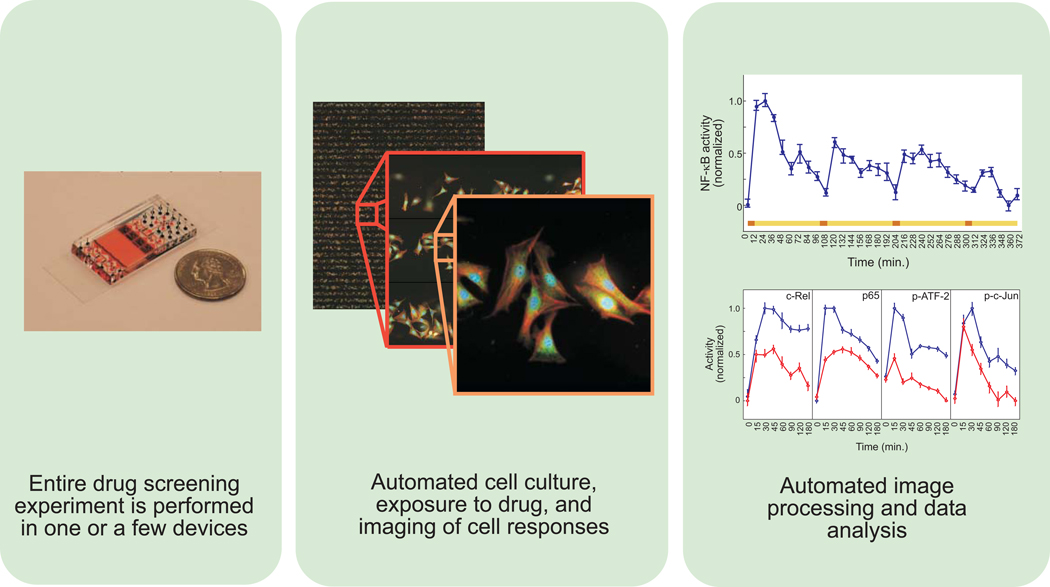
Figure 2. Features of a microfluidic-based high content screening system.
Left panel, depiction of device next to a U.S. quarter of a dollar coin. Highly miniaturized microfluidic devices may enable large scale drug screening experiments to be performed in a single device. Middle panel, high content images of cells stimulated and stained within a microfluidic device. Cell culture, exposure to stimuli, and staining and imaging of cell responses can all be performed in microfluidic devices. Right panel, images of individual cells are processed and analyzed to determine drug effects. Top graph shows NF-κB responses to periodic stimulus. The colored bar depicts the input time course consisting of sequential pulses of tumor necrosis factor (TNF) in red with intervening washouts in yellow. The bottom graph shows TNF responses of the indicated transcription factors in the presence (red) or absence (blue) of the drug SC-514. Inhibitory effects are seen on the NF-κB pathway (c-Rel and p65) as well as JNK signaling (ATF-2 and c-Jun). Images and graphs were originally published in Molecular Cellular Proteomics 19, and are used with permission.
As in traditional HCS assays, the primary device readout is the localization and concentration of signaling proteins as revealed by specific antibodies, or mRNA, as visualized using specific in situ hybridization probes (Fig. 2). Important ancillary readouts obtained for each cell include cell shape, cell density, distribution, as well as other counterstained structures such as DNA and the cytoskeleton. Differential responses of cells exposed to different conditions are obtained by examining the distributions of these readouts across hundreds of cells exposed to the same condition, which allows statistically significant comparisons.
The microfluidic device is a highly versatile tool for high content screening. In particular, the single device design enables measurements of responses at the single cell level to receptor ligands and/or small molecule inhibitors applied in a variety of patterns. One such experiment is measuring signaling events in a detailed nonlinearly-spaced time-course, for example, to capture both rapid early signaling and slower late signaling. We performed this type of experiment to characterize signaling dynamics in the TNF-NF-κB pathway, observing biphasic NF-κB activity consistent with experiments performed previously using population-level measurements19. We also used our rich dataset, with its single cell resolution, to demonstrate that asynchronous oscillations observed in cells overexpressing fluorescent protein-labeled NF-κB are inconsistent with observations of genetically unmodified wildtype cells, particularly in the mean and variance of the distribution of the individual cell responses 22.
Another experiment that can be easily implemented in the device is measuring multiple signaling responses to multiple stimuli. This can be done by actuating specific valves to partition the chambers into fluidically-isolated groups, and performing different experiments in each of the groups in parallel. In one such experiment, we measured the response of A549 lung cancer cells to TNF in the presence or absence of SC-514, an inhibitor of a key kinase, IKK, in the TNF-NF-κB pathway19. We found that, despite in vitro enzymatic data indicating selectivity of the drug, SC-514 had off-target effects on Jun kinase (JNK) signaling (Fig. 2, right panel). More broadly, this experiment demonstrates how cellular responses to drugs and small molecule inhibitors may be assayed in microfluidic devices.
Finally, another type of experiment that can be easily implemented in the device is measuring responses to complex temporal inputs. Signal transduction is often investigated through experiments in which cells are exposed to step inputs of stimuli, but this is rarely representative of the in vivo stimulus. Rather, cells experience time-varying dynamic stimuli, such as gradually decreasing stimuli (e.g., drug pharmcokinetics resulting from metabolism) or periodic stimuli (e.g., repetitive drug dosing, or episodic disease). To demonstrate how the device can be used to measure responses to such complex temporal inputs, we mimicked repeated episodes of inflammation, which is a hallmark of immunologic diseases such as asthma, by exposing cells to periodic pulses of TNF. We observed, in a single device, that the resulting amplitude of NF-κB activity decreased for each pulse of TNF, suggesting that the signaling pathway partially adapts to repeated stimuli possibly through receptor downregulation19 (Fig. 2, right panel). As described below, this experiment demonstrates how complex stimuli representing more physiologic stimuli may be easily and accurately reproduced and studied using microfluidic devices.
3. Advantages of HCS performed in a microfluidic format
The various advantages of using microfluidic-based technology, such as the device described in section 2, for performing HCS are discussed in this section. These advantages include cost savings and efficient use of primary cells driven by miniaturization, improved accuracy and generation of complex stimuli driven by enhanced fluid control, and ease of integration driven by compatibility with automated systems.
3.1 Miniaturization
It is estimated that existing HCS systems have only been adopted by 12–15% of the available market and that one of the key factors hindering broad market adoption is the cost per sample 23. Our prototype microfluidic device uses ~150-fold lower amount of costly drug libraries and chemical media than the best available conventional HCS technology, which may enable savings of up to ~$1–2 per datapoint on these reagents alone (Fig. 3, top panel). A similar reduction in the number of cells needed for microfluidic experimentation can lead to additional reductions in the time and expense needed to scale-up for an HCS-based screen. The aggregate savings driven by microfluidic-based miniaturization may thereby enable the use of HCS in high throughput drug screening of medium to large scale libraries consisting of 50K-2M compounds, which is not economical with other technologies.
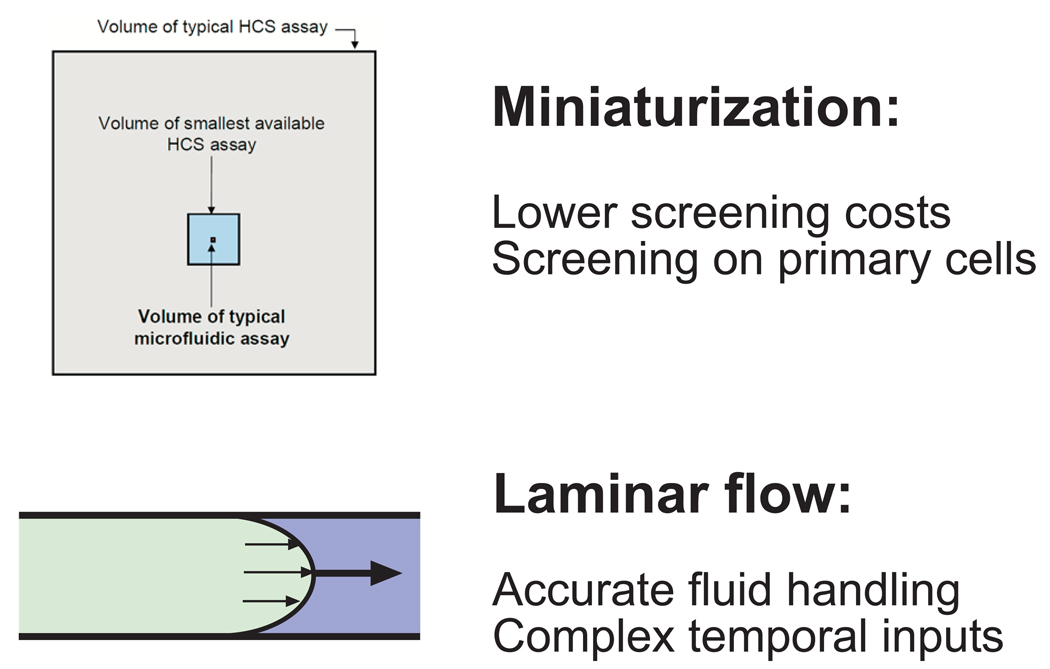
Figure 3. Summary of main advantages of microfluidics for HCS.
Top panel, areas of the shaded squares indicate, to scale, the relative volumes of typical HCS assays, miniaturized HCS assays, and microfluidic assays, for a single sample. The extreme miniaturization of microfluidics drives lower screening costs and enables high throughput screening on primary cells. Bottom panel, depicts complete fluid exchange in microscopic channels due to laminar flow. This microfluidic property enables highly accurate fluid handling and enables experimentation with highly complex time-varying cellular inputs.
A second key problem that miniaturization solves is efficient experimentation on small numbers of valuable primary cells. In comparison to cell line-based screening, experiments using primary cells hold the promise of identifying more clinically relevant hits, which is frequently cited as a highly desired and critical criterion for the adoption of cell-based screening assays 23. The extremely low consumption of cells by microfluidic devices allows large drug screens to be performed on small primary cell samples with no or minimal sub-culturing, thus reducing the inter-assay variability resulting from using different primary cell samples.
3.2 Enhanced fluid control
Since it is difficult to accurately exchange extremely small fluid volumes in a serial fashion (e.g., multiple washes of a single well with micro- to nano-liters of fluid), miniaturization of multi-well assays typically comes at the cost of increased assay variability24. The subsequent loss of robustness and reproducibility further limits the adoption of multi-well-based HCS for high throughput drug screening.
In contrast, liquids in microscopic channels flow in a highly predictable laminar fashion. This property of microfluidic flows ensures that replacement of one fluid by another is complete, thereby preventing contamination by residual amounts of the previous fluid and eliminates the need for multiple washes (Fig. 3, bottom panel). Indeed, we have demonstrated that this can lead to at least 5-fold lower intra-assay variability than that of multi-well assays19, which can lead to substantially improved data quality with sharply reduced false positives or negatives.
3.3 Control over stimuli applied to cells
The laminar nature of fluid flow ensures that mixing of chemicals usually occurs by molecular diffusion only. Therefore, the flow parameters and chemical concentration profiles can be controlled with high precision, e.g., kept constant over extended periods of time and quickly modified when needed. This enables accurate reproduction of complex time-varying dynamic stimuli, such as gradually decreasing or periodic stimuli, which may better mimic pharmacokinetic drug profiles in the body. In comparison, such experiments would be cumbersome or impossible to perform accurately in a multi-well format, as it would require dozens of rapid washes and fluid manipulations. Complete removal of each fluid would be difficult to ensure, leading to increased experimental error. In a microfluidic device, however, laminar flow ensures rapid, complete, and reproducible fluid exchange. This technological advantage of microfluidic devices leads to even greater improvements in accuracy as compared to the multi-well format for such experiments.
3.4 Compatibility with automation, integration, and scaling
Microfluidic device designs, such as the one highlighted in section 2, are highly parallelized and easily scalable, which enable further quantum leaps in the throughput and materials usage efficiency of cell-based screens. These features could allow, for example, screening of an entire drug library on a single sample of primary cells. Additionally, experiments are run in the device in a completely automated fashion, thus reducing human error, labor, and time. In principle, microfluidic devices are compatible with existing screening equipment, such as fluorescence microscopes and image analysis software, which will further facilitate adoption of the technology and its integration into existing drug screening infrastructure.
4. Expert opinion
High content screening is a promising technology that is being increasingly adopted by the pharmaceutical industry as well as academic researchers for drug discovery and screening. This is particularly because of the physiological relevance of these assays, as well as the multi-dimensional data readouts, including phenotypic readouts like cell morphology and cytotoxicity, and fluorescence-based readouts like protein localization and expression. Currently, HCS has been focused on hit prioritization and lead development in smaller, focused libraries and in secondary screens. However, HCS has still not attained its potential for broader usage, particularly in primary drug screens, which could be achieved through decreased assay costs, increased assay reliability and flexibility, as well as more physiologically relevant assay results.
The authors, who have demonstrated proof-of-principle of microfluidic-based HCS, suggest that microfluidics enables much broader adoption and wider usage of HCS. In particular, the miniaturization inherent in microfluidics can lead to orders of magnitude cost savings due to reduced consumption of costly drug libraries and numerous other biochemical reagents. Perhaps most importantly, the miniaturization of microfluidics allows the efficient use of primary cells, which are increasingly being favored over cell lines, in order to obtain more physiologically relevant results. Microfluidics allows the exposure of these cells to varied stimuli without the need for prior sub-culturing, which can potentially alter their response to drugs.
Additionally, the laminar nature of fluid flow at micron scales allows complete fluid exchange without extra washing steps that are typically required of multi-well assays to yield reliable results. These steps in turn can be time-consuming and potentially stressful to cells. These difficulties are amplified in high density multi-well formats which thus sacrifice accuracy for miniaturization. In contrast, the enhanced fluid control within microfluidic devices circumvents the need for such trade-offs. As a result, microfluidic devices, such as the one highlighted in the article, enable researchers to study dynamic stimuli, e.g. to mimic time-varying physiological cellular exposures to these stimuli, in order to obtain more relevant cellular responses.
Microfluidic-based HCS systems currently function well in the research environment. To successfully implement the technology in an industrial environment, some additional development will be needed, particularly with regards to integration with existing drug screening infrastructure. We anticipate that these developments will occur rapidly and that microfluidic-based approaches will become increasingly important in HCS applications, by making them cost-effective and hence potentially applicable to larger and earlier screens, as well as providing physiologically relevant data to spur drug discovery.
Acknowledgments
Declaration of Interest
The authors acknowledge support from the National Institutes of Health (CA131920). R.C. acknowledges support from the Medical Scientist Training Program at Johns Hopkins University. The authors are co-founders of a company that plans to commercialize technology described in Section 2, but the authors did not receive any payments to produce this article.
Bibliography
- 1.Lang P, Yeow K, Nichols A, Scheer A. Cellular imaging in drug discovery. Nat Rev Drug Discov. 2006;5:343–356. doi: 10.1038/nrd2008. [DOI] [PubMed] [Google Scholar]
- 2.Haney SA, LaPan P, Pan J, Zhang J. High-content screening moves to the front of the line. Drug Discov Today. 2006;11:889–894. doi: 10.1016/j.drudis.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 3.Denner P, Schmalowsky J, Prechtl S. High-content analysis in preclinical drug discovery. Comb Chem High Throughput Screen. 2008;11:216–230. doi: 10.2174/138620708783877780. [DOI] [PubMed] [Google Scholar]
- 4.Soleilhac E, Nadon R, Lafanechere L. High-content screening for the discovery of pharmacological compounds: advantages, challenges and potential benefits of recent technological developments. 5:135. doi: 10.1517/17460440903544456. [DOI] [PubMed] [Google Scholar]
- 5.Korn K, Krausz E. Cell-based high-content screening of small-molecule libraries. Curr Opin Chem Biol. 2007;11:503–510. doi: 10.1016/j.cbpa.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 6.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 7.Beebe DJ, Mensing GA, Walker GM. Physics and applications of microfluidics in biology. Annu Rev Biomed Eng. 2002;4:261–286. doi: 10.1146/annurev.bioeng.4.112601.125916. [DOI] [PubMed] [Google Scholar]
- 8.Haeberle S, Zengerle R. Microfluidic platforms for lab-on-a-chip applications. Lab Chip. 2007;7:1094–1110. doi: 10.1039/b706364b. [DOI] [PubMed] [Google Scholar]
- 9.Roman GT, Kennedy RT. Fully integrated microfluidic separations systems for biochemical analysis. J Chromatogr A. 2007;1168:170–188. doi: 10.1016/j.chroma.2007.06.010. discussion 169. [DOI] [PubMed] [Google Scholar]
- 10.Bilitewski U, Genrich M, Kadow S, Mersal G. Biochemical analysis with microfluidic systems. Anal Bioanal Chem. 2003;377:556–569. doi: 10.1007/s00216-003-2179-4. [DOI] [PubMed] [Google Scholar]
- 11.Spurgeon SL, Jones RC, Ramakrishnan R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS One. 2008;3:e1662. doi: 10.1371/journal.pone.0001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue Q, Wainright A, Gangakhedkar S, Gibbons I. Multiplexed enzyme assays in capillary electrophoretic single-use microfluidic devices. Electrophoresis. 2001;22:4000–4007. doi: 10.1002/1522-2683(200110)22:18<4000::AID-ELPS4000>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 13.Dunne J, Reardon H, Trinh V, Li E, Farinas J. Comparison of on-chip and off-chip microfluidic kinase assay formats. Assay Drug Dev Technol. 2004;2:121–129. doi: 10.1089/154065804323056468. [DOI] [PubMed] [Google Scholar]
- 14.El-Ali J, Sorger PK, Jensen KF. Cells on chips. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 15.Warrick J, Meyvantsson I, Ju J, Beebe DJ. High-throughput microfluidics: improved sample treatment and washing over standard wells. Lab Chip. 2007;7:316–321. doi: 10.1039/b613350a. [DOI] [PubMed] [Google Scholar]
- 16.Chen DL, Ismagilov RF. Microfluidic cartridges preloaded with nanoliter plugs of reagents: an alternative to 96-well plates for screening. Curr Opin Chem Biol. 2006;10:226–231. doi: 10.1016/j.cbpa.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clausell-Tormos J, Lieber D, Baret JC, El-Harrak A, Miller OJ, et al. Droplet-based microfluidic platforms for the encapsulation and screening of Mammalian cells and multicellular organisms. Chem Biol. 2008;15:427–437. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Huebner A, Bratton D, Whyte G, Yang M, Demello AJ, et al. Static microdroplet arrays: a microfluidic device for droplet trapping, incubation and release for enzymatic and cell-based assays. Lab Chip. 2009;9:692–698. doi: 10.1039/b813709a. [DOI] [PubMed] [Google Scholar]
- 19.Cheong R, Wang CJ, Levchenko A. High content cell screening in a microfluidic device. Mol Cell Proteomics. 2009;8:433–442. doi: 10.1074/mcp.M800291-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheong R, Wang CJ, Levchenko A. Using a microfluidic device for high-content analysis of cell signaling. Sci Signal. 2009;2 doi: 10.1126/scisignal.275pl2. pl2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 22.Barken D, Wang CJ, Kearns J, Cheong R, Hoffmann A, et al. Comment on "Oscillations in NF-kappaB signaling control the dynamics of gene expression". Science. 2005;308:52. doi: 10.1126/science.1107904. author reply 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cell-Based Assays for Drug Discovery. Trimark Publications; 2009. [Google Scholar]
- 24.Dunn DA, Feygin I. Challenges and solutions to ultra-high-throughput screening assay miniaturization: submicroliter fluid handling. Drug Discov Today. 2000;5:84–91. doi: 10.1016/s1359-6446(00)00064-7. [DOI] [PubMed] [Google Scholar]