Abstract
Much attention has focused on DNA condensation because of its fundamental biological importance. The recent discovery of new roles for RNA duplexes demands efficient packaging of dsRNA for therapeutics. Here we report measurements of short DNA and RNA duplexes in the presence of trivalent ions. Under conditions where UV spectroscopy indicates condensation of DNA duplexes into (insoluble) precipitates, RNA duplexes remain soluble. SAXS results suggest that the differing surface topologies of RNA and DNA may be crucial in generating the attractive forces that result in precipitation.
The attraction of like-charged objects is an important theme in polymer physics, biology and biotechnology. It is remarkable that, despite its uniform, large negative charge, double stranded DNA precipitates from dilute solution when even small numbers of multivalent ions are introduced [1]. Much effort has been expended investigating the nature of this multivalent ion-induced attraction because of its relevance to DNA packaging, either in viruses [2] or for applications in non-viral gene delivery [3]. Here, we extend these studies to duplex RNA. Recent attention has focused on dsRNA because of its role in RNA interference (RNAi) [4]. In this process, low quantities of short RNA duplexes set into motion a molecular machine that exerts powerful control over gene expression. The nucleotide sequence encoded by the short duplex is used to target and destroy mRNA containing a complementary sequence. RNAi is an ideal vehicle for novel therapeutic applications by targeting and silencing specific genes. The ability to tightly package (condense) numerous, short RNA duplexes is an important prerequisite for optimal design of these next-generation therapeutics [5].
There is no universally accepted explanation of the physical origin of like-charge attraction. Because mean field theories, such as those based on the Poisson Boltzmann equation, do not predict attractive forces, there is intense theoretical interest in developing more sophisticated models. Numerous mechanisms (see recent reviews [6] and references within) have been proposed to explain ion-induced attraction of dsDNA helices, including counter-ion correlations, models that account for ion-bridging, attraction resulting from hydration forces, or from precisely coordinated patterns of charge distributions which form an electrostatic zipper.
With the sudden interest in short dsRNA, we have undertaken a biophysical study comparing DNA condensation to RNA condensation. Using ionic conditions that cause short DNA helices to aggregate, we have attempted to identify similar phases in RNA. Surprisingly, dsRNA resists condensation. The strikingly different behavior of these identically charged systems provides important clues about the physics of like charge attraction.
Although complementary strands of RNA or DNA easily combine to form stable, double helices, chemical differences between RNA and DNA drive RNA helices into the A-form, while DNA helices assume the B-form. The former is shorter and wider, with a deep major groove that is of the order of the radius of the helix. By comparison, the Bform helix is more cylindrical, and the depth of the major and minor grooves is more uniform. The linear charge density of the A-form helix is -2|e| per 2.8 Å, while that of the B-form helix is -2|e| per 3.4 Å. Previous work shows that monovalent and divalent counterions penetrate into the major grooves of the RNA, more fully compensating the overall negative charge of the molecule [7]. As a result, screening of the large negative charge of the RNA duplex occurs at lower bulk ionic strength than for DNA. If attraction were solely determined by the degree of charge compensation, one might expect RNA to condense more readily than DNA; we observe the opposite.
The small trivalent ion Cobalt hexammine (Co-hex, Co(NH3)63+) is one of the most powerful condensing agents of DNA [8, 9]. This ion has radius of 3 Å and a nearly spherical surface charge distribution. Even small quantities can precipitate dsDNA from dilute solution at room temperature. Co-hex has also been used in RNA folding studies [10].
Here, we examine the role of trivalent Co-hex ions in RNA and DNA charge screening efficiency and condensation. We use two established experimental techniques—UV absorption and Small Angle X-ray Scattering (SAXS)—to probe the condensation of nucleic acid duplexes from dilute solution, driven by the addition of small quantities of Co-hex. Identical experiments were carried out on both DNA and RNA duplexes.
To measure the condensing power of Co-hex, 25bp DNA and RNA were dialyzed against buffer containing 20 mM NaCl and 1 mM NaMOPS at pH 7, respectively. This low monovalent salt background ensures that added Co-hex will be maximally effective in condensing the sample [8]. Calibrated amounts of Co-hex, between 0 ~ 6 mM in solution, were added to each tube, and the mixed solutions were stored at 4°C for 2 hours. Subsequently, each tube was centrifuged at 10,000 rpm (~8000 g) for 10 min e.g. [8]. The supernatant was collected and absorbance at 260 nm was measured for each sample in a UV spectrophotometer (Cary 50 Bio, Varian, Inc., Walnut Creek, CA). The fraction of precipitated nucleic acid can be calculated by direct measurement of the change in UV absorption of the supernatant. Since the A-helical form is shorter than the B-helical form, we compared 25bp RNA with both 25 and 16bp DNA, to control for any length-dependent effects of condensation.
Synchrotron small angle x-ray scattering (SAXS) reports the strength of inter-molecular interactions. The presence of either repulsive or attractive forces between particles results in distinctive modulation of the scattering profiles at the lowest angles [11]. These experiments were carried out at C1 station at Cornell High Energy Synchrotron Source (CHESS). The experimental setup is described in Ref. [12]. Single strand DNA and RNA oligomers were purchased from Integrated DNA Technologies (Coralville, IA) and Dharmacon Inc. (Chicago, IL), respectively. DNA samples for SAXS studies were prepared as described in Refs. [11, 13] using equilibrium dialysis to establish a fixed bulk ion concentration. All SAXS buffers contain 100 mM NaCl in addition to varying amounts of Co-hex. Identical protocols were employed in preparing RNA samples. Because the competitive association of Co-hex to DNA is a strong function of NaCl concentration [8], the increased concentration of monovalent ions used in SAXS relative to UV studies allows measurements of DNA in solution over a broader range of Co-hex concentrations, and enables a more detailed comparison of Co-hex interactions with DNA as opposed to RNA.
Figure 1 shows results of UV absorption measurements of RNA and DNA as a function of (added) Co-hex. These curves clearly indicate that dsRNA precipitation by Co-hex is much less favorable than dsDNA precipitation. For trivalent ion concentrations below 1 mM, almost no RNA precipitates, in contrast to DNA. In the presence of 4 mM Co-hex, 85% of 25bp DNA molecules condense while ~80% of RNA molecules remain in the supernatant. Only for Co-hex concentrations in excess of 10 mM do we measure significant RNA condensation.
Figure 1.
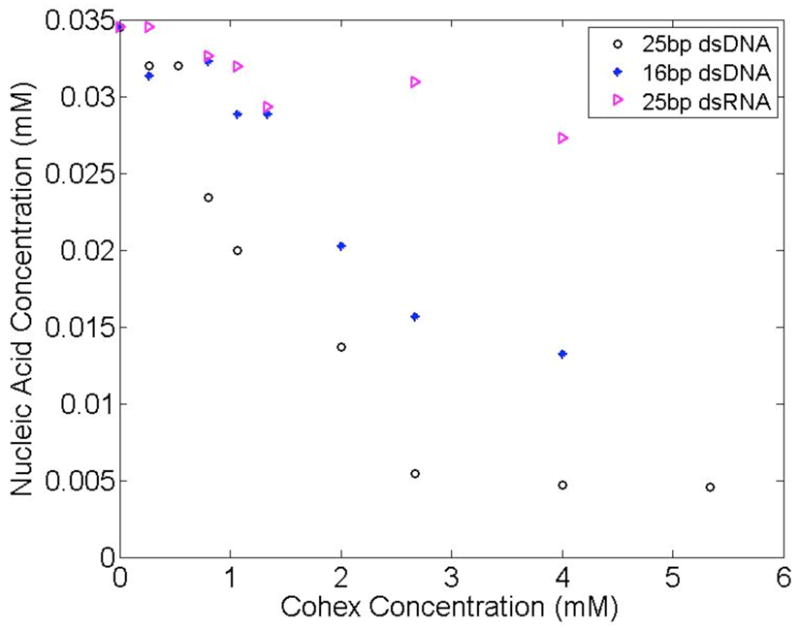
The concentration of DNA and RNA molecules in the supernatant as a function of [Co-hex] is calculated from UV absorption. Short, 16 and 25 bp DNA molecules are more easily condensed than 25 bp RNA. The 16 bp DNA, used as a control, indicates that the changing length of the double stranded nucleic acid has a smaller effect in generating condensation than the type of nucleic acid used.
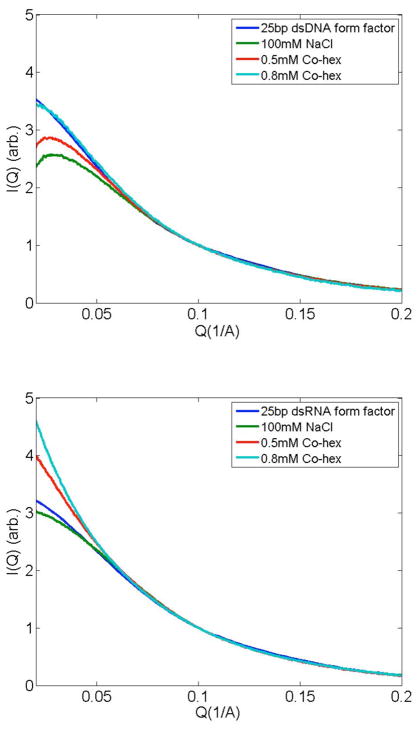
One possible explanation of these data is that many fewer Co-hex ions bind to RNA than to DNA: the negative charge of RNA is not effectively screened by these counterions. To test for this eventuality, SAXS was used to measure inter-duplex interactions [11]. SAXS profiles of solutions containing DNA and RNA were measured under carefully controlled ionic conditions, beginning at 100 mM Na+, where weak repulsion between nucleic acids is measured [7]. For these studies, the duplex concentration is maintained at 0.6 mM, more than an order of magnitude below the regime where liquid crystalline behavior is expected e.g. [14]. Since Co-hex is a trivalent ion, it very effectively competes with monovalent Na [12]. Its strong electrostatic attraction to the DNA enhances localized screening of the duplex charge [15] and dramatically reduces electrostatic repulsion between neighboring duplexes. Because Co-hex condenses DNA so efficiently, the higher NaCl concentration present in the SAXS studies ensures that samples can be prepared without aggregation. SAXS studies carried out on mixed phase samples, containing both soluble DNA and aggregates, are difficult to interpret [13]. In 100 mM NaCl, the onset of aggregation occurs in DNA with about 1 mM (free) Co-hex [12]. Figure 2 shows the results of SAXS studies on DNA (Fig. 2a) and RNA (Fig 2b) as a function of free Co-hex concentration [12]. To evaluate the magnitude of inter-particle interference, the high angle (or Q) regions of scattering profiles are matched with the form factor [11, 16], which represents the scattering of an isolated duplex. Repulsion/attraction between particles is indicated by a decrease/increase of the scattering profile relative to the form factor at the lowest angle [11].
Figure 2.
To assess interparticle interactions, solution SAXS profiles of (a) DNA and (b) RNA samples in Cobalt hexamine are compared with the form factor, the scattering from apparently non-interacting molecules (at infinite dilution). Only marginal repulsion between DNA molecules is measured when [Co-hex] ~ 0.8 mM. Comparatively, the sharp and smooth low Q upturn in (b) can be interpreted as end-to-end stacking of RNA molecules when Co-hex is present at concentrations above 0.5 mM (see supplementary material, Ref. [7] for a detailed discussion of this effect). The calculated DNA and RNA form factors (shown) match the experimentally measured form factors [7].
In the absence of Co-hex (100 mM NaCl), SAXS profiles of both nucleic acids indicate clear repulsion, consistent with previous work [7, 11]. The diverging behavior of DNA (Fig 2a) and RNA (Fig 2b) becomes apparent when small amounts of Co-hex are introduced. Scattering profiles for DNA in 100 mM NaCl plus 0.5 mM Co-hex still decrease at the lowest angles, consistent with repulsive forces. Around 0.8 mM Co-hex electrostatic interactions between duplexes are nearly neutralized. (Small amounts of Co-hex have dramatic effects on DNA. Near ~ 0.8 mM Co-hex, inter-DNA interactions rapidly change from repulsive to attractive. One concern might be potential changes to the amplitude of the scattering resulting from the closer association of Co-hex (replacing Na) in the ion cloud. The replacement of a few Na ions with Co-hex ions, will lead to a negligible change in scattering amplitude, compared to that from the nucleic acid itself and its hydration shell.)
Figure 2b shows an identical experimental series, performed with dsRNA instead of dsDNA. As expected, RNA duplexes repel in the presence of 100 mM NaCl. In 100 mM NaCl plus 0.5 mM Co-hex, where repulsion between DNAs is still evident, the SAXS profiles of RNA duplexes display a strong increase in low angle scattering, consistent with end-to-end stacking. To stack, two helices must come into close contact, thus the appearance of end-to-end stacking signals significant reduction in electrostatic repulsion, consistent with our previous observation that ions more effectively screen dsRNA than dsDNA [7, 17]. However, as opposed to precipitates, which are insoluble, these end-to-end stacked molecules remain soluble and are readily detected by solution SAXS. Thus, replacement of Na with Co-hex ions results in RNA duplex association via end-to-end stacking. This association mode is validated by comparing experimental to computed scattering profiles, Figure 3 (see also [7, 16]). For comparison, scattering profiles of side-by-side packed duplexes have been computed and are also shown in the figure. (The latter arrangements were selected because short DNA duplexes form close packed hexagonal arrays in the condensed phase [13], consistent with side-by-side arrangements.). End-to-end stacking of short nucleic acid duplexes has also been observed in work carried out by others [14, 17]. The seemingly conflicting results of UV absorption and SAXS measurement lead to the well-defined fundamental question—if, as the SAXS data suggest, RNA’s charge is more effectively screened than DNA’s, why is DNA more susceptible to precipitation by Co-hex than RNA? To answer this question, we must consider how the ions bind to the nucleic acid.
Figure 3.
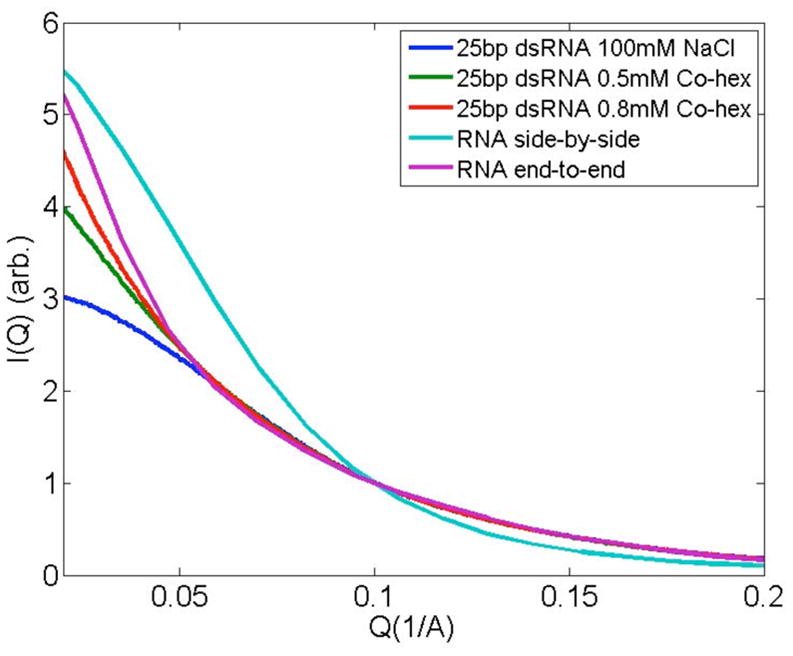
Solution SAXS data of RNA samples in NaCl or Co-hex are displayed along with scattering profiles that simulate RNA duplex association in different modes. The purple and cyan curves represent the configurations of RNAs, stacked either end-to-end or placed side-by-side to mimic relative placement in hexagonal arrays, respectively. The end-to-end stacking model (supplementary material Ref. [7]) is in better agreement with experimental SAXS profiles, though clearly not all duplexes participate in stacking interactions.
Computations of the potential around DNA or RNA duplexes [18] show that the major groove of A-RNA has a higher negative potential than the minor groove while the opposite is true for B-DNA. These different potentials have a profound impact on the spatial distribution of ions around RNA or DNA. Comparison of experimentally determined ion-distributions with models (based on the non-linear Poisson-Boltzmann equation, which are valid for monovalent ions) confirms that counterion distributions reflect these differences. Monovalent ions are localized to the RNA major groove, while they are more uniformly distributed around DNA [7]. In contrast to monovalent ions, experimental studies of Co-hex suggest that this trivalent ion prefers to bind in the major groove of both nucleic acids. For B-DNA, support for this binding pattern comes from NMR [19], capillary electrophoresis [20] and x-ray crystallographic [21] studies. Notably, the Guanines in the DNA major groove provide a preferential binding site for Co-hex [19]. Other important factors in determining binding sites may include the observed dehydration of Co-hex ions around DNA [22]. Although fewer studies have focused on Co-hex binding to RNA, solution NMR studies [23] find Co-hex ions buried deep within the major groove of a short stem loop. We therefore propose that the observed differences in condensation arise from the dramatically different geometries of the underlying nucleic acid structures. This picture is consistent with two models for condensation. In the first, competition between inter and intra molecular ion bridging explains why condensation forces (inter molecular bridging [24, 25]) and charge screening efficiency (intra molecular binding [25]) should be anti-correlated. Therefore, one possible explanation of the resistance of RNA to condensation arises from the more favorable binding of Co-hex to the RNA major groove compared to DNA. Second, in the electrostatic zipper model [26], the surface of the nucleic acid presents a pattern of alternating positive and negative charges. This alternating pattern can result in attraction if adjacent molecules pack so that opposing charged surfaces are in contact. There may be a significant difference in RNA+ion and DNA+ion surface charge due to differences in geometry. For example, the depth of the DNA major groove is ~8 Å, comparable to the 6 Å diameter of Co-hex molecule: Co-hex molecules bound in the DNA major groove remain “accessible” from outside. The DNA molecules can be condensed when the surface charge patterns are electrostatically in register with each other. In contrast, although RNA’s identical negative charge is also strongly screened, the trivalent ions have the potential to bury themselves too deep within the major groove to be “visible” at the surface.
Either geometric model of nucleic acid association is consistent with results from both absorption and scattering experiments. We note that the hydration structure of DNA and counterions also play an important role in DNA condensation [27], and could lead to a measurable difference in RNA and DNA condensation behavior since the RNA surface is more polar [28] and hence more hydrated, than the DNA surface.
Both SAXS and absorption measurements lead us to propose that the interaction modes of nucleic acids depend on the geometric details of charge arrangement in each system, highlighting the important role of molecular structure in condensation. Under conditions where DNA precipitates readily, the well-buried Co-hex ions inside the major groove of RNA contribute to charge neutralization but ultimately lend aggregation resistance to RNA duplexes. These readily testable results should provide the basis for further molecular dynamic (MD) simulations of multivalent-ion mediated interactions between like-charged nucleic acids and may provide guidance to overcome the many challenges associated with packaging duplex RNA for therapeutic applications.
Acknowledgments
We thank K. Finkelstein for experimental assistance. This research was supported by the NSF, the NIH and the NBTC at Cornell. This work is based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS) which is supported by the NSF and the NIH/NIGMS. We also made use of the Cornell NanoScale Facility, a member of the National Nanotechnology Infrastructure Network, which is supported by the NSF. and CNF at Cornell. CHESS is supported by NSF and NIH/NIGMS.
References
- 1.Bloomfield VA. Biopolymers. 1997;44:269. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 2.Hud NV, Vilfan ID. Annu Rev Biophys Biomol Struct. 2005;34:295. doi: 10.1146/annurev.biophys.34.040204.144500. [DOI] [PubMed] [Google Scholar]
- 3.Boussif O, Lezoualch F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proc Natl Acad Sci USA. 1995;92:7297. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 5.Bouxsein NF, McAllister CS, Ewert KK, Samuel CE, Safinya CR. Biochemistry. 2007;46:4785. doi: 10.1021/bi062138l. [DOI] [PubMed] [Google Scholar]; Grimm D, Kay MA. J Clin Invest. 2007;117:3633. doi: 10.1172/JCI34129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kornyshev AA, Lee DJ, Leikin S, Wynveen A. Rev Mod Phys. 2007;79:943. [Google Scholar]; Wong GCL, Pollack L. Annu Rev Phys Chem. 2010;61:171. doi: 10.1146/annurev.physchem.58.032806.104436. [DOI] [PubMed] [Google Scholar]
- 7.Pabit SA, Qiu XY, Lamb JS, Li L, Meisburger SP, Pollack L. Nucleic Acids Res. 2009;37:3887. doi: 10.1093/nar/gkp257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelta J, Livolant F, Sikorav JL. J Biol Chem. 1996;271:5656. doi: 10.1074/jbc.271.10.5656. [DOI] [PubMed] [Google Scholar]
- 9.Widom J, Baldwin RL. J Mol Biol. 1980;144:431. doi: 10.1016/0022-2836(80)90330-7. [DOI] [PubMed] [Google Scholar]
- 10.Heilman-Miller SL, Thirumalai D, Woodson SA. J Mol Biol. 2001;306:1157. doi: 10.1006/jmbi.2001.4437. [DOI] [PubMed] [Google Scholar]
- 11.Qiu XY, Kwok LW, Park HY, Lamb JS, Andresen K, Pollack L. Phys Rev Lett. 2006;96:138101. doi: 10.1103/PhysRevLett.96.138101. [DOI] [PubMed] [Google Scholar]; Qiu XY, Andresen K, Kwok LW, Lamb JS, Park HY, Pollack L. Rev Lett Phys. 2007;99:038104. doi: 10.1103/PhysRevLett.99.038104. [DOI] [PubMed] [Google Scholar]
- 12.Andresen K, Qiu XY, Pabit SA, Lamb JS, Park HY, Kwok LW, Pollack L. Biophys J. 2008;95:287. doi: 10.1529/biophysj.107.123174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu XY, Andresen K, Lamb JS, Kwok LW, Pollack L. Phys Rev Lett. 2008;101:228101. doi: 10.1103/PhysRevLett.101.228101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakata M, Zanchetta G, Chapman BD, Jones CD, Cross JO, Pindak R, Bellini T, Clark NA. Science. 2007;318:1276. doi: 10.1126/science.1143826. [DOI] [PubMed] [Google Scholar]
- 15.Rouzina I, Bloomfield VA. Journal of Physical Chemistry. 1996;100:4292. [Google Scholar]
- 16.Li L, Pabit SA, Lamb JS, Park HY, Pollack L. Appl Phys Lett. 2008;92:223901. doi: 10.1063/1.2937402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zanchetta G, Bellini T, Nakata M, Clark NA. J Am Chem Soc. 2008;130:12864. doi: 10.1021/ja804718c. [DOI] [PubMed] [Google Scholar]
- 18.Chin K, Sharp KA, Honig B, Pyle AM. Nat Struct Biol. 1999;6:1055. doi: 10.1038/14940. [DOI] [PubMed] [Google Scholar]
- 19.Robinson H, Wang AHJ. Nucleic Acids Res. 1996;24:676. doi: 10.1093/nar/24.4.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouameur AA, Tajmir-Riahi HA. J Biol Chem. 2004;279:42041. doi: 10.1074/jbc.M406053200. [DOI] [PubMed] [Google Scholar]
- 21.Nunn CM, Neidle S. J Mol Biol. 1996;256:340. doi: 10.1006/jmbi.1996.0090. [DOI] [PubMed] [Google Scholar]
- 22.Kankia BI, Buckin V, Bloomfield VA. Nucleic Acids Res. 2001;29:2795. doi: 10.1093/nar/29.13.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kieft JS, Tinoco I. Structure. 1997;5:713. doi: 10.1016/s0969-2126(97)00225-6. [DOI] [PubMed] [Google Scholar]
- 24.Dai L, Mu YG, Nordenskiold L, van der Maarel JRC. Phys Rev Lett. 2008;100:118301. doi: 10.1103/PhysRevLett.100.118301. [DOI] [PubMed] [Google Scholar]; Delacruz MO, Belloni L, Delsanti M, Dalbiez JP, Spalla O, Drifford M. J Chem Phys. 1995;103:5781. [Google Scholar]
- 25.Nyrkova IA, Semenov AN. Soft Matter. 2009;5:979. doi: 10.1039/d4sm00188e. [DOI] [PubMed] [Google Scholar]
- 26.Kornyshev AA, Leikin S. Phys Rev Lett. 1999;82:4138. [Google Scholar]
- 27.Todd BA, Parsegian VA, Shirahata A, Thomas TJ, Rau DC. Biophys J. 2008;94:4775. doi: 10.1529/biophysj.107.127332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egli M, Portmann S, Usman N. Biochemistry. 1996;35:8489. doi: 10.1021/bi9607214. [DOI] [PubMed] [Google Scholar]