Summary
Large populations of bacteria live on leaf surfaces and these phyllosphere bacteria can have important effects on plant health. However, we currently have a limited understanding of bacterial diversity on tree leaves and the inter- and intra-specific variability in phyllosphere community structure. We used a bar-coded pyrosequencing technique to characterize the bacterial communities from leaves of 56 tree species in Boulder, Colorado, USA, quantifying the intra- and inter-individual variability in the bacterial communities from 10 of these species. We also examined the geographic variability in phyllosphere communities on Pinus ponderosa from several locations across the globe. Individual tree species harboured high levels of bacterial diversity and there was considerable variability in community composition between trees. The bacterial communities were organized in patterns predictable from the relatedness of the trees as there was significant correspondence between tree phylogeny and bacterial community phylogeny. Inter-specific variability in bacterial community composition exceeded intra-specific variability, a pattern that held even across continents where we observed minimal geographic differentiation in the bacterial communities on P. ponderosa needles.
Introduction
The phyllosphere, the microbial habitat found on the surface of leaves, may be one of the largest microbial habitats on earth, with terrestrial leaf surface area estimated to exceed 108 km2 globally (Morris and Kinkel, 2002). The phyllosphere is a unique and dynamic habitat, with phyllosphere communities subjected to irregular, and sometimes relatively large, changes in temperature, UV radiation, relative humidity and leaf wetness (Hirano and Upper, 2000; Lindow and Brandl, 2003; Whipps et al., 2008). Despite these environmental constraints, microbes flourish on leaf surfaces. Although fungi and archaea are known to colonize leaves, bacteria are numerically dominant in the phyllosphere environment (Andrews and Harris, 2000; Lindow and Brandl, 2003) and these leaf-dwelling bacteria can have either neutral, negative or positive influences on their host plants by serving as pathogens or preventing leaf colonization by pathogens (Kishore et al., 2005). Phyllosphere bacteria are also important in that they likely represent an important source of bacteria to the atmosphere (Lighthart, 1997) and they may play key roles in nutrient cycling by fixing nitrogen (Jones, 1970; Freiberg, 1998).
Despite their potential importance, we know surprisingly little about the diversity and biogeography of phyllosphere bacterial communities. Most previous work has used traditional culture-based methods to describe the bacterial inhabitants of the phyllosphere, focusing primarily on aerobic plant pathogens (Lindow and Brandl, 2003). However, since culture-based studies usually detect only a small fraction of the microbial diversity present in environmental samples (Pace, 1997), they are likely to underestimate the full extent of bacterial diversity on leaf surfaces. The few studies that have used culture-independent methods to characterize phyllosphere bacterial communities suggest that leaf surfaces harbour many hundreds of unique bacterial taxa with bacterial community composition varying across plant species (Yang et al., 2001; Lambais et al., 2006; Delmotte et al., 2009). However, we do not know the relative importance of plant location versus species identity in structuring phyllosphere communities. By comparing phyllosphere communities on multiple plant species from a single location, we can test the null hypothesis that plants in close proximity are likely to be exposed to similar microbial inocula and thus plant location rather than species identity per se may have the more important influence on phyllosphere community structure.
More generally, the phyllosphere represents a unique environment for testing ecological principles, as demonstrated by recent work on bacterial succession (Redford and Fierer, 2009), given that the bacterial communities can be sampled in a discrete and hierarchical manner (e.g. individual leaves, trees and tree species) across time and space. From research on plant and animal biogeography, we would expect that, within a given habitat type, communities located in close proximity will be more similar to one another than communities that are geographically distant (Lomolino et al., 2006). We would expect a similar phenomenon to occur in phyllosphere bacterial communities; bacterial communities associated with a single tree species would be expected to become more dissimilar as the geographic distance between the trees increases. These differences could arise due to constraints on microbial dispersal, differences in leaf characteristics (structural, phenological or physiological), or differences in climatic conditions (Whipps et al., 2008; Redford and Fierer, 2009), but the net effect would be a positive correlation between geographic distance and the phylogenetic ‘distance’ between bacterial communities on leaf surfaces. However, since few studies have compared phyllosphere communities from the same plant species across a range of geographic distances, we do not know how geographic distance may influence the structure of these microbial communities.
We designed this study to determine how plant species identity and geographic location influence the biogeography of phyllosphere communities. Specifically, we addressed three questions: How does intra-specific variability in phyllosphere communities compare with inter-specific variability across a range of geographic scales? Do distinct plant species harbour distinct phyllosphere communities? and, if so, can the bacterial phyllosphere community structure be predicted from tree species phylogeny? We characterized and compared the bacterial communities on 56 tree species from one location (a university campus in Colorado, USA) to determine if these communities are indeed plant-species specific and to gain a more comprehensive understanding of the types of bacteria inhabiting the phyllosphere of various tree species. Ten of these 56 tree species were selected for more intensive sampling to assess the degree of variability in bacterial communities from samples collected from a single individual and from individuals of the same species. We also sampled a single species, Pinus ponderosa, from a number of locations with pairwise distances ranging from 10 m to > 10 000 km apart in order to determine the relative influence of geographic location on bacterial phyllosphere community composition. Bacterial community structure in each of the 174 collected samples was determined using a barcoded pyrosequencing approach, obtaining more than 600 16S rRNA gene sequences per sample.
Results
General characteristics of phyllosphere communities
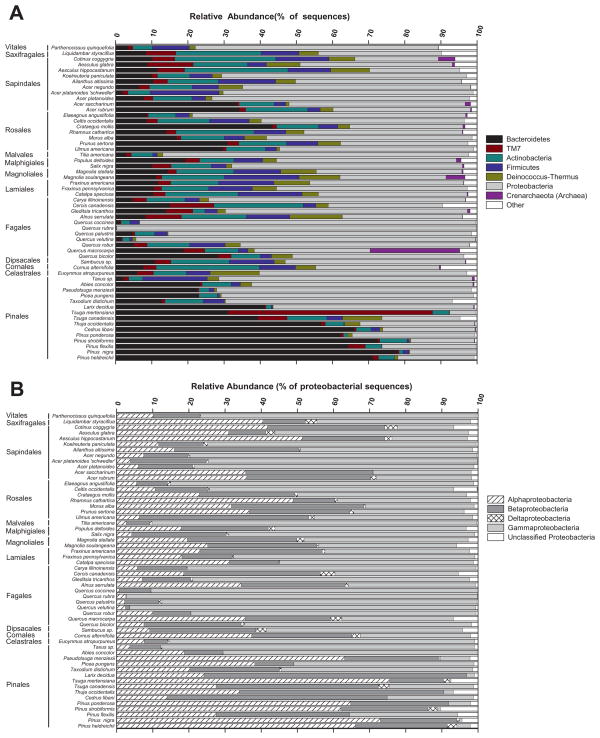
Across all samples, we obtained 115 394 quality sequences (average read length = 240 bp) and a minimum of 600 sequences per sample (range 600–1500 sequences per sample, median of 936 sequences). Of these sequences, 109 434 (94.8%) could be classified for a total of 5476 unique bacterial operational taxonomic units (OTUs) at the 97% sequence similarity level across all samples and an average of 252 OTUs per sample. The single most abundant OTU, a representative of the TM7 lineage, only accounted for 5% of the sequences obtained from all samples, and no OTUs were shared across all samples. We found 25 different bacterial phyla across all samples with the most abundant groups within the Proteobacteria (24.5%, 16.4% and 7.9% of sequences in the Alpha-, Beta- and Gammaproteobacteria sub-phyla respectively), Bacteroidetes (22.5% of sequences), Actinobacteria (9.0% of sequences), TM7 (9.0% of sequences) and Firmicutes (5.3% of sequences) phyla. The most common group of bacteria found on the leaves was Sphingobacteriales from the Bacteroidetes lineage, which represented 21.3% of all sequences. Additional details on the taxonomic structure of the phyllosphere bacterial communities can be found in the supplementary material Tables S1 and S2.
Bacterial abundances as determined by 4,6-diamidino-2-phenylindole (DAPI) ranged from 2.4 × 104 cells cm−2 (Juniperus scopulorum) to 6.0 × 105 cells cm−2 (Cedrus libani) with an average of 2.1 × 105 cells cm−2 (Table S1). There was no significant difference between bacterial abundances on gymnosperms versus angiosperms (t34 = 0.97, P = 0.17) and no single order of trees had consistently higher bacterial abundances than any other order (Table S1).
Interspecies differences
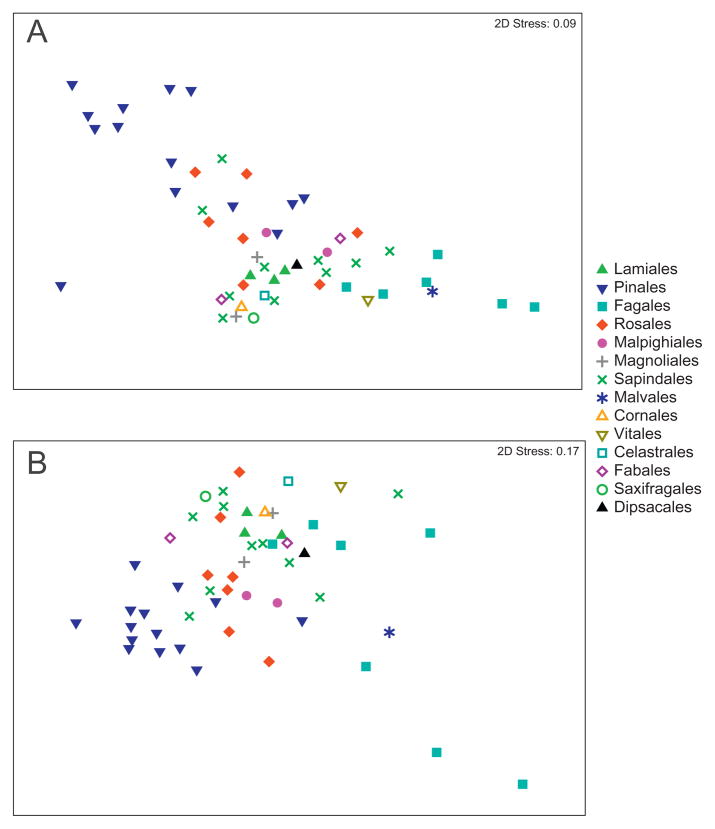
Among the 56 tree species sampled from this single location, we found a high degree of variability in the taxonomic structure of the phyllosphere communities (Figs 1 and 2). In general, Bacteroidetes and Betaproteobacteria were more common on gymnosperms (the Celastrales and Pinales orders), while Actinobacteria and Gammaproteobacteria were more common on angiosperms (Fig. 1, Table S1). The pairwise phylogenetic distances between the bacterial communities from the 56 tree species were calculated using the Unifrac metric (Lozupone et al., 2006) which effectively measures the phylogenetic overlap between the taxa represented in any pair of communities. Different tree species harboured unique bacterial communities (Fig. 2) with all pairwise P values < 0.05 based on the UniFrac significance test (Lozupone et al., 2006). The gymnosperm communities clustered together, but did not necessarily cluster to the exclusion of the angiosperm communities and there was some clustering by plant order, but many orders had overlapping phyllosphere communities (Fig. 2). However, there was a significant correlation between bacterial community phylogeny and tree species phylogeny (r = 0.51, P < 0.001), indicating that, although weak, there was some association between the evolutionary history of the trees and the bacterial communities on the tree leaves. This association was confirmed by the weak, but significant, influence of tree order identity on bacterial community composition (ANOSIM Global R = 0.36, P < 0.001).
Fig. 1.
Relative abundances of bacterial phyla and sub-phyla in each of the phyllosphere communities from the ‘inter-species’ study: (A) bacterial phyla (B) only proteobacterial sub-phyla. Note that B only shows the percentages of the various proteobacterial sub-phyla relative to the total abundance of Proteobacteria on each tree. We also note that in this figure we are only focusing on the interspecies variation as each symbol represents data from a single individual tree (see Fig. 3 and the associated section of the Results for a description of how intraspecies variation in bacterial community composition compares to interspecies variation across this study site).
Fig. 2.
NMDS plot illustrating differences between bacterial communities on 56 tree species. Pairwise community distances determined using the weighted Unifrac algorithm (A) and the Kulczynski distance metric (B). Figure S3 shows this same plot with labels indicating the specific tree species represented by each point.
The diversity of the phyllosphere bacterial communities varied widely among plant orders (Table S3). The Malvales and Pinales orders had the lowest average diversity levels with the Cornales and Dipsacales having the highest diversity levels (with diversity estimated using either the number of unique OTUs or Faith’s PD metric). There was no correlation between community diversity and bacterial abundance (r = 0.06, P = 0.4).
Intraspecies variability
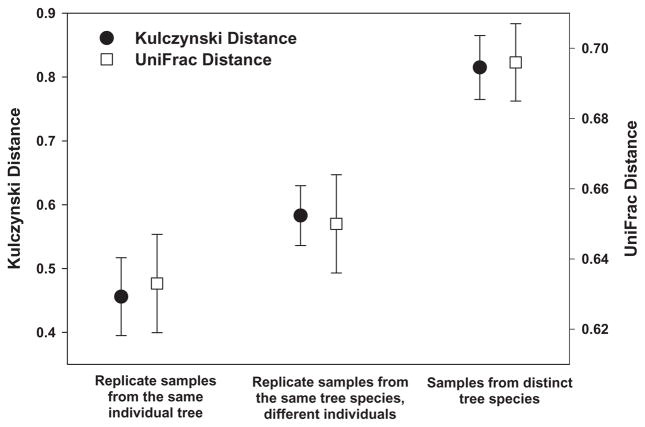
Ten of the 56 tree species were selected for more intensive sampling to assess the variability in phyllosphere communities within and between individual trees of the same species. The degree of variability among samples collected from a single individual, among individuals of the same species and among individuals of different species is illustrated in Fig. 3. While there was variability between samples from the same individual and between individuals of the same species, this intra-individual and intraspecies variability in phyllosphere communities was far lower than the variability between samples from different tree species, with samples from the same species generally clustering together (Fig. S1).
Fig. 3.
Phylogenetic and OTU-based distances (Unifrac and Kulczynski distances respectively) between bacterial communities on an individual tree, on different individual trees of the same species, and on different tree species. Distances calculated from the 10 tree species that were examined to determine with intra-and inter-individual replicate samples. Note that this figure represents results from trees sampled at the same location (the University of Colorado campus) on the same day. Bars represent 95% confidence intervals around the mean.
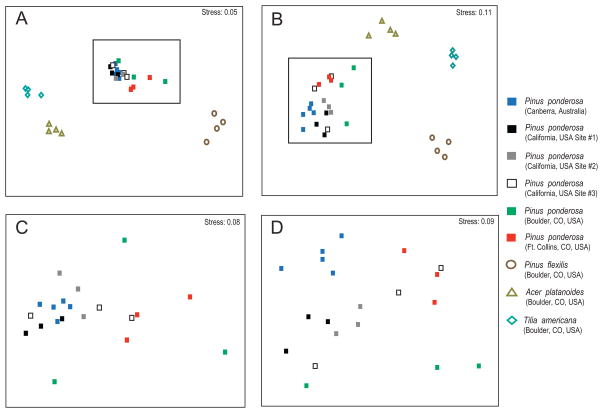
Within P. ponderosa, there was remarkably little influence of geographic location on community composition, even across thousands of kilometres. This is evident in Fig. 4 which shows that the phyllosphere communities on P. ponderosa trees did not cluster by location, and this lack of an influence of geographic location on phyllosphere community composition was confirmed by Mantel tests relating geographic distance to weighted Unifrac distances (r = 0.08, P = 0.5). However, when compared with replicates of different species, the P. ponderosa phyllosphere communities were clearly distinct from those on other representative species (even a closely related species, Pinus flexilis), regardless of geographic location (Fig. 4A and B).
Fig. 4.
NMDS plots based on weighted UniFrac distances (A and C) and Kulczynski distances (panels B and D). Panels A and B show the Pinus ponderosa samples collected from sites in Australia and the USA along with samples representing other tree species (the P. ponderosa samples are within the dashed boxes). These two panels show that bacterial communities on P. ponderosa are relatively similar to one another regardless of geographic location (i.e. more similar to one another than to the bacteria on other tree species). Panels C and D focus just on the P. ponderosa samples in order to better show that sampling location has little to no effect on the observed differences between P. ponderosa- associated bacterial communities.
Discussion
We found high levels of bacterial diversity in the phyllosphere with the molecular analyses revealing far greater diversity than previously recognized from cultivation-based surveys. We identified many of the same bacterial taxa found in other culture-independent studies of the phyllosphere, including high relative abundances of lineages within the Bacteroidetes, Actinobacteria and Proteobacteria phyla (Yang et al., 2001; Jackson et al., 2006; Lambais et al., 2006; Delmotte et al., 2009). Also of note are the numbers of Deinococcus-Thermus and TM7 sequences that were found on nearly every tree, suggesting that these poorly studied bacterial phyla may be more common in phyllosphere communities than previously recognized, perhaps because of their resistance to UV radiation. However, it is important to highlight that the full extent of phyllosphere diversity is likely to be even higher than was captured with this survey (even though we averaged > 900 sequences per sample) as the rarefaction curves did not asymptote (Fig. S2) indicating that we have not surveyed all of the rare lineages in the individual samples. Likewise, because our primer set was designed to screen out chloroplast DNA, we could not capture any cyanobacteria that may live on the leaves.
The bacterial communities inhabiting the leaves of a given species varied within and across individuals of that species; however, there was far more variability in bacterial community structure across tree species than within species (Fig. 3). This finding supports results from other studies showing pronounced interspecies variability in phyllosphere communities (Yang et al., 2001; Lambais et al., 2006; Whipps et al., 2008). This high degree of variability in bacterial communities between trees found at the same location and the observation that the dominant bacterial taxa are very distinct from those commonly found in soil (Lauber et al., 2009) or in air samples (Bowers et al., 2009) suggests that the phyllosphere bacteria are not simply passive inhabitants of the leaf surface that are deposited on leaves. Rather, these results suggest a more intimate relationship between trees and their phyllosphere bacteria with tree species having a strong direct or indirect influence on structure and composition of their leaf- associated bacterial communities.
The shifts in phyllosphere community composition across the tree species were, to a certain extent, predictable from species identity as trees that were more closely related harboured bacterial communities that were more phylogenetically similar, a pattern primarily driven by the differences in the communities found on gymnosperms and angiosperms (Figs 1 and 2). Leaves of gymnosperms and angiosperms are distinct in many respects and it is not clear from this study which specific aspects of leaf structure, chemistry and physiology explain the observed differences in phyllosphere communities on these major tree taxa or the overall interspecies patterns evident in Fig. 1 and 2. We hypothesize that the interspecies differences in phyllosophere communities may be related to specific leaf characteristics not measured here, such as fine scale cuticle structure and composition, leaf age, leaf chemical composition and/or leaf volatile organic compound emissions, but determining the relative importance of these various factors will require more detailed examination.
The results from our cross-site survey of P. ponderosa leaf communities provide some unique insights into the structure and biogeography of phyllosphere communities. Ponderosa pine communities are fundamentally similar to one another regardless of geographic location, even across continental scales (Fig. 4). Although geographic distance and the associated dispersal constraints may have an important influence on the distribution of specific microbes (Cho and Tiedje, 2000), such dispersal constraints do not appear to have a major influence on overall patterns of bacterial community assembly. Geographic distance, in and of itself, had little influence on these phyllosphere communities with more variation at individual sites than between trees located thousands of kilometers apart. The characteristics of the leaves themselves (albeit unmeasured characteristics) appear to have a greater influence on what types of microbes can thrive on a particular tree species than the climatic conditions or geographic location. Although many studies have shown that environmental conditions can have important effects on phyllosphere community structure (Lindow and Brandl, 2003; Redford and Fierer, 2009), our data are consistent with the notion that plant species identity has an verarching influence on the structure of the phyllosphere community.
Conclusions
Bacteria living in the phyllosphere, like other microbial habitats (Martiny et al., 2006; Ramette and Tiedje, 2007; Fierer, 2008), exhibit predictable biogeographic patterns. Specifically, we find that interspecies variability exceeds intraspecies variability and there is a reasonable correlation between tree phylogeny and bacterial community composition that is likely driven by differences in leaf characteristics that remain undetermined. However, the general patterns are, to some degree, distinct from those observed in most plant and animal communities as the bacterial communities within an individual habitat type (the P. ponderosa phyllosphere) do not become more phylogenetically distinct with increasing geographic distance, even across thousands of kilometres. This supports the speculation (Finlay, 2002; Fenchel, 2003) that dispersal constraints may often be less important in structuring the biogeography of microbial communities than the biogeography of most plant and animal communities.
Experimental procedures
Sample collection – interspecies variability
Leaves were collected from 56 tree species representing 14 different plant orders (Fig. 1, Table S4). All samples were collected on the same day (July 15, 2008) from a 35 hectare area on the University of Colorado campus in Boulder, Colorado, USA (40°0′N, 105°16′W, elevation 1655 m). The site is flat and the inter-tree spaces are typically occupied by either irrigated lawns or buildings. A sterile plastic bag (10 × 20 cm) was filled with ~50 g of leaves collected from a single representative tree of each species. Undamaged leaves were randomly selected from around the canopy at the same height on each tree (1–2 m above the ground surface) as there is likely to be variation in leaf surface bacterial communities with canopy height (Kinkel, 1997). Leaves were weighed then washed with 1:50 diluted leaf wash solution by placing 100 ml of the wash solution in the bag with the leaves and shaking for 60 s (Kadivar and Stapleton, 2003). The wash solution was filtered through sterile glass wool into two 50 ml centrifuge tubes and centrifuged at 2200 g for 15 min at 4°C. DNA was extracted from the resulting pellets using the MoBio PowerSoil DNA Kit following the manufacturer’s instructions (MoBio Laboratories, Carlsbad, CA, USA).
Sample collection – intraspecies variability
To determine how the variability in phyllosphere bacterial community composition within individual trees and among individual trees of the same species compared with the variability among trees of different species we selected 10 of the 56 tree species for more comprehensive sampling: Cercis canadensis, Catalpa speciosa, Fraxinus pennsylvanica, Tilia americana, Picea pungens, P. flexilis, Celtis occidentalis, Acer platanoides, Abies concolor and Aesculus hippocastanum (Table S4). The sampling scheme described above was repeated three times on a single individual tree for each of these 10 species, and also for two additional individuals of each of the 10 species that were all located within the 35 hectare area on the University of Colorado campus. These samples were processed using the same methods described above.
Sample collection – geographic variability
In order to determine how the bacterial phyllosphere communities of a single tree species varied across larger spatial scales, we took samples of Pinus ponderosa leaves from three individuals on the University of Colorado (CU) campus, three individuals from the foothills near Fort Collins, CO (~100 km north of CU), three individuals from each of three separate locations in northern California, USA (~2000 km away from CU), and five individuals from Canberra, Australia (~14 000 km away from CU). See Table S4 for the coordinates of each sampling location. Samples were collected as described above and were shipped to the CU campus at 4°C where they were washed and extracted. To compensate for the time spent in transit from the collection sites to the University of Colorado and any effect this transit time may have on the bacterial communities, all samples were stored for 5 days after collection at 4°C before being processed. Samples from three other tree species on the CU campus (including a congener, P. flexilis) were collected at the same time and handled in an identical manner to compare the variability in P. ponderosa-associated communities across space to the variability across selected tree species.
Determination of bacterial abundances, diversity and community composition
In order to determine the relative abundance of phyllosphere microbes on the collected leaves, we counted individual bacterial cells in aliquots of the leaf wash solution using the DAPI method described previously (Bowers et al., 2009) with cell numbers reported per unit leaf surface area. We determined leaf surface area by tracing three representative leaves or leaflets per tree species and weighing the tracing paper. For the tree species with needle-shaped leaves, we used calipers to measure the dimensions of the leaves and then estimated the surface area of the leaves assuming that individual needles are roughly cylindrical.
We used a barcoded pyrosequencing procedure targeting bacterial 16S rRNA genes to analyse the diversity and composition of the bacteria in each of the collected samples on a single 454 Life Sciences Genome Sequencer FLX (Roche) run at the Environmental Genomics Core Facility at the University of South Carolina. The procedure was identical to that described previously (Fierer et al., 2008; Bowers et al., 2009; Lauber et al., 2009), except we used a primer pair that does not amplify chloroplast DNA. The forward primer contained the 454 Life Sciences primer B sequence, the bacterial primer 799f (Chelius and Triplett, 2001) and a two-base linker sequence (‘AG’). The reverse primer contained the 454 Life Sciences primer A sequence, a unique 12 bp error-correcting Golay barcode used to tag each PCR product (Fierer et al., 2008), the ‘universal’ bacterial primer 1115r (Reysenbach and Pace, 1995), and a ‘GT’ linker sequence inserted between the barcode and the rRNA primer. PCR reactions were carried out in triplicate and after visualization, purification and amplicon quantification, the amplicons from all samples (120 samples in total) were combined in equimolar ratios into a single tube. Additional details on this pyrosequencing procedure can be found in Lauber and colleagues (2009) and Fierer and colleagues (2008).
Sequences were processed and analysed following the procedures described previously (Fierer et al., 2008; Hamady et al., 2008; Lauber et al., 2009). To determine the amount of dissimilarity (distance) between any pair of bacterial communities, we employed both a phylogenetic metric [weighted UniFrac (Lozupone and Knight, 2005; Lozupone et al., 2006; Lozupone et al., 2007)] and a taxonomic metric [Kulczynski distance (Cha, 2007)]. UniFrac distances are based on the fraction of branch length shared between any pair of communities within a phylogenetic tree constructed from the 16S rRNA gene sequences from all communities being compared. A relatively small UniFrac distance implies that two communities are compositionally similar, harbouring lineages sharing a common evolutionary history (Lozupone et al., 2006). In weighted UniFrac, branch lengths are weighted based on the relative abundances of lineages within communities (Lozupone et al., 2007). The Kulczynski distance metric ignores phylogeny and is simply based on those OTUs that are shared between any pair of samples (with OTUs defined at the 97% sequence similarity level). We used the analysis of similarities (ANOSIM) function in PRIMER (Clarke and Gorley, 2006) to test for differences in community composition among groups of samples. For the ‘interspecies’ study, we compared the correlation between tree phylogeny and the phylogenetic structure of the bacterial communities on the trees using a Mantel test as implemented in PRIMER. The pairwise phylogenetic distances between each of the 56 tree species was determined using the ‘phylomatic’ function in Phylocom (Webb et al., 2008).
We used two indices to compare community-level diversity between the samples from the 56 different plant species. We compared the number of unique OTUs and Faith’s phylodiversity index [Faith’s PD (Faith, 1992)] in order to compare taxonomic and phylogenetic diversity levels, respectively, across samples. For both diversity metrics, we used a randomly selected subset of 750 sequences per sample in order to compensate for differences in sampling effort between samples.
Supplementary Material
Acknowledgments
We would like to thank Chris Lauber, Heather Hamilton, Katie Eilers, Scott Felgner and Kelly Ramirez for help with sample collection and the laboratory analyses. We thank Micah Hamady for his assistance with sequence analyses and Joe Jones at the University of South Carolina for his help with the pyrosequencing. This work was funded by grants from the National Science Foundation and the National Geographic Society to N.F and grants from the National Institutes of Health to R.K.
References
- Andrews JH, Harris RF. The ecology and biogeography of microorganisms of plant surfaces. Annu Rev Phytopathol. 2000;38:145–180. doi: 10.1146/annurev.phyto.38.1.145. [DOI] [PubMed] [Google Scholar]
- Bowers R, Lauber C, Wiedinmyer C, Hamady M, Hallar A, Fall R, et al. Characterization of airborne microbial communities at a high elevation site and their potential to act as atmospheric ice nuclei. Appl Environ Microbiol. 2009;75:5121–5130. doi: 10.1128/AEM.00447-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha S. Comprehensive survey on distance/similarity measures between probability density functions. Int J Math Models Meth Appl Sci. 2007;1:300–307. [Google Scholar]
- Chelius MK, Triplett EW. The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb Ecol. 2001;41:252–263. doi: 10.1007/s002480000087. [DOI] [PubMed] [Google Scholar]
- Cho JC, Tiedje JM. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl Environ Microbiol. 2000;66:5448–5456. doi: 10.1128/aem.66.12.5448-5456.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K, Gorley R. PRIMER. Plymouth, UK: PRIMER-E Ltd; 2006. [Google Scholar]
- Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, et al. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci USA. 2009;106:16428–16433. doi: 10.1073/pnas.0905240106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- Fenchel T. Biogeography for bacteria. Science. 2003;301:925–926. doi: 10.1126/science.1089242. [DOI] [PubMed] [Google Scholar]
- Fierer N. Microbial biogeography: patterns in microbial diversity across space and time. In: Zengler K, editor. Accessing Uncultivated Microorganisms: From the Environment to Organisms and Genomes and Back. Washington, DC, USA: ASM Press; 2008. pp. 95–115. [Google Scholar]
- Fierer N, Hamady M, Lauber C, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Freiberg E. Microclimatic parameters influencing nitrogen fixation in the phyllosphere in a Costa Rican premontane rain forest. Oecologia. 1998;117:9–18. doi: 10.1007/s004420050625. [DOI] [PubMed] [Google Scholar]
- Hamady M, Walker J, Harris J, Gold N, Knight R. Error-correcting barcoded primers allow hundreds of samples to be pyrosequenced in multiplex. Nature Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano SS, Upper CD. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae – a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev. 2000;64:624–653. doi: 10.1128/mmbr.64.3.624-653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EF, Echlin HL, Jackson CR. Changes in the phyllosphere community of the resurrection fern, Polypodium polypodioides, associated with rainfall and wetting. FEMS Microbiol Ecol. 2006;58:236–246. doi: 10.1111/j.1574-6941.2006.00152.x. [DOI] [PubMed] [Google Scholar]
- Jones K. Nitrogen fixation in phyllosphere of Douglas Fir, Pseudotsuga-Douglasii. Ann Bot. 1970;34:239–244. [Google Scholar]
- Kadivar H, Stapleton A. Ultraviolet radiation alters maize phyllosphere bacterial diversity. Microb Ecol. 2003;45:353–361. doi: 10.1007/s00248-002-1065-5. [DOI] [PubMed] [Google Scholar]
- Kinkel L. Microbial population dynamics on leaves. Annu Rev Phytopathol. 1997;35:327–347. doi: 10.1146/annurev.phyto.35.1.327. [DOI] [PubMed] [Google Scholar]
- Kishore GK, Pande S, Podile AR. Biological control of late leaf spot of peanut (Arachis hypogaea) with chitinolytic bacteria. Phytopathology. 2005;95:1157–1165. doi: 10.1094/PHYTO-95-1157. [DOI] [PubMed] [Google Scholar]
- Lambais MR, Crowley DE, Cury JC, Bull RC, Rodrigues RR. Bacterial diversity in tree canopies of the Atlantic forest. Science. 2006;312:1917–1917. doi: 10.1126/science.1124696. [DOI] [PubMed] [Google Scholar]
- Lauber C, Knight R, Hamady M, Fierer N. Soil pH as a predictor of soil bacterial community structure at the continental scale: a pyrosequencing-based assessment. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighthart B. The ecology of bacteria in the alfresco atmosphere. FEMS Microbiol Ecol. 1997;23:263–274. [Google Scholar]
- Lindow SE, Brandl MT. Microbiology of the phyllosphere. Appl Environ Microbiol. 2003;69:1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomolino M, Riddle B, Brown J. Biogeography. Sunderland, MA, USA: Sinauer Assoc; 2006. [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac –an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Kelley S, Knight R. Quantitative and qualitative b diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown J, Colwell R, Fuhrman J, Green J, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Morris C, Kinkel L. Fifty years of phyllosphere microbiology: significant contributions to research in related fields. In: Lindow S, Hecht-Poinar E, Elliott V, editors. Phyllosphere Microbiology. St. Paul, MN, USA: APS Press; 2002. pp. 365–375. [Google Scholar]
- Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–739. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- Ramette A, Tiedje J. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb Ecol. 2007;53:197–207. doi: 10.1007/s00248-005-5010-2. [DOI] [PubMed] [Google Scholar]
- Redford AJ, Fierer N. Bacterial succession on the leaf surface: a novel system for studying successional dynamics. Microb Ecol. 2009;58:189–198. doi: 10.1007/s00248-009-9495-y. [DOI] [PubMed] [Google Scholar]
- Reysenbach A, Pace N. Reliable amplification of hyperthermophilic Archaeal 16s rRNA genes by the polymerase chain reaction. In: Robb F, editor. Archaea: A Laboratory Manual. New York, NY, USA: Cold Spring Harbor Laboratory Press; 1995. pp. 101–107. [Google Scholar]
- Webb CO, Ackerly DD, Kembel SW. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics. 2008;24:2098–2100. doi: 10.1093/bioinformatics/btn358. [DOI] [PubMed] [Google Scholar]
- Whipps JM, Hand P, Pink D, Bending GD. Phyllosphere microbiology with special reference to diversity and plant genotype. J Appl Microbiol. 2008;105:1744–1755. doi: 10.1111/j.1365-2672.2008.03906.x. [DOI] [PubMed] [Google Scholar]
- Yang CH, Crowley DE, Borneman J, Keen NT. Microbial phyllosphere populations are more complex than previously realized. Proc Natl Acad Sci USA. 2001;98:3889–3894. doi: 10.1073/pnas.051633898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.