Abstract
Maintenance of genome stability during cell division depends on establishing correct attachments between chromosomes and spindle microtubules. Correct, bi-oriented attachments are stabilized, while incorrect attachments are selectively destabilized. This process relies largely on increased phosphorylation of kinetochore substrates of Aurora B kinase at misaligned versus aligned kinetochores. Current models explain this differential phosphorylation by spatial changes in the position of substrates relative to a constant pool of kinase at the inner centromere. However, these models are based on studies in aneuploid cells. We show that normal diploid cells have a more robust error correction machinery. Aurora B is enriched at misaligned centromeres in these cells, and the dynamic range of Aurora B substrate phosphorylation at misaligned versus aligned kinetochores is increased. These findings indicate that in addition to Aurora B regulating kinetochore-microtubule binding, the kinetochore also controls Aurora B recruitment to the inner centromere. We show that this recruitment depends on both activity of Plk1, a kinetochore-localized kinase, and activity of Aurora B itself. Our results suggest a feedback mechanism in which Aurora B both regulates and is regulated by chromosome attachment to the spindle, which amplifies the differential phosphorylation of kinetochore substrates and increases the efficiency of error correction.
Results and Discussion
Proper chromosome segregation during cell division is essential to maintain genome stability. The centromere is the chromosomal locus that directs this process and is the site of formation in mitosis of the kinetochore that mediates attachment to the microtubule-based spindle [1, 2]. Prior to segregation, sister kinetochores are bound by microtubules emanating from opposite spindle poles (biorientation), which is achieved through a trial-and-error process. Correct kinetochore-microtubule attachments exert tension across the centromere and are stabilized, while those that lack tension are selectively destabilized by the action of the Aurora B kinase, which phosphorylates kinetochore targets such as the KNL-1/Mis12/Ndc80 complex (KMN) components to reduce microtubule binding [3–6]. The effectiveness of this trial and error process should depend on the magnitude of the kinetochore switch from phosphorylation to dephosphorylation, which determines the differential stability of correct and incorrect attachments. Current models for how this switch functions are based on the position of Aurora B, along with its binding partners in the chromosome passenger complex (CPC), at the inner centromere. The CPC localizes to the chromatin between sister kinetochores. Bi-oriented sister kinetochores are under tension and spatially separated from the kinase at the inner centromere. Therefore, even when kinase activity is constant, phosphorylation of kinetochore substrates is reduced to stabilize correct attachments [7]. This model is based on experiments in aneuploid cell lines, such as HeLa and U2OS, which may have a less effective error correction machinery compared to cells that maintain a normal chromosome complement.
Normal diploid cells have a more robust error correction machinery and enriched Aurora B at misaligned centromeres
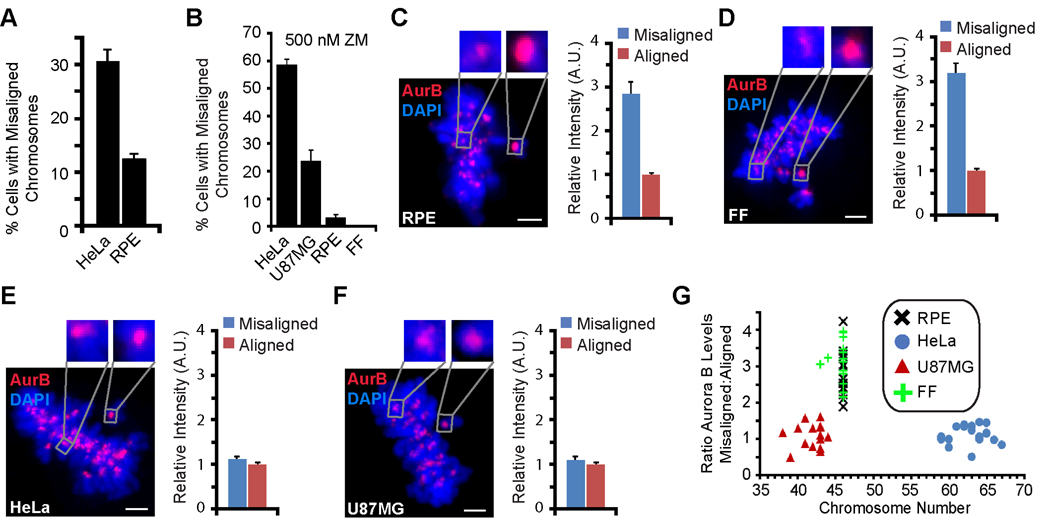
To compare the efficiency of error correction in different cell lines, we used an established assay to accumulate monopolar cells by reversible chemical inhibition of kinesin-5 using monastrol [8]. Such treatment generates a large number of attachment errors (i.e. both sister kinetochores attached to the single spindle pole), which are corrected when monastrol is removed and the spindle becomes bipolar. This error correction pathway requires Aurora B-mediated destabilization of incorrect attachments [9]. We measured the number of cells containing misaligned chromosomes 45 min after monastrol withdrawal and found that HeLa cells are greater than two times more likely to have misaligned chromosomes than diploid retinal pigment epithelial (RPE) cells (31%, HeLa; 12%, RPE) (Figure 1A). To test whether the Aurora B error correction pathway functions differently in these cell lines, we measured the sensitivity to partial Aurora B inhibition using a small molecule inhibitor of Aurora B kinase activity, ZM447439 (ZM) [10]. At 500 nM ZM, ~60% of HeLa cells contain misaligned chromosomes one hour after monastrol withdrawal as compared to only ~5% in RPE cells (Figures 1B and S1A–D). In addition, diploid primary fetal fibroblasts (FF) are insensitive to ~500 nM ZM, whereas this treatment causes aneuploid U87MG glioblastoma cells to have substantially more mitotic errors (Figure 1B and S1E). These results demonstrate that RPE and FF cells have a more robust, Aurora B-dependent error correction machinery compared to HeLa and U87MG cells.
Figure 1. Efficient mitotic error correction, resistance to Aurora B inhibition, and enrichment of Aurora B at misaligned centromeres in healthy, diploid cells but not in aneuploid cells.
(A) The percentage of HeLa and RPE cells containing misaligned chromosomes was quantified at 45 min following monastrol withdrawal. Error bars represent standard error of the mean.
(B) The percentage of each cell type containing misaligned chromosomes with an intermediate dose of ZM (500 nM) at one hour following monastrol withdrawal. Error bars represent standard error of the mean.
(C–F) Representative images of RPE (C), FF (D), HeLa (E), and U87MG (F) cells stained for Aurora B with misaligned chromosomes and a clearly discernable metaphase plate are shown. Insets show 4X magnified views of the boxed area. Quantitation of Aurora B levels is shown for each cell line at misaligned (blue) and aligned (red) chromosomes. Error bars represent standard error of the mean. Scale bars = 2 µm.
(G) Each cell line is plotted as Aurora B enrichment on the centromeres of misaligned chromosomes versus the chromosome number per cell (each data point represents a single cell).
Because of the importance of Aurora B localization for the error correction mechanism, we compared endogenous Aurora B staining in the diploid and aneuploid cell lines. Aurora B localizes to the inner centromere in all cases, but it is dramatically enriched (~three-fold) at misaligned centromeres compared to aligned centromeres in RPE and FF cells (Figure 1C,D). We found a similar enrichment in another diploid fibroblast cell line (Figure S1F), but not in HeLa or U87MG cells (Figure 1E,F). One possible cause for the difference in Aurora B behavior between aneuploid and diploid cell lines is a fixed pool size of Aurora B protein and a variable number of chromosomes in each cell. Indeed, the pool size of Aurora B protein has been reported to be similar between the HeLa, RPE, and U87MG cell lines [11], and we also found that FF cells have similar amounts of Aurora B protein (Figure S1G). Despite the similar levels of Aurora B found in all four of the cells lines we examined, both HeLa (super-diploid) and U87MG (pseudo-/sub-diploid) lack Aurora B enrichment on misaligned chromosomes that we observe in the diploid cell lines (RPE and FF)(Figure 1G). These findings strongly suggest that differences in chromosome number, or ‘chromosome load’, of each cell is not the basis for the absence of Aurora B enrichment on the centromeres of misaligned chromosomes in HeLa and U87MG cells.
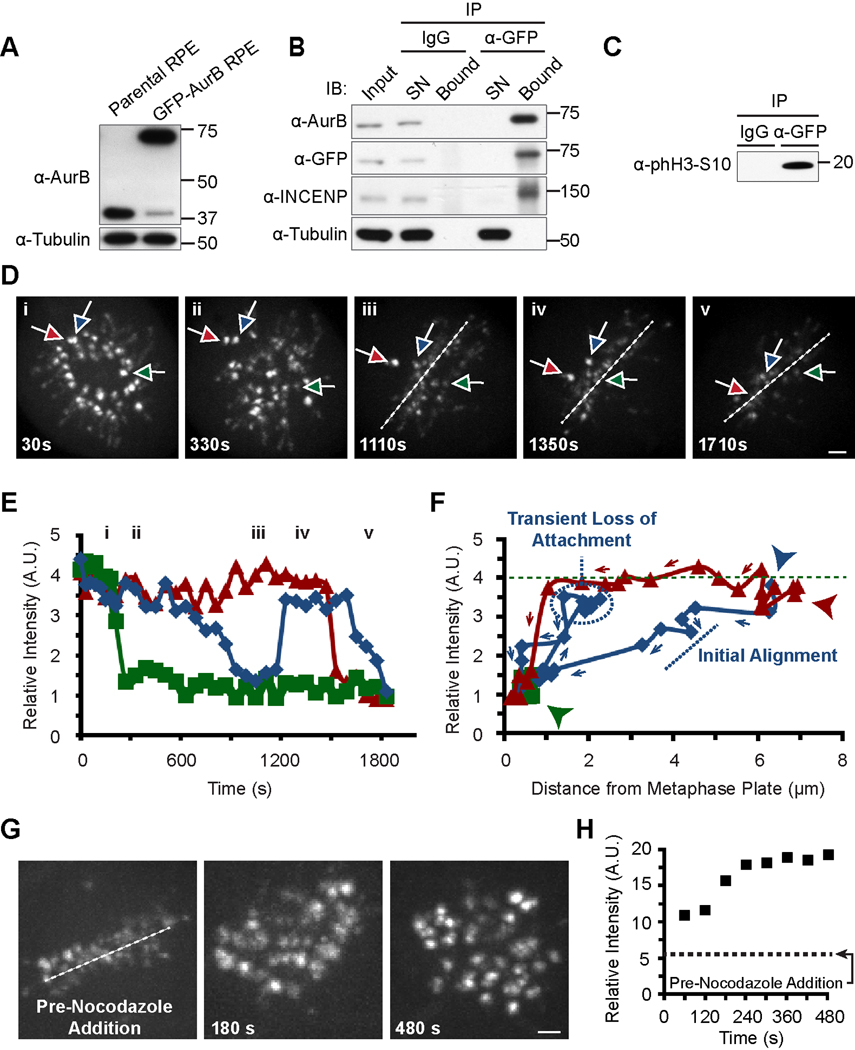
To study the dynamics of Aurora B recruitment to centromeres in relation to chromosome alignment status, we generated an RPE cell line stably expressing GFP-tagged Aurora B. Multiple lines of evidence suggest that the fusion protein is functional: 1) endogenous Aurora B is heavily down-regulated in the cells that express sufficient levels of GFP-Aurora B to replace the endogenous pool, so that the vast majority of Aurora B expressed is the GFP-tagged version (Figure 2A), 2) immunoprecipitation copurifies endogenous inner centromere protein (INCENP), the closest partner of Aurora B in the CPC [12, 13], to the extent that it depletes detectable INCENP from the cell lysate (Figure 2B), and 3) the immunoprecipitated Aurora B phosphorylates histone H3 on serine 10 (Figure 2C), a well known substrate for Aurora B [14]. We then used the monastrol washout assay because it allowed us to examine many examples of misaligned and aligned chromosomes simultaneously and to track Aurora B levels in real time as chromosomes align. We found a three- to four-fold enrichment of GFP-Aurora B at misaligned centromeres (Figure 2D–F), similar to our result for endogenous Aurora B in fixed cells (Figure 1C). By tracking individual centromeres, the live cell studies revealed a switch-like mechanism based on attachment status, where Aurora B levels sharply drop upon proper alignment (Figure 2D, Movie S1). Furthermore, the switch is reversible, as Aurora B levels drop when a centromere aligns, rise again when the same centromere loses attachment, evidenced by an excursion of ~2 µm from the spindle equator, and then drop again when the chromosome aligns a second time (Figures 2D [blue arrow] and 2E,F [blue data points]). We find a tight temporal coupling between alignment status (either initial alignment, subsequent misalignment, or realignment) and Aurora B enrichment on individual centromeres (Figures 2D–F). We also find a similar alignment-coupled reduction of Aurora B levels at mitotic centromeres in the absence of any chemical perturbation (Figure S2A,B; Movie S2). Furthermore, the loss of all stable kinetochore/microtubule attachments following the addition of the microtubule depolymerizing agent, nocodazole, leads to global Aurora B centromere enrichment within 1–4 minutes (Figures 2G,H and S2C; Movie S3).
Figure 2. Aurora B levels at centromeres drop upon chromosome alignment but rapidly increase if proper attachments are lost.
(A) Replacement of the vast majority of endogenous Aurora B in RPE cells with a stably expressed GFP-tagged version.
(B) Co-immunoprecipitation of GFP-Aurora B and endogenous INCENP. IB indicates immunoblot. SN indicates supernatant.
(C) GFP-Aurora B immunoprecipitation contains active Aurora B kinase that potently phosphorylates recombinant histone H3 on serine 10. Molecular weights (kDa) are indicated in panels A, B and C.
(D) Individual images from live cell imaging following monastrol washout in GFP-Aurora B-expressing RPE cells (see Movie S1). Three centromeres exhibiting different alignment kinetics are tracked with colored arrows. Time represents duration following withdrawal of monastrol. Dashed lines show the position of the metaphase plate that was first discernable at ~500 s following monastrol washout.
(E) Quantification of Aurora B levels over time at same three centromeres tracked in panel D using the same color scheme. Roman numerals indicate time points of still images shown in panel D.
(F) Plots of fluorescence intensity for each centromere versus distance from metaphase plate for each centromere with coloring as in panels D and E. The first timepoint shown in this panel is 510 s (indicated by the three arrowheads with colors corresponding to each chromosome), the first time point at which the metaphase plate is discernable. Note that the quickly aligning chromosome (green) has already reached the metaphase plate by 510 s and remains aligned throughout the remainder of data acquisition. The green dashed line indicates the initial fluorescence intensity of GFP-Aurora B on the quickly aligning chromosome. Arrows indicate the direction of the time dimension with data points representing each time increment measured (see panel E).
(G) Individual timepoints of a metaphase cell prior to and following (with the indicated timepoints) nocodazole addition (see Movie S3). Scale bar = 2 µm in panels D and G.
(H) Quantification of Aurora B levels at all centromeres from panel G, in aggregate. Aurora B levels prior to nocodazole addition are indicated with a dashed line.
CPC enrichment at misaligned centromeres amplifies the preferential phosphorylation of kinetochore substrates
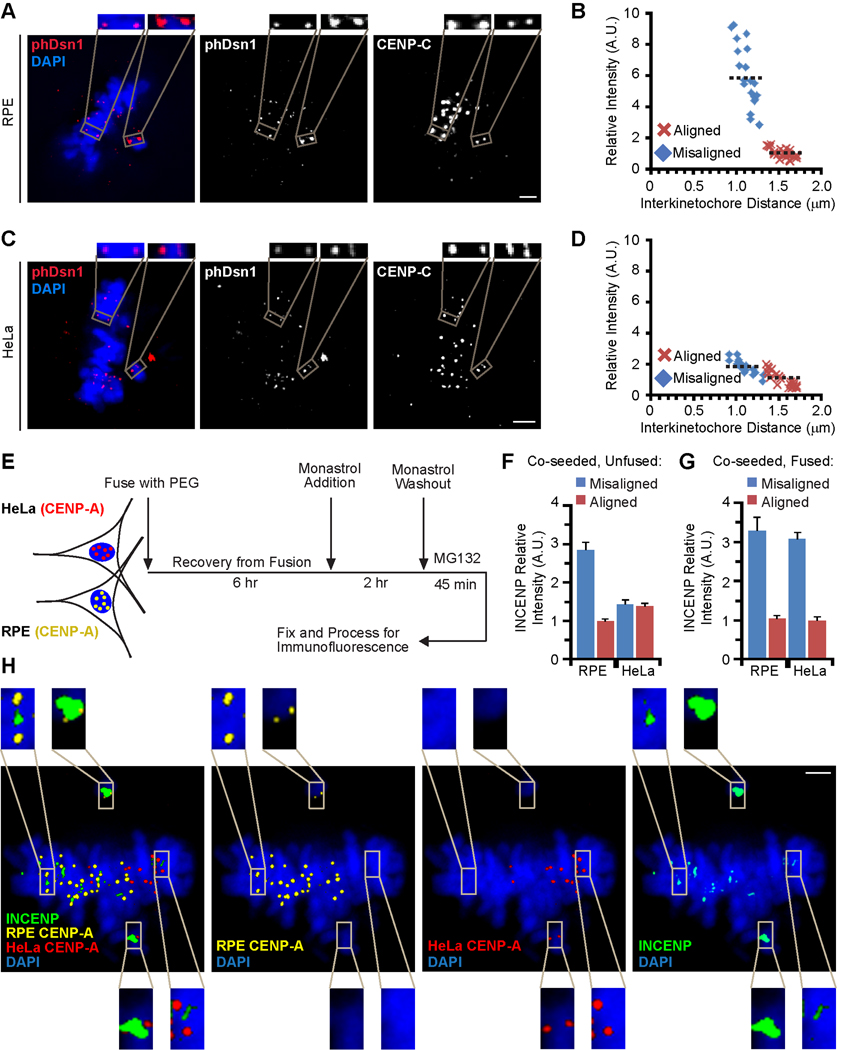
To test whether Aurora B enrichment at misaligned centromeres leads to increased phosphorylation of kinetochore substrates, we measured phosphorylation of Dsn1, a component of the KMN network and an established Aurora B substrate involved in microtubule binding [6], at Ser100. Using a previously characterized phospho-specific antibody [6], we measured the increase of phosphorylation on Dsn1 at Ser100 on misaligned kinetochores relative to aligned kinetochores at the metaphase plate after monastrol washout. The dynamic range of this differential phosphorylation is substantially greater in RPE cells (~6-fold; Figure 3A,B) than in HeLa cells using this assay (~2-fold; Figure 3C,D) or what was previously reported in unperturbed HeLa cells (1.4-fold; [6]). Taken together, these findings indicate that Aurora B recruitment amplifies the increased phosphorylation of kinetochore substrates at misaligned centromeres, which is a crucial part of the error correction mechanism.
Figure 3. Aurora B enrichment at misaligned chromosomes leads to increased phosphorylation of Dsn1 and is a dominant feature of RPE cytoplasm.
(A–D) RPE (panels A and B) and HeLa (panels C and D) cells were subjected to monastrol washout, fixed and stained for immunofluorescence. Representative images are shown in panels A and C and quantification is shown in panels B and D of levels of Dsn1 phosphorylated at Ser100. CENP-C is used as a kinetochore marker. In all cases, the relative intensity of the target at aligned chromosomes is normalized to 1 A.U. The insets are 2X magnified views of the boxed area. Black dashed lines (B and D) indicate the mean of relative intensity for each population.
(E) Scheme for cell fusion experiments starting with HeLa and RPE cells stably expressing CENP-A fusion proteins (HA-tagged in HeLa and YFP-tagged in RPE) that mark the cell line of origin for every chromosome in our analysis.
(F) Quantification of INCENP levels on adjacent co-seeded (but unfused) cells imaged on the same coverslip. Error bars represent standard error of the mean.
(G) Quantification of INCENP levels on misaligned and aligned chromosomes originating from the indicated cell line in fused RPE:HeLa cells. Error bars represent standard error of the mean.
(H) Image of a fused RPE:HeLa cell. The insets are 3X magnified views of the boxed area. Scale bars = 2 µm in panels A, C, and H.
The increased Dsn1 phosphorylation could be due solely to kinase enrichment on unaligned centromeres, or kinase activation could also contribute as suggested in some models. To test this possibility, we first generated a phospho-specific antibody against the C-terminal TSS motif of human INCENP (Figure S3A,B). This motif is both an Aurora B substrate and a crucial part of the mechanism of kinase activation [13, 15, 16], and is therefore a useful marker for kinase activation. We found that phospho-INCENP staining is enriched ~3-fold at misaligned centromeres of RPE cells (Figure S3C,E), consistent with Aurora B enrichment (Figure 1C). Total INCENP protein levels are also enriched to a similar extent (Figure S3D,F). The strongly correlating localization of INCENP and Aurora B is expected since the CPC is a single functional module where all four components transit together [16–18]. Furthermore, the quantitative similarity between INCENP and phospho-INCENP enrichment suggests that Aurora B is recruited to misaligned centromeres but not further activated.
Basal CPC recruitment to the inner centromere involves local chromatin modifications including phosphorylation of histone H3 on Thr3 by the haspin kinase [19–21] and phosphorylation of histone H2A on Thr120 by the Bub1 kinase [19, 22]. We find that phH3-T3 is not enriched on misaligned chromosomes of RPE cells that contain high levels of Aurora B (Figure S3G,J–L). On the other hand, phH2A-T120 is heavily enriched on the centromeres of misaligned chromosomes in RPE cells, corresponding to the centromeres with high levels of Aurora B (Figure S3H,M–O). Neither phH2A-T120 nor Aurora B is enriched on the centromeres of the misaligned chromosomes of HeLa cells (Figure S3I,P–R). The correlation between Aurora B levels and the amount of phH2A-T120 staining suggests a potential link between this particular chromatin modification and the specific recruitment of Aurora B to the centromeres of the chromosomes requiring its mitotic error correction activity.
CPC enrichment on the centromeres of misaligned chromosomes is dominant in fused cells
Our findings suggest either that changes to centromeres in aneuploid cells render them unable to enrich the CPC on misaligned chromosomes, or that aneuploid cells have lost a diffusible/exchangeable factor(s) that contributes to CPC enrichment. To distinguish between these possibilities, we fused RPE cells stably expressing YFP-CENP-A with HeLa cells stably expressing HA-CENP-A (Figure 3E). CENP-A in flies and humans is exclusively targeted to centromeres at mitotic exit and the G1 phase of the cell cycle [23, 24], so in all of the mitotic cells that we monitor within 9 hours of cell fusion, each centromere is loaded with a tagged CENP-A that indicates the cell line of origin on every chromosome. For cells that are co-seeded without inducing fusion, we measured INCENP levels on adjacent cells on the same coverslip and found enrichment on misaligned chromosomes only in the RPE cells (Figures 3F and S3S), mirroring our findings in earlier experiments that compared cells imaged on separate coverslips (Figure S3D,F). In fused cells, however, all centromeres showed equivalently robust recruitment of INCENP (Figure 3G,H) and Aurora B (Figure S3T), regardless of whether the chromosome originated from HeLa or RPE cells. Thus, the deficiency in HeLa cells in recruiting high levels of the CPC to misaligned chromosomes is ameliorated by the cytoplasm of a healthy, diploid RPE cell.
Plk1 and Aurora B activities are required for Aurora B enrichment
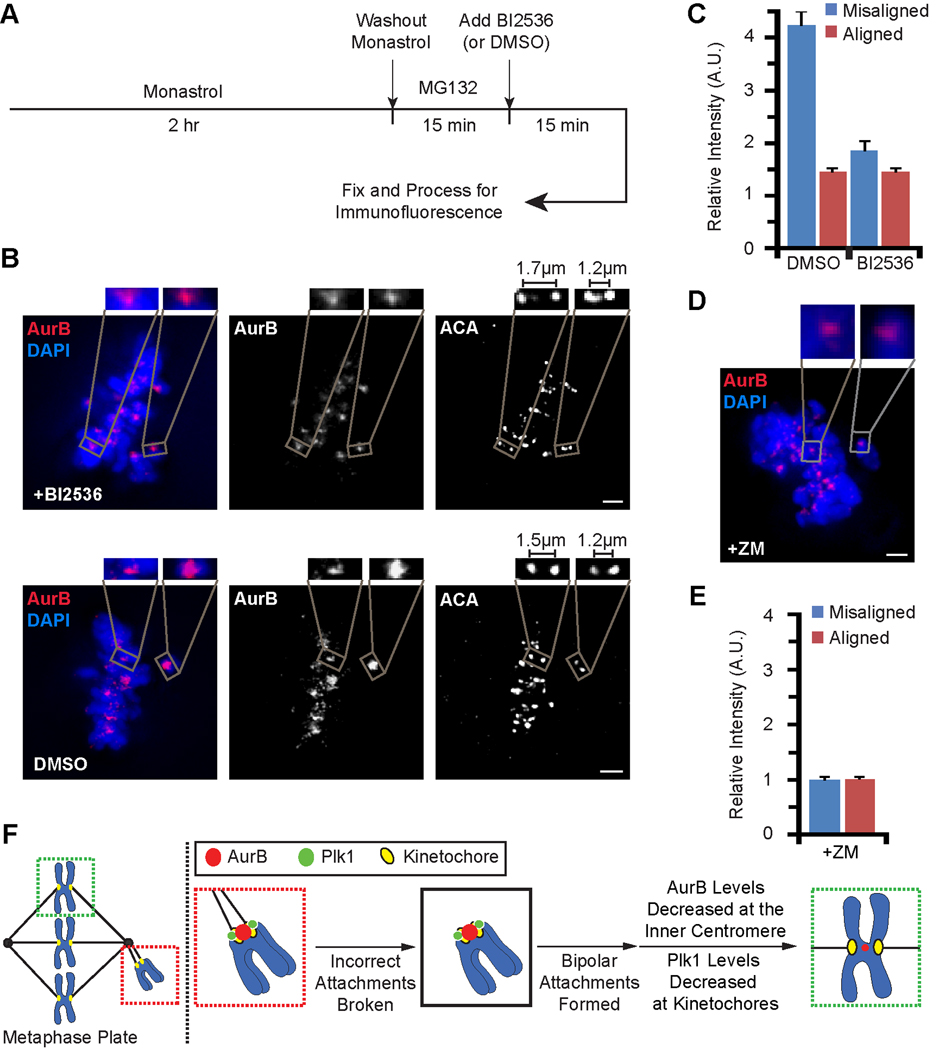
It is well established that Aurora B at the inner centromere signals to the outer kinetochore to regulate microtubule attachments. Our results suggest that there is also signaling in the opposite direction, as the kinetochore attachment state controls Aurora B recruitment to the inner centromere. Several kinetochore components are enriched at kinetochores early in mitosis and removed from each chromosome upon alignment at the spindle equator. We focused on kinases that exhibit this behavior, as potential regulators that might modulate Aurora B levels at the inner centromere. Both Plk1 and Mps1 kinases are attractive candidates because of their dynamic kinetochore localization [25–27] and known interactions with CPC components [28–31]. To test if either Plk1 or Mps1 is required for enrichment of Aurora B on misaligned centromeres, we took advantage of the temporal control possible with chemical inhibitors: BI2536 [32] for Plk1 and Reversine for Mps1 [31]. Using these inhibitors allowed us to examine only 15 min of inhibition in the monastrol washout assay (Figure 4A), a brief time window that minimizes effects on kinetochore microtubules, for example those that arise after prolonged BI2536 treatment [27, 33]. We compared aligned centromeres, which are stretched by bipolar attachment to the spindle (i.e., interkinetochore distances of 1.5–1.7 µm) to misaligned centromeres. Inhibition of Plk1 (Figure 4B) but not Mps1 (Figure S4A–D) largely eliminates the Aurora B enrichment on misaligned centromeres (Figure 4C). BI2536 treatment did not prevent H3-T3 or H2A-T120 phosphorylation at centromeres (Figure S4E,F), indicating that Plk1 inhibition does not lead to loss of the histone phosphorylations known to recruit the CPC.
Figure 4. Plk1 and Aurora B activity are each required for enrichment of Aurora B at misaligned centromeres.
(A) Scheme for testing a requirement for Plk1 activity in modulating the centromere enrichment of Aurora B.
(B) Immunofluorescence images of RPE cells subjected to scheme depicted in A. Interkinetochore distances measured from centroid-to-centroid of ACA staining. Insets are 2.5X magnified views of the boxed area. Line scan quantification and further enlargement of Aurora B staining of the vehicle-only control (dimethyl sulfoxide (DMSO)) are shown in Figure S4I.
(C) Quantification of the experiment in panel B. Error bars represent standard error of the mean.
(D and E) RPE cells were incubated in monastrol for two hours, followed by subsequent monastrol washout in the presence of the Aurora B kinase inhibitor ZM. Representative image and quantification of Aurora B levels are shown. Relative intensity at aligned chromosomes is normalized to 1 A.U. Insets are 4X magnified views of the boxed area. Error bars represent standard error of the mean. Scale bar = 2 µm for panels B and D.
(F) Diagram showing the targeting behavior of Plk1 and centromere enrichment behavior of Aurora B during error correction and subsequent chromosome alignment. See text for details.
Since Aurora B, itself, is proposed to be a key sensor of chromosome attachment status, we tested if its kinase activity is required for its own enrichment on misaligned chromosomes. An earlier report in X. laevis cultured cells found that centromeres with distorted Ndc80 foci, interpreted as merotelic attachments, recruit higher levels of Aurora B in a manner that is independent of Aurora B kinase activity [34]. We treated RPE cells with the Aurora B inhibitor ZM and found that Aurora B levels are no longer enriched at misaligned chromosomes in both fixed and living cells (Figures 4D,E and S4G,H; Movie S4). Thus, in normal diploid mammalian cells, Aurora B relies on its kinase activity to drive its own accumulation specifically at the centromeres that require error correction.
Conclusions
Our findings in normal diploid cells indicate that the Aurora B-based mechanism to destabilize erroneous connections between kinetochores and spindle microtubules utilizes dynamic modulation of the levels of the kinase in a chromosome autonomous fashion. The requirement for both Aurora B and Plk1 activity in the enrichment of Aurora B at the centromere suggests a model for positive feedback from kinetochores to the inner centromere (Figure 4F). Upon formation of bi-oriented kinetochore-microtubule attachments, Plk1 dissociates from the kinetochore and Aurora B levels rapidly drop to avoid destabilizing the new correct attachments. The rapid response of Aurora B levels to both chromosome alignment and then subsequent misalignment (Figure 2D–H) is reminiscent of the kinetochore autonomous enrichment of mitotic checkpoint components, such as Mad1 and Mad2, with removal upon formation of proper attachments but re-targeting to kinetochores if attachments are subsequently broken [25, 35, 36]. In the case of Aurora B, a basal pool of the kinase persists even upon proper chromosome biorientation (Figure 1C). We suggest that retention of a basal pool of the CPC serves three purposes. First, it is required for the structural integrity of the kinetochore [37]. Second, the well-established ‘passenger’ behavior of the CPC requires centromere localization of a fraction of the CPC to deliver Aurora B to the spindle mid-zone after anaphase onset, where it is needed in late mitosis to direct cytokinesis [38, 39]. Third, because Aurora B kinase activity is required for its own enrichment at misaligned centromeres (Figures 4C and 4D), a basal pool is probably required, since its complete removal upon biorientation would be incompatible with its re-enrichment in the instance that initial proper connections are lost and/or erroneous attachments are gained.
We favor a simple model for Aurora B sensing of erroneous attachments that utilizes two key properties of centromeric chromatin: 1) an increase in the spatial separation from the site of Aurora B enrichment from its kinetochore substrates that is only achieved upon proper chromosome biorientation [7] and 2) the dynamic regulation of the level of Aurora B kinase at centromeres that we describe in this study. The two properties work together to generate a very large dynamic range of Aurora B phosphorylation of kinetochore target sites (six-fold on Ser100 on Dsn1; Figure 3A,B). If spatial separation is prevented by artificial targeting of the kinase nearer to its kinetochore targets, Aurora B silencing is not possible [7]. If the regulation of Aurora B levels at centromeres is absent, the dynamic range of Aurora B activity at kinetochores is substantially narrowed (e.g. 1.9-fold change in phospho-Dsn1 between misaligned and aligned chromosomes in HeLa cells [Figure 3C,D; [6]] compared to 6-fold in RPE cells [Figure 3A,B]). The presence of the modulatory mechanism that adjusts Aurora B levels at each centromere correlates, in RPE and FF cells, with a higher efficiency of error correction and increased robustness to perturbations of Aurora B kinase activity (Figure 1). Normal diploid RPE cells rapidly progress from the onset of mitosis to sister-chromatid separation at the beginning of anaphase [40], relative to aneuploid cell lines such as HeLa cells. Our data supports the notion that the Aurora B feedback pathway that culminates in enriching the kinase at the centromeres of misaligned chromosomes is a key contributor to efficient mitoses that progress error free. By supplying high levels of the kinase only at the centromeres that require destabilization of kinetochore/spindle attachments, initial erroneous chromosome attachments are rapidly corrected. Upon correct attachment, the corresponding reduced levels of the kinase and the increased distance from its kinetochore targets switches the centromere to a mode that is stabilized until all chromosomes are properly aligned and the cell progresses to anaphase. Failure to regulate Aurora B levels may contribute to the increased aneuploidy frequently observed in cancer cells.
Highlights.
Healthy, diploid cells efficiently correct spindle/kinetochore attachment errors
The levels of Aurora B are dynamically modulated at each centromere during mitosis
Enrichment of Aurora B on misaligned chromosomes expands its dynamic range
The activities of Plk1 and Aurora B are each required for enrichment of Aurora B
Supplementary Material
Acknowledgements
We thank A. Choo, M. Soo, G. Dreyfuss, I. Cheeseman, and Y. Watanabe for the kind gifts of reagents. We also thank D. Liu and E. Wang for many helpful discussions throughout these experiments, Y. Watanabe and J. Higgins for advice with phosphospecific histone antibodies, D. Foltz and T. Stukenberg for suggesting experiments, and T. Yen and I. Cheeseman for comments on the manuscript. This work was supported by grants from the National Institutes of Health (GM082989 to B.E.B and GM083988 to M.A.L.), the Searle Scholars Program (to M.A.L.), a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund (to B.E.B.), and a Rita Allen Foundation Scholar Award (to B.E.B.). T.P. is supported by the University of Pennsylvania Structural Biology Training Grant (National Institutes of Health grant GM08275). K.J.S. is supported by a predoctoral fellowship from the American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 2.Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr. Opin. Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJR, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 4.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 5.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 6.Welburn JPI, Vleugel M, Liu D, Yates JR, Lampson MA, Fukagawa T, Cheeseman IM. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D, Vader G, Vromans MJM, Lampson MA, Lens SMA. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 9.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 10.Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr. Biol. 2009;19:1937–1942. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams RR, Eckley DM, Vagnarelli P, Wheatley SP, Gerloff DL, Mackay AM, Svingen PA, Kaufmann SH, Earnshaw WC. Human INCENP colocalizes with the Aurora-B/AIRK2 kinase on chromosomes and is overexpressed in tumour cells. Chromosoma. 2001;110:65–74. doi: 10.1007/s004120100130. [DOI] [PubMed] [Google Scholar]
- 13.Sessa F, Mapelli M, Ciferri C, Tarricone C, Areces LB, Schneider TR, Stukenberg PT, Musacchio A. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell. 2005;18:379–391. doi: 10.1016/j.molcel.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 14.Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H, Sen S, Allis CD, Sassone-Corsi P. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bishop JD, Schumacher JM. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B Kinase stimulates Aurora B kinase activity. J. Biol. Chem. 2002;277:27577–27580. doi: 10.1074/jbc.C200307200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honda R, Körner R, Nigg EA. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell. 2003;14:3325–3341. doi: 10.1091/mbc.E02-11-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolton MA, Lan W, Powers SE, McCleland ML, Kuang J, Stukenberg PT. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol. Biol. Cell. 2002;13:3064–3077. doi: 10.1091/mbc.E02-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gassmann R, Carvalho A, Henzing AJ, Ruchaud S, Hudson DF, Honda R, Nigg EA, Gerloff DL, Earnshaw WC. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 2004;166:179–191. doi: 10.1083/jcb.200404001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamagishi Y, Honda T, Tanno Y, Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330:239–243. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- 20.Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JMG. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330:231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K-ichiro, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 23.Jansen LET, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENPA and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 25.Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 2000;150:1233–1250. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahonen LJ, Kallio MJ, Daum JR, Bolton M, Manke IA, Yaffe MB, Stukenberg PT, Gorbsky GJ. Polo-like kinase 1 creates the tension-sensing 3F3/2 phosphoepitope and modulates the association of spindle-checkpoint proteins at kinetochores. Curr. Biol. 2005;15:1078–1089. doi: 10.1016/j.cub.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 27.Lénárt P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters J-M. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 28.Goto H, Kiyono T, Tomono Y, Kawajiri A, Urano T, Furukawa K, Nigg EA, Inagaki M. Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nat. Cell Biol. 2006;8:180–187. doi: 10.1038/ncb1350. [DOI] [PubMed] [Google Scholar]
- 29.Chu Y, et al. Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J Mol Cell Biol. 2010 doi: 10.1093/jmcb/mjq037. published online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jelluma N, Brenkman AB, McLeod I, Yates JR, Cleveland DW, Medema RH, Kops GJPL. Chromosomal instability by inefficient Mps1 auto-activation due to a weakened mitotic checkpoint and lagging chromosomes. PLoS ONE. 2008;3:e2415. doi: 10.1371/journal.pone.0002415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santaguida S, Tighe A, D’Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steegmaier M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr. Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 33.Peters U, Cherian J, Kim JH, Kwok BH, Kapoor TM. Probing cell-division phenotype space and Polo-like kinase function using small molecules. Nat. Chem. Biol. 2006;2:618–626. doi: 10.1038/nchembio826. [DOI] [PubMed] [Google Scholar]
- 34.Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr. Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 35.Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 36.Bomont P, Maddox P, Shah JV, Desai AB, Cleveland DW. Unstable microtubule capture at kinetochores depleted of the centromere-associated protein CENP-F. EMBO J. 2005;24:3927–3939. doi: 10.1038/sj.emboj.7600848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S-T, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooke CA, Heck MM, Earnshaw WC. The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J. Cell Biol. 1987;105:2053–2067. doi: 10.1083/jcb.105.5.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terada Y, Tatsuka M, Suzuki F, Yasuda Y, Fujita S, Otsu M. AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J. 1998;17:667–676. doi: 10.1093/emboj/17.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z, Loncarek J, Khodjakov A, Rieder CL. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat. Cell Biol. 2008;10:748–751. doi: 10.1038/ncb1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.