Abstract
Background.
Frailty is a late-life syndrome of vulnerability to adverse health outcomes characterized by a phenotype that includes muscle weakness, fatigue, and inflammatory pathway activation. The identification of biologically relevant pathways that influence frailty is challenged by its biological complexity and the necessity in separating disease states from the syndrome of frailty. As with longevity research, genetic analyses may help to provide insights into biologically relevant pathways that contribute to frailty.
Methods.
Based on current understanding of the physiological basis of frailty, we hypothesize that variation in genes related to inflammation and muscle maintenance would associate with frailty. One thousand three hundred and fifty-four single-nucleotide polymorphisms were genotyped across 134 candidate genes using the Illumina Genotyping platform, and the rank order by strength of association between frailty and genotype was determined in a cross-sectional study.
Results.
Although no single-nucleotide polymorphism reached study-wide significance after controlling family-wise false-discovery rate at 0.05, single-nucleotide polymorphisms within the 5-methyltetrahydrofolate-homocysteine methyltransferase (MTR), Caspase 8 (CASP8), CREB-binding protein (CREBBP), lysine acetyltransferase 2B (KAT2B), and beta-transducin repeat containing (BTRC) loci were among those strongly associated with frailty.
Conclusions.
The apoptosis– and transcription regulation–related pathways highlighted by this preliminary analysis were consistent with prior gene expression studies in a frail mouse model and provide useful etiological insights for future biological studies of frailty.
Keywords: Frailty, Candidate genes, Apoptosis
FRAILTY is a late-life syndrome of vulnerability to adverse health outcomes and mortality that is characterized by muscle weakness, weight loss, and fatigue (1,2). The biological basis of frailty is unknown, but data from population studies of community-dwelling older adults demonstrate strong associations between frailty and serum inflammatory mediators, such as interleukin-6, C-reactive protein, and white blood cell count, and between frailty and hormones important in muscle mass maintenance, including IGF-1, DHEA-S, and cortisol (3–5). Although studies show that skeletal muscle strength and inflammatory pathway activation are heritable traits (6,7), few studies have evaluated the genetic risk for frailty. Additionally, evidence for nonlinear effects of multisystem dysregulation underlying the vulnerability and phenotype of frailty has led to a search for potential factors that might initiate dysregulation in multiple systems (8). Processes that span multiple physiological systems and tissues including apoptosis, cellular senescence, and cellular homeostasis are increasingly being implicated in a number of frailty- and aging-related phenotypes (9,10). Given this biological plausibility, we initiated this study to explore the influence of candidate genetic variants on frailty.
METHODS
Population
Previously collected DNA and clinical information regarding frailty from the Women’s Health and Aging Studies (WHAS) I and II were utilized for this investigation (11,12). WHAS was designed to study the natural history and evolution of disability in a representative community-dwelling population of older women from the Baltimore, Maryland metropolitan area. The design of these studies and their combination into a single data set have been previously described (1,2,13). Frailty was measured using a previously validated 5-point scale that includes measurements of grip strength, walking speed, weight loss, fatigue, and physical activity questions (1,12). Women were evaluated at baseline for the following frailty measures: (a) shrinking, defined as either body mass index less than 18.5 kg/m2 or greater than 10% loss of body weight since age 60; (b) weakness, defined as the lowest quintile of grip strength of the dominant hand assessed with a hand-held JAMAR dynamometer (model BK-7498; Fred Sammons Inc, Burr Ridge, IL); (c) poor endurance and energy, defined as self-report of being either more tired or weaker than usual in the past 30 days; (d) slowness, defined as the lowest quintile in time to complete a 4-m walk; and (e) low activity level, defined by the lowest quintile of self-report of weekly activity determined by a subset of questions from the Minnesota Leisure Activity Questionnaire. Those who met three of five criteria were considered frail, those that met one or two were considered prefrail, and those with none were considered robust or not frail. Three hundred and forty-nine Caucasian women aged 70–79 years from the combined baseline WHAS I and II populations who had genomic DNA available and provided consent for genetic studies were included in the current cross-sectional study.
Candidate Gene Selection and Genotyping
A broad set of candidate genes was initially selected based on roles in the physiological systems most likely to influence frailty, namely skeletal muscle and inflammation. These genes were entered into a manually curated protein interaction database, Human Protein Reference Database (14). Proteins that represented important interaction points between the endophenotypic pathways were identified and added to the candidate list if greater than three interactions per seed protein were identified. A total of 134 genes were selected to move forward for single-nucleotide polymorphism (SNP) analysis. An initial list of SNP markers in each gene or within 10 kb flanking either side of the candidate genes was created from two sources, (a) all HapMap Phase 1–genotyped SNPs (public release #16) and (b) heterozygous validated SNPs from the dbSNP database (Human Genome Build 33), dbSNP Build 116). SNP selection occurred prior to the emergence of common SNP tagging tools and was thus carried out based on custom criteria and programming at the time as follows: SNP inclusion in this study was prioritized based on minor allele frequency (MAF > 0.05), Illumina design scores (>0.6), and coverage based on pairwise linkage disequilibrium estimates (LD) (D’ > 0.8) using the solid spine of LD definition in Haploview (15). SNP spacing was no more than 20 kb within LD blocks and no more than 5 kb outside LD blocks. Lastly, all available coding SNPs were included for each candidate gene. Tag SNPs were selected using HapMap CEU data to achieve the best possible coverage. Genotyping was performed on the Illumina custom GoldenGate 1536 SNP panel (Illumina, Inc., Foster City, CA).
Statistical Analyses
To determine the strength of association between each SNP marker and three-level frailty phenotype, an additive genetic model was assumed and multinomial logistic regression analyses adjusting for age were performed. A likelihood ratio test was implemented comparing the model with SNP genotype as a covariate (full model) to the model without SNP genotype (reduced model). To adjust for multiple comparisons, we controlled the family-wise false-discovery rate at 0.05 (16).
To determine whether frailty SNP associations were related to particular frailty components versus the syndrome itself, we also performed logistic regression analyses using each of the five binary components of frailty as the outcome variable in separate analyses while adjusting for age. The likelihood ratio test was used to compare the full model with the reduced model for each of the five components. We sought to identify variants that were common to all five components of frailty, assuming there are common underlying genetic pathways that contribute to frailty physiology. For each SNP, we used Fisher’s method (17) to combine the p values obtained from logistic regression using the five components of frailty as outcomes and obtained the p value for the combined test statistic.
RESULTS
Descriptive statistics for the 349 women genotyped are shown in Table 1. There are less than 2.3% of missing values occurred in the five components measurements and were treated as zero. After removal of 43 SNPs due to MAF < 0.01 in our sample and 23 SNPs due to deviation from Hardy–Weinberg equilibrium (p < .01), 1,354 SNPs remained for analysis. The average fraction of SNPs tagged per gene compared with Phase I HapMap data at r2 > .8 was 67% (range from 17% to 100%). The top 20 SNPs by strength of association with frailty are given in Table 2. Genes represented in this list include methionine synthase (MTR), Caspase 8 (CASP8), fibronectin (FN1), CREB-binding protein (CREBBP), glutathione transferase zeta 1 (GSTZ1), lysine acetyltransferase 2B (KAT2B), T-cell lymphoma invasion and metastasis 1 (TIAM1), signal transducer and activator of transcription 1 (STAT1), transcobalamin II (TCN2), beta-transducin repeat containing (BTRC), and vitronectin (VTN).
Table 1.
Descriptive Statistics for 349 Women’s Health and Aging Studies I and II Women
| Mean age, y | 74.2 |
| Number | |
| Nonfrail, n (%) | 32 (9.2) |
| Prefrail, n (%) | 165 (47.3) |
| Frail, n (%) | 152 (43.6) |
| Weakness, n (%) | 77 (22.6) |
| Slowness, n (%) | 98 (28.2) |
| Weight loss, n (%) | 38 (11.1) |
| Fatigue, n (%) | 54 (15.5) |
| Low physical activity, n (%) | 65 (18.9) |
Table 2.
Top 20 SNPs Associated With Frailty
| Gene (total SNPs genotyped) | SNP Marker | MAF | Odds Ratio (95% CI) | LRT, p Value |
| MTR (19) | rs10925235* | 0.36 (A) | 1.78 (0.97–3.27) | .0011 |
| rs2297967* | 0.36 (A) | 1.89 (1.03–3.49) | .0015 | |
| rs10802569* | 0.37 (G) | 0.56 (0.31–1.03) | .0024 | |
| rs4659725 | 0.36 (C) | 1.89 (1.03–3.47) | .0014 | |
| rs1770449 | 0.36 (G) | 0.52 (0.29–0.97) | .0015 | |
| rs1050993 | 0.36 (A) | 1.73 (0.96–3.15) | .0027 | |
| CASP8 (14) | rs3769827* | 0.46 (G) | 1.63 (0.94–2.81) | .0014 |
| rs6747918* | 0.49 (G) | 0.79 (0.46–1.36) | .0032 | |
| rs2037815 | 0.50 (G) | 0.77 (0.45–1.33) | .0037 | |
| rs6745051* | 0.50 (C) | 1.31 (0.76–2.25) | .0058 | |
| FN1 (15)* | rs7567647 | 0.25 (A) | 4.20 (1.69–10.39) | .0016 |
| CREBBP (17) | rs129968 | 0.35 (A) | 2.98 (1.48–5.99) | .0038 |
| GSTZ1 (7) | rs2287396 | 0.18 (A) | 0.49 (0.25–0.97) | .0046 |
| KAT2B (22) | rs2929408 | 0.17 (A) | 0.47 (0.23–0.93) | .0079 |
| TIAM1 (107) | rs2833383 | 0.23 (A) | 0.87 (0.47–1.60) | .0088 |
| STAT1 (10) | rs1400657 | 0.11 (C) | 3.01 (1.44–6.29) | .0089 |
| TCN2 (10) | rs740234 | 0.20 (G) | 0.36 (0.14–0.94) | .0100 |
| BTRC (33) | rs10883642 | 0.46 (A) | 0.48 (0.27–0.85) | .012 |
| FN1 (15) | rs10883631 | 0.46 (A) | 0.49 (0.28–0.86) | .014 |
| VTN (3) | rs2227729 | 0.07 (G) | 2.38 (1.13–5.01) | .013 |
Notes: BTRC = beta-transducin repeat containing; CASP8 = Caspase 8; CREBBP = CREB-binding protein; CI = confidence interval; FN1 = fibronectin; GSTZ1 = glutathione transferase zeta 1; KAT2B = lysine acetyltransferase 2B; LRT = likelihood ratio test; MAF = minor allele frequency; MTR = 5-methyltetrahydrofolate-homocysteine methyltransferase; SNPs = single-nucleotide polymorphisms; STAT1 = signal transducer and activator of transcription 1; TCN2 = Transcobalamin II; TIAM1 = T-cell lymphoma invasion and metastasis 1; VTN = vitronectin.
SNPs that likely represent the same signal within a gene due to linkage disequilibrium (r2 > .98).
In the MTR gene, 6 SNP markers are reported among top 20 SNPs, among these, rs10925235, rs2297967, and rs10802569 might represent the same signal due to LD (|r2| > 0.98). Similarly, among the four SNP markers within the CASP8 gene, rs3769827, and rs6747918 might represent the same signal due to LD (|r2| > 0.99). After adjusting for multiple comparisons using family-wise false-discovery rate at 0.05, none of these SNPs passed the study-wide significance level for association with frailty.
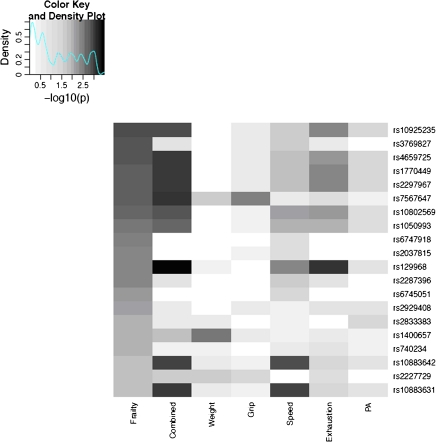
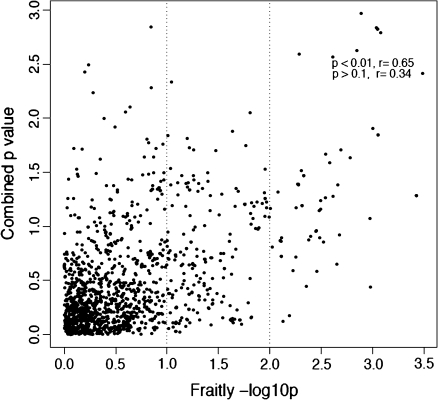
Associations between the 20 SNPs most associated with frailty and each of the five frailty components are shown in the heat map presented in Figure 1. These results illustrate that there is no single frailty component driving the genetic signals observed among each of the top 20 frailty SNP associations; indeed, when we combine the p values from each individual component, stronger genetic signals appear for most of these SNPs (Figure 1, column 2). In addition, the top ranking genetic signals from combined p values are consistent with the top association rankings for frailty and high correlation between the p values for frailty and for five-component combined p values is observed. As shown in the scatter plot (Figure 2), the correlation (r) for the two −log10 p values is .65 for the SNP markers that are highly associated with frailty (p value < .01). For the weaker SNP signals (p value > .1), we did not observe this consistency as the correlation decreases to .34. Although it seems that exhaustion status contributes the most significant signal to the combined p value, the same conclusion can be observed when we removed the p value of exhaustion status when pooling the p values (Supplementary Figure 1).
Figure 1.
Heat map of ranked association p values for single-nucleotide polymorphisms (SNPs) in Table 1. First column: frailty association p values; second column: combined p values of frailty component variables; subsequent columns: individual phenotypic components of frailty. Each row represents one SNP with darker shades indicating increased strength of association compared with lighter shades.
Figure 2.
Scatter plot of −log10 p value of individual single-nucleotide polymorphism association with frailty and combined p value.
DISCUSSION
In this study, we present genetic data from an exploratory candidate gene study in community-dwelling older women. Although the associations do not reach statistical significance after adjustment for multiple comparisons, the results support and are consistent with theory and phenotypic evidence that frailty is a clinical syndrome. It provides additional genetic support for the clinical theory and evidence reported in the literature that frailty is a syndrome which is a combination of these five components as a group and not driven by any one single component (1,2,18).
The syndromic aspects of frailty are supported by observations of association across multiple components and the consistent suggestive significant p values when the five frailty component p values are pooled together for the most frailty-associated SNPs. This is similar to other frailty-related evaluations where aggregate analyses are far more correlated with adverse outcomes than any single measure (2).
These findings also provide preliminary corroborative information on biological pathways that may be operant in frailty. Of the 134 candidate genes and 1,354 SNPS studied in this analysis, 11 genes were represented with the most strongly associated 20 SNPs. Of these genes, many are involved in apoptotic and transcription regulation pathways rather than inflammation per se. For example, Caspase 8 is a crucial protein that facilitates the later stages of apoptosis at the level of mitochondria (19). CREBBP plays important roles in homeostasis by coupling chromatin remodeling to transcription factor recognition and also acts as a scaffold to stabilize additional protein interactions related to inflammation, apoptosis, and many other transcription complexes (20). KAT2B is thought to regulate the balance between cell cycle arrest and apoptosis in hypoxia by modulating the activity and protein stability of both p53 and HIF-1alpha (21). Methionine synthase (MTR) facilitates the remethylation of homocysteine to methionine; reduced MTR activity has been associated with hypomethylation and increased DNA damage (22).
Given that this was a candidate gene study and that only genes that had strong rationale were included in the list, the top genes make biological sense. However, it is worth noting that none of the SNPs within inflammatory and muscle-related genes chosen initially for these analyses were among the top 20 associated SNPs. Rather, the findings were mostly in genes identified through the protein interaction analyses; these helped to identify genes that acted as bridges between pathways or were important hub proteins in both systems. Interestingly, the function of the highest ranking genes is related to the regulation of apoptosis, biosynthesis, and transcriptional regulation rather than inflammation and muscle maintenance per se. This is consistent with previous gene expression studies in the skeletal muscle of a frail mouse model that showed a significant upregulation in expression of genes related to apoptosis and a downregulation in genes related to biosynthesis and transcriptional regulation (23). In addition, other investigators have suggested that apoptosis and the development of cellular senescence is crucial to the development of frailty and aging processes and phenotypes across multiple tissues (9,10). Furthermore, Sharpless and DePinho (24) have recently reviewed data that suggest that heritable intrinsic events slow the renewal of stem cells and tissues and that apoptosis may accelerate the replicative decline of aging stem cells and tissues that underlie frailty and disease development . Although certainly not conclusive, our data provide preliminary genetic evidence that is consistent with emerging evidence that proteins in apoptotic pathways and in altered transcriptional and biosynthetic pathways may play a role in the development of frailty and late-life decline.
FUNDING
This project was funded by the National Institutes of Health, National Institute on Aging, grants R01 AG027236, by N01 AG12112 and R01 AG11703 for Women's Health and Aging Studies I and II, and by P30AG021334 Johns Hopkins Older Americans Independence Center.
SUPPLEMENTARY MATERIAL
Supplementary material can be found at: http://biomed.gerontologyjournals.org/
References
- 1.Bandeen-Roche K, Xue Q, Ferrucci L, et al. Phenotype of frailty: characterization in the Women's Health and Aging Studies. J Gerontol. 2006;61(3):260–261. doi: 10.1093/gerona/61.3.262. [DOI] [PubMed] [Google Scholar]
- 2.Fried LP, Tangen C, Walston J, et al. Frailty in Older Adults: evidence for a phenotype. J Gerontol Med Sci. 2001;56A(3):M1–M11. doi: 10.1093/gerona/56.3.m146. [DOI] [PubMed] [Google Scholar]
- 3.Leng S, Chaves P, Koenig K, Walston J. Serum interleukin-6 and hemoglobin as physiological correlates in the geriatric syndrome of frailty: a pilot study. J Am Geriatr Soc. 2002;50(7):1268–1271. doi: 10.1046/j.1532-5415.2002.50315.x. [DOI] [PubMed] [Google Scholar]
- 4.Varadhan R, Walston J, Cappola AR, Carlson MC, Wand GS, Fried LP. Higher levels and blunted diurnal variation of cortisol in frail older women. J Gerontol A Biol Sci Med Sci. 2008;63(2):190–195. doi: 10.1093/gerona/63.2.190. [DOI] [PubMed] [Google Scholar]
- 5.Walston J, McBurnie MA, Newman A, et al. Frailty and activation of the inflammation and coagulation systems with and without clinical morbidities: results from the Cardiovascular Health Study. Arch Intern Med. 2002;162:2333–2341. doi: 10.1001/archinte.162.20.2333. [DOI] [PubMed] [Google Scholar]
- 6.de Craen AJ, Posthuma D, Remarque EJ, van den Biggelaar AH, Westendorp RG, Boomsma DI. Heritability estimates of innate immunity: an extended twin study. Genes Immun. 2005;6(2):167–170. doi: 10.1038/sj.gene.6364162. [DOI] [PubMed] [Google Scholar]
- 7.Frederiksen H, Gaist D, Petersen HC, et al. Hand grip strength: a phenotype suitable for identifying genetic variants affecting mid- and late-life physical functioning. Genet Epidemiol. 2002;23(2):110–122. doi: 10.1002/gepi.1127. [DOI] [PubMed] [Google Scholar]
- 8.Fried LP, Xue QL, Cappola AR, et al. Nonlinear multisystem physiological dysregulation associated with frailty in older women: implications for etiology and treatment. J Gerontol A Biol Sci Med Sci. 2009;64:1049–1057. doi: 10.1093/gerona/glp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campisi J, Sedivy J. How does proliferative homeostasis change with age? What causes it and how does it contribute to aging? J Gerontol A Biol Sci Med Sci. 2009;64:164–166. doi: 10.1093/gerona/gln073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marzetti E, Leeuwenburgh C. Skeletal muscle apoptosis, sarcopenia and frailty at old age. Exp Gerontol. 2006;41(12):1234–1238. doi: 10.1016/j.exger.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Fried LP, Bandeen-Roche K, Chaves PH, Johnson BA. Preclinical mobility disability predicts incident mobility disability in older women. J Gerontol Med Sci. 2000;55(1):M43–M52. doi: 10.1093/gerona/55.1.m43. [DOI] [PubMed] [Google Scholar]
- 12.Guralnik JM, Fried LP, Simonsick EM, Kasper JD, Lafferty ME. The Women's Health and Aging Study: Health and Social Characteristics of Older Women with Disability. 95–4009. Bethesda, MD: National Institute on Aging; 1995. [Google Scholar]
- 13.Fried LP, Young Y, Rubin G, Bandeen-Roche K. Self-reported preclinical disability identifies older women with early declines in performance and early disease. J Clin Epidemiol. 2001;54(9):889–901. doi: 10.1016/s0895-4356(01)00357-2. [DOI] [PubMed] [Google Scholar]
- 14.Peri S, Navarro JD, Amanchy R, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13(10):2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–252. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 16.Storey JD. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Stat. 2003;31(6):2013–2035. [Google Scholar]
- 17.Fisher RA. Questions and Answers #14. Am Stat. 1948;2(5):30–31. [Google Scholar]
- 18.Fried LP, Hadley EC, Walston JD, et al. From bedside to bench: research agenda for frailty. Sci Aging Knowledge Environ. 2005;2005(31):e24. doi: 10.1126/sageke.2005.31.pe24. [DOI] [PubMed] [Google Scholar]
- 19.Kantari C, Walczak H. Caspase-8 and bid: caught in the act between death receptors and mitochondria. Biochim Biophys Acta. 2011;1813(4):558–563. doi: 10.1016/j.bbamcr.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 20.Tergaonkar V, Perkins ND. p53 and NF-kappaB crosstalk: IKKalpha tips the balance. Mol Cell. 2007;26(2):158–159. doi: 10.1016/j.molcel.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Xenaki G, Ontikatze T, Rajendran R, et al. PCAF is an HIF-1alpha cofactor that regulates p53 transcriptional activity in hypoxia. Oncogene. 2008;27(44):5785–5796. doi: 10.1038/onc.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishikawa H, Ishikawa T, Miyatsu Y, Kurihara K, Fukao A, Yokoyama K. A polymorphism of the methionine synthase reductase gene increases chromosomal damage in peripheral lymphocytes in smokers. Mutat Res. 2006;599(1–2):135–143. doi: 10.1016/j.mrfmmm.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Walston J, Fedarko N, Yang H, et al. The physical and biological characterization of a frail mouse model. J Gerontol. 2008;63(4):391–398. doi: 10.1093/gerona/63.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8(9):703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]