Abstract
We evaluated imaging plates (IPs) and the DITABIS Micron scanner for their use in recording images of negatively stained single-particle specimens and electron diffraction patterns of two-dimensional crystals. We first established the optimal imaging and read-out conditions for images of negatively stained single-particle specimens using the signal-to-noise ratio of the images as the evaluation criterion. We found that images were best recorded on IPs at a magnification of 67 000×, read out with a gain setting of 20 000 and a laser power setting of 30% with subsequent binning over 2 × 2 pixels. Our results show that for images of negatively stained specimens, for which the resolution is limited to ∼20 Å, IPs are a good alternative to EM film. We also compared IPs with a 2K × 2K Gatan charge-coupled device (CCD) camera for their use in recording electron diffraction patterns of sugar-embedded two-dimensional crystals. Diffraction patterns of aquaporin-0 recorded on IPs and with the CCD camera showed reflections beyond 3 Å and had similar RFriedel as well as Rmerge values. IPs can thus be used to collect diffraction patterns, but CCD cameras are more convenient and remain the best option for recording electron diffraction patterns.
Keywords: imaging plates, negative staining, single-particle electron microscopy, two-dimensional crystals, electron diffraction
Introduction
Photographic film is the traditional medium used to record images in electron microscopy (EM). To make such images available for computational processing, the negatives have to be removed from the microscope after exposure to the electron beam, chemically developed and fixed, and digitized with a scanner. For projects that require digital image processing, the use of photographic film is thus tedious and time-consuming, and the chemical processing of the negatives adds additional variation to the data. EM imaging has been revolutionized with the introduction of charge-coupled device (CCD) cameras, with which images can be recorded directly in digital format. The chips used in CCD cameras have become increasingly bigger over time. While 8K × 8K chips are now available, these are not widespread due to their high cost. Chip sizes of 2K × 2K and 4K × 4K are sufficient, however, for many applications, and CCD cameras have now become the predominant recording medium for the collection of electron tomographic tilt series and electron diffraction patterns. In contrast, the smaller size of images recorded with 2K × 2K or 4K × 4K CCD cameras as compared to photographic film is a drawback in single-particle EM studies, in which thousands or even tens of thousands of particles have to be imaged.
Imaging plates (IPs) have also been evaluated as an alternative to film and CCD cameras for recording EM images. IPs have initially been developed for medical applications, such as X-ray radiography [1], but were later also used to collect X-ray diffraction patterns in structural biology [2,3]. The first evaluation of IPs for EM imaging was reported over 20 years ago [4,5]; improved designs for read-out devices have been evaluated [6], and a commercial system has become available. IPs use a layer of tiny crystals to locally store high-energy radiation. The crystals, consisting of doped barium fluoride, are embedded in a blue resin. This energy-storing layer is sandwiched between a protective layer on top and a supporting polyester layer below. A thin metal plate provides mechanical strength for the IPs. When exposed to an electron beam, the electrons excite the crystals to a semi-stable state. While the image information due to the excitation of the crystals decays over the course of a few days, it is stable for several hours. By illumination with a highly focused red laser beam, the crystals can be excited again and stimulated to release the stored information as blue luminescence (390 nm). This signal is collected with special confocal mirror optics, detected by a photomultiplier tube, electronically amplified, and finally converted to a digital value. The intensity of the blue light directly correlates with the electron dose received during imaging under the electron microscope. Since both the recording and read-out processes are fully reversible, IPs can be used many times without deterioration of the image quality. Exposure to white light for ∼15 min causes all the stored excitation to be released and is done to regenerate the IPs between uses.
IPs have properties that make them an interesting alternative to EM film. As the excitation is stored within the crystal luminescence centers of the IPs, the number of available storage cells is virtually unlimited. IPs are thus never saturated under the imaging conditions used in EM and their dynamic range is only limited by the read-out device. The blue color of the resin absorbs the red laser light while perfectly transmitting the blue luminescence signal. These features, in combination with the highly focused laser beam, which has a spot diameter of ∼5 μm, yield highly localized information. According to the manufacturer, the sensitivity of IPs is also about 10 times higher than that of EM film, because each electron produces a number of photons (http://www.ditabis.de/iptech/iptech.html). The number of produced photons shows good linearity with the electron dose. The linearity and wide dynamic range of IPs make them a particularly promising medium for recording electron diffraction patterns of two-dimensional (2D) crystals.
IPs are used in the same way as photographic film. They are loaded into the camera, exposed in the electron microscope and then removed from the instrument. IPs do not have to be chemically developed, however, making them more environmentally friendly. In addition, IPs can be loaded into the camera in the presence of light, making them more convenient to use than EM film, and their life-time is only limited by physical damage, making them also a cheaper imaging solution than EM film. While IPs do not need to be developed, they have to be read out with a scanner, similar to the digitization of developed EM films. The image is thus not immediately available as in the case of CCD cameras, but the additional time needed to read out the IPs is somewhat compensated by the larger image area, approximately the same as that of EM film, particularly when compared to CCD cameras with only a 2K × 2K chip.
A very careful comparison of the properties of IPs with those of EM film and CCD cameras has already been published [7]. The intent of this paper is to share our findings in optimizing the practical use of the commercially available IP system from DITABIS Digital Biomedical Imaging System AG (Pforzheim, Germany). In particular, we describe our results in the use of the commercially available IPs to record images of negatively stained single-particle specimens and electron diffraction patterns of sugar-embedded 2D crystals. We also report the imaging and scanning conditions that gave the best results for such studies.
Materials and methods
Protein samples and specimen preparation
Human diferric transferrin–transferrin receptor (Tf–TfR) complex was prepared as described in [8] and the 20S proteasome from Thermoplasma acidophilum was prepared as described in [9]. The protein preparations were negatively stained as described in [10].
Double-layered aquaporin-0 (AQP0) 2D crystals were prepared as described in [11], except that the crystals were produced with E. coli polar lipids (Avanti, Alabaster, AL) rather than dimyristoyl phosphatidyl choline. Samples were prepared for EM by mixing crystals with an equal volume of 20% glucose and applying the mixture to a molybdenum grid covered with a thin carbon film. Grids were blotted to remove excess material, transferred to the electron microscope using a cryo-specimen holder, and cooled to liquid nitrogen temperature prior to data collection.
Electron microscopy and image processing
Images of negatively stained specimens were collected with an FEI Tecnai T12 electron microscope operated at an acceleration voltage of 120 kV. Images were recorded from the same specimen area on Kodak SO-163 film at a magnification of 52 000× and on IPs at magnifications of 30 000×, 67 000× and 110 000×. All images were recorded under strict low-dose conditions (15 electrons Å−2). Electron diffraction patterns of double-layered AQP0 2D crystals were collected with an FEI Tecnai F20 electron microscope equipped with a field emission gun and operated at an acceleration voltage of 200 kV as previously described [11]. Diffraction patterns of untilted 2D crystals were recorded for 30 s on IPs and with a 2K × 2K CCD camera (Gatan Megascan 795, 30 μm pixel size) using a camera length of 3000 mm and a selected area aperture of 70 μm.
Images recorded on EM film were developed for 12 min in a full-strength Kodak D-19 developer at 20°C. The IP system used in this study is commercially available from DITABIS Digital Biomedical Imaging System AG (Pforzheim, Germany). It consists of IPs that are loaded into the film holders and then into the EM camera, the DITABIS Micron scanner used to read out the data from the IPs (the scanner used in this study has a minimum step size of 15 μm) and a light box used to erase the IPs (supplementary Fig. 1, available at http://jmicro.oxfordjournals.org/). IPs were scanned with different combinations of gain and laser power settings to optimize the read-out conditions. The optimized gain and laser power settings were then used to evaluate the best combination of EM magnification and pixel binning.
Boxer, part of the EMAN software package [12], was used to interactively select the particles from the images taken on IPs at a magnification of 110 000×. The coordinates were then converted and used to select exactly the same particles from the images taken on IPs at different magnifications and the images taken on film. The SPIDER software package [13] was used to calculate class averages over 10 cycles of multi-reference alignment and K-means classification.
The performance of IPs compared to film was evaluated based on the signal-to-noise ratio (SNR) rather than the modulation transfer function (MTF) or detective quantum efficiency (DQE). The same approach was used before to evaluate the performance of a 4K × 4K CCD camera [14]. Curves of the SNR were generated using the program CTFIT, which is part of the EMAN package [12]. Briefly, all the boxed particles from an image were used to calculate an average power spectrum, and the circularly averaged Fourier intensity was plotted with the program CTFIT. In parallel, the contrast transfer function (CTF) curve was simulated by adjusting the background noise, amplitude contrast, defocus value, and experimental B factor to produce the best visual fit with the experimental curve [15]. The SNR was then calculated as
where CTF is the contrast transfer function, E is the estimated overall amplitude decay function (envelop function) for a given electron micrograph, N is the background (noise) level derived from the four parameters in CTFIT [12,15] and s is the spatial frequency. The envelop function E and the experimental factor B are related by the equation [15]
Only the peak after the first zero transition in the one-dimensional power spectrum was used to fit the simulated curve to the experimental data. Therefore, only this peak (in the resolution range between 22 Å and 14 Å) was considered for the comparison of SNR curves from different images. For all images (except images recorded on IPs at 30 000× without binning), the simulated curves could be fit well to the experimental data (supplementary Fig. 2, available at http://jmicro.oxfordjournals.org/), suggesting that the derived estimates for the SNRs should provide a reliable criterion for the image quality.
The electron diffraction patterns recorded with the CCD camera were processed using the program XDP [16] according to established protocols. Diffraction patterns recorded on IPs were recorded from the same sample using the same electron dose. IPs were scanned with the DITABIS Micron scanner using gain and laser power settings of 10 000 and 30%, respectively. The IPs were scanned using two data channels, which generate two 16-bit images with one image being read out with a 33-fold higher gain setting. The two images were then automatically combined into a single, high-dynamic-range 32-bit image. The scanned images with a size of 5744 × 5066 pixels, corresponding to the entire imaging area of the IP, were then cropped to 4096 × 4096 pixels and converted to a 16-bit image prior to processing in XDP. Intensities were extracted from ∼80 diffraction patterns recorded on each medium. The best 12 diffraction patterns of each data set (15%), as judged by their RFriedel values and data completeness, were merged without tilt angle refinement using the MRC programs [17]. After merging to 3 Å resolution, only the seven diffraction patterns (8.8%) with the lowest Rmerge values were kept in the final data set.
Results
Optimization of the read-out conditions
In addition to the pixel size used for digitization, which depends on the used scanner and is usually set to the smallest available size, two major parameters control the quality of images read out with the DITABIS imaging plate scanner: the gain setting, which determines how many counts are created for a given electron dose and the laser power setting. Scanning of IPs with a laser power setting of 100% will produce the highest signal. However, the reflected light will also reach its maximum value with the laser power set at 100%, causing an increase in the noise level of the images. Furthermore, according to the manufacturer, correction for the reflected light works better at lower laser power settings. Finally, gain and laser power settings affect each other. We therefore tested different gain and laser power settings to identify the combination that yields the highest quality of digital images, i.e. images with the highest SNR. As test images, we used images of the negatively stained Tf–TfR complex. The images were recorded on IPs with the same EM settings that we routinely use to collect images on photographic film (acceleration voltage of 120 kV, defocus of −1.5 μm, electron dose of 15 electrons Å−2). In our laboratory, we routinely collect images on film at a magnification of 52 000×, scan them with a step size of 7 μm with our Zeiss SCAI scanner and then average 3 × 3 pixels, resulting in a pixel size of 4.04 Å. To obtain a comparable pixel size with IPs, we recorded the images on IPs at a magnification of 67 000×, read them out with a step size of 15 μm (the smallest step available on our DITABIS Micron scanner) and binned the images over 2 × 2 pixels to produce a pixel size of 4.48 Å.
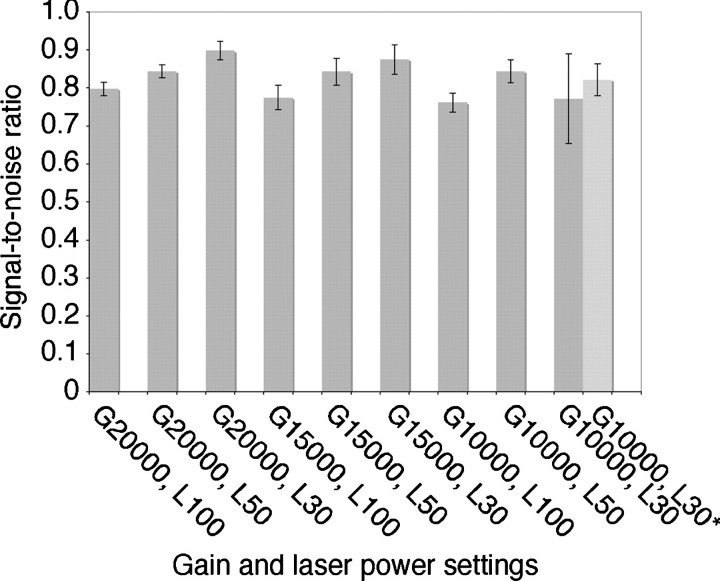
To optimize the scan parameters, we tested gain settings of 20 000, 15 000 and 10 000 and laser power settings of 100%, 50% and 30%. For each combination, five images were scanned using a single 16-bit data channel and used to calculate spectral SNR curves (representative curves are shown in supplementary Fig. 3, available at http://jmicro.oxfordjournals.org/). Figure 1 shows the SNR variations between sets of images read out with the various scan settings. For gain settings of 20 000 and 15 000, the best SNRs were consistently obtained with a laser power setting of 30%, while the best SNR for a gain setting of 10 000 was obtained with a laser power setting of 50%. One of the images read out with a gain setting of 10 000 and a laser power setting of 30% showed an unusually low SNR. Including this image in the calculation resulted in a lower mean SNR and a larger standard deviation (Fig. 1, G10000 + L30). These values became more comparable to the other conditions, however, when this particular image was excluded from the calculation (Fig. 1, G10000 + L30*). Comparison of the mean SNRs shows that under our imaging conditions, the highest SNR was obtained with the highest gain and the lowest laser power that we tested (Fig. 1, G20000 + L30). Based on these results, a gain of 20 000 and a laser power of 30% were chosen as the default settings for reading out IP images of negatively stained single particles.
Fig. 1.
Influence of the gain and laser power settings used to read out IPs on the SNR of the images. Each column represents the mean SNR for sets of five images scanned with the same combination of gain (G) and laser power (L) settings. The error bars indicate the standard deviations. One of the five images read out with a gain setting of 10 000 and a laser power setting of 30% showed a very low SNR compared to the other images in the same group, resulting in an unusually high standard deviation (G10000, L30). Removal of this image reduced the standard deviation (G10000, L30*).
Optimization of imaging magnification and pixel binning
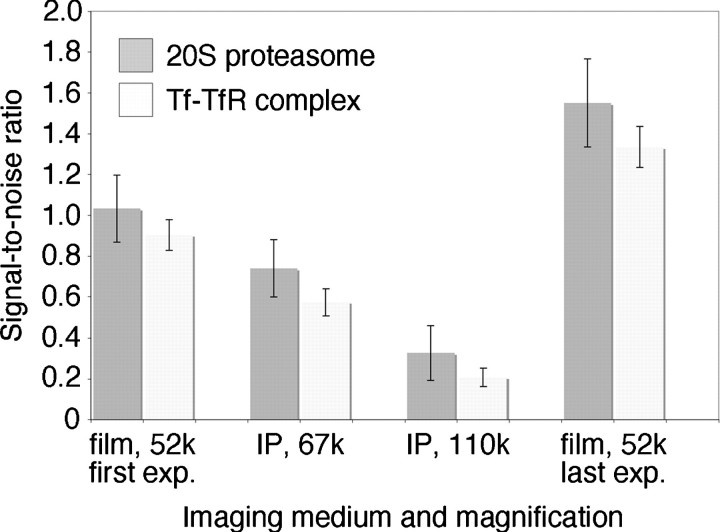
As stated above, we typically record images of negatively stained specimens with a Tecnai T12 electron microscope (Cs = 2 mm) at an acceleration voltage of 120 kV and a magnification of 52 000× with a defocus value of about −1.5 μm. These conditions bring the first zero transition of the CTF to a spatial frequency of about 1/(20 Å), which is approximately the resolution limit introduced by the negative stain. The negatives are then scanned with a Zeiss scanner using a step size of 7 μm, and 3 × 3 pixels are subsequently averaged to produce a final pixel size of 4.04 Å on the specimen level. To compare the quality of the IP images in terms of SNR, we chose the EM magnification and binning that would generate approximately the same final pixel size. The EM magnification and binning we chose were 30 000× with no binning (pixel size of 5.0 Å), 67 000× with 2 × 2 binning (pixel size of 4.48 Å) and 110 000× with 3 × 3 binning (pixel size of 4.09 Å) (Table 1). As test specimens, we used negatively stained 20S proteasome and Tf–TfR complex. First, an image was taken on photographic film at 52 000×, then the same specimen area was imaged on the IP at magnifications of 30 000×, 67 000× and 110 000×, before imaging the specimen again on photographic film at 52 000× (photographic film and IPs were loaded into the camera in this order). After scanning and binning of the images, particles were interactively selected from the images taken at 110 000× on the IP (5096 Tf–TfR complex particles from 20 images and 1190 20S proteasome particles from 22 images). These coordinates were converted to select the same particles from the images taken under other imaging conditions. The particles in each data set were aligned and classified into 50 classes. Supplementary Fig. 4 (available at http://jmicro.oxfordjournals.org/) shows representative image areas and Fig. 2 shows class averages obtained under the different imaging conditions. Visually, there is little difference between the different imaging conditions, with the exception of the IP images recorded at 30 000× without binning. Under these imaging conditions, the raw images appeared blurrier and the averages showed less clear fine structure. To assess the quality of the images in a more quantitative way, we calculated spectral SNR curves for all the images, except for those taken at 30 000× with no binning (representative SNR curves are shown in supplementary Fig. 5, available at http://jmicro.oxfordjournals.org/). The noise level in these images was so high that no signal was discernable after the first zero transition in the one-dimensional power spectrum, which made it impossible to reliably fit CTF curves and to determine the SNRs. Figure 3 shows the mean SNRs and standard deviations for the sets of recorded images. Under the used imaging conditions, both data sets recorded on photographic film, the first and the last images taken in each series, show the best SNR values, with the last images displaying a better SNR than the first images. Comparison of the images recorded on IPs show that images taken at a magnification of 67 000× with 2-fold binning have significantly better SNRs than those taken at 110 000× with 3-fold binning.
Table 1.
Imaging conditions
| Imaging | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| sequence | |||||
| Recording | EM film | IP | IP | IP | EM film |
| medium | |||||
| Magnification | 52 000× | 30 000× | 67 000× | 110 000× | 52 000× |
| Scanning step | 7 | 15 | 15 | 15 | 7 |
| size (μm) | |||||
| Binning | 3 × 3 | – | 2 × 2 | 3 × 3 | 3 × 3 |
| Pixel size of the | 4.04 | 5.00 | 4.48 | 4.09 | 4.04 |
| final image (Å) |
Fig. 4.
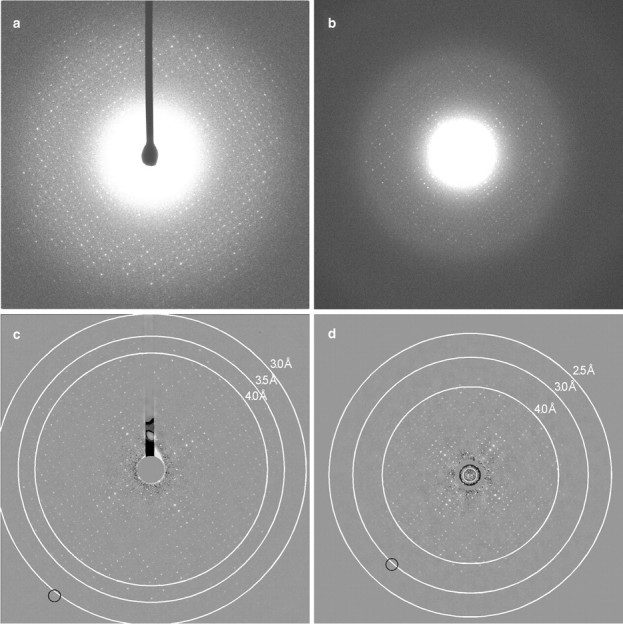
Electron diffraction patterns of double-layered aquaporin-0 2D crystals recorded on IPs and with a CCD camera. (a) Electron diffraction pattern recorded on a Gatan 2K × 2K CCD camera using a beam stop. (b) Electron diffraction pattern recorded on an IP without beam stop. (c) Electron diffraction pattern shown in panel (a) after background subtraction. The (9, 20) reflection is circled (∼3.0 Å). (d) Electron diffraction pattern shown in panel (b) after background subtraction. The (3, 23) reflection is circled (∼2.9 Å).
Fig. 2.
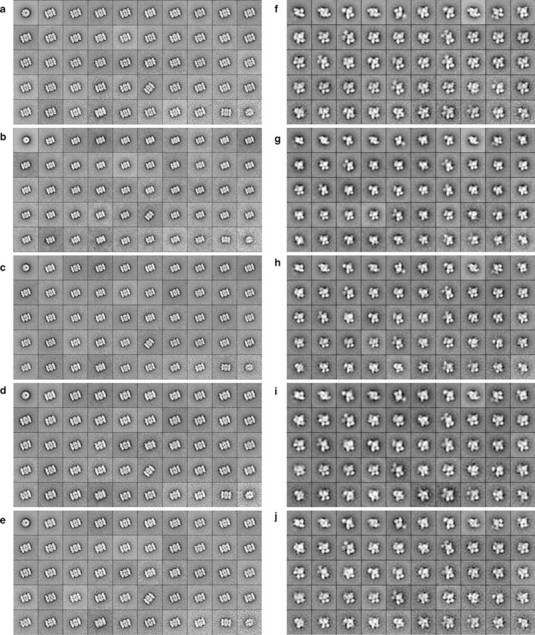
Class averages obtained with images of the same specimen areas recorded on film and on imaging plates at different magnifications and using different binnings. (a–e) 20S proteasome recorded first on film at 52 000× with a binning of 3 (a), then on IPs at 30 000× without binning (b), then on IPs at 67 000× with a binning of 2 (c), then on IPs at 110 000× with a binning of 3 (d) and finally on film again at 52 000× with a binning of 3 (e). The side length of the individual class averages is 38.8 nm (a and e), 48 nm (b), 43 nm (c) and 39.3 nm (d). (f–j) Tf–TfR complex recorded first on film at 52 000× with a binning of 3 (f), then on IPs at 30 000× without binning (g), then on IPs at 67 000× with a binning of 2 (h), then on IPs at 110 000× with a binning of 3 (i), and finally on film again at 52 000× with a binning of 3 (j). The side length of the individual class averages is 25.9 nm (f and j), 32 nm (g), 28.7 nm (h) and 26.2 nm (i). All the class averages in panels (a)–(e) and panels (f)–(j) contain the same particles. The particles appear bright on a dark background.
Fig. 3.
Influence of the imaging conditions on the SNR of the images. The columns represent the mean SNRs of images of 20S proteasome and Tf–TfR complex obtained under the various imaging conditions. The error bars indicate the standard deviations. The data are presented in the order in which the data were acquired: first on film at 52 000× with a binning of 3 (film, 52k, first exp.), then on IPs at 67 000× with a binning of 2 (IP, 67k), then on IPs at 110 000× with a binning of 3 (IP, 110k) and finally on film again at 52 000× with a binning of 3 (film, 52k, last exp.). Images taken on IPs at 30 000× without binning were too noisy to be analyzed and are therefore not shown in this graph.
Electron diffraction
The quality of electron diffraction patterns recorded on IPs and with a CCD camera were compared using double-layered AQP0 2D crystals as a test specimen. Similar crystals have previously been used to determine the atomic structure of AQP0 [11,18]. The patterns recorded with the CCD camera were 2048 × 2048 pixels in size, the dimensions of the actual CCD chip, and a beam stop was used to prevent damage to the CCD chip by the direct beam. A representative diffraction pattern collected with the CCD camera is shown in Fig. 4a. With a camera length of 3000 mm, the edge of the diffraction pattern corresponds to a resolution of ∼3 Å (Fig. 4c). In the case of IPs, the scanner rather than the size of the plates determines the dimensions of the read-out diffraction patterns. With a step size of 15 μm, the size of a diffraction pattern is 5744 × 5066 pixels. Although we cropped the diffraction patterns to 4096 × 4096, the maximum file size that can be processed with current software, the size of the diffraction pattern recorded on IPs was four times the size of those recorded with the 2K × 2K CCD camera. Since IPs cannot be oversaturated, no beam stop was used to record diffraction patterns on IPs. A representative diffraction pattern collected on an IP is shown in Fig. 4b. With a camera length of 3000 mm, the edge of the diffraction pattern corresponds to a resolution of ∼2.5 Å (Fig. 4d).
After background subtraction, diffraction spots beyond 3 Å resolution could be seen in diffraction patterns recorded both on IPs and with the CCD camera (circles in Fig. 4c and d). The merging statistics from the best seven diffraction patterns are presented in Table 2. The mean RFriedel and Rmerge values are similar for the patterns recorded on the two media (10.8% and 22.9% for the CCD camera and 10.5% and 23.3% for IPs, respectively). However, the Rmerge values vary over a much larger range for diffraction patterns recorded on IPs, suggesting that there may be more noise in diffraction patterns recorded on IPs. In addition, although part of the diffraction pattern is obstructed by the beam stop in diffraction patterns collected with the CCD camera, ∼100 (∼19%) more reflection intensities could be extracted from these patterns than from those recorded on IPs.
Table 2.
Merging statistics for electron diffraction patterns recorded on IPs and with a 2K × 2K CCD camera
| Diffraction pattern | Rmerge | RFriedel | Reflections |
|---|---|---|---|
| indexed | |||
| IP000008 | 0.179 | 0.104 | 551 |
| IP000034 | 0.213 | 0.087 | 604 |
| IP000042 | 0.230 | 0.103 | 497 |
| IP000049 | 0.214 | 0.114 | 504 |
| IP000050 | 0.241 | 0.099 | 524 |
| IP000071 | 0.293 | 0.120 | 521 |
| IP000075 | 0.283 | 0.110 | 529 |
| IP average | 0.232 | 0.105 | 533 |
| Standard deviation | 0.040 | 0.010 | 35.9 |
| CCD000006 | 0.217 | 0.086 | 713 |
| CCD000041 | 0.234 | 0.128 | 698 |
| CCD000042 | 0.217 | 0.109 | 640 |
| CCD000043 | 0.214 | 0.164 | 570 |
| CCD000044 | 0.256 | 0.095 | 638 |
| CCD000051 | 0.230 | 0.103 | 609 |
| CCD000052 | 0.229 | 0.077 | 582 |
| CCD average | 0.229 | 0.108 | 636 |
| Standard deviation | 0.015 | 0.027 | 54.5 |
Discussion
Based on the superior properties of IPs stated by the manufacturer, IPs would have the potential to be ideal for collecting electron microscopic data. Our results indicate, however, that this is not the case under the imaging conditions we used in this study. Nevertheless, under the appropriate imaging and read-out conditions, images of negatively stained single particles taken on IPs are of a quality comparable to that of images recorded on photographic film.
Use of IPs for imaging negatively stained single particles
Under the conditions we use to image negatively stained single particles, images recorded on film and digitized and binned to yield a pixel size of 4.04 Å had better SNRs than any of the images recorded on IPs and read out and binned to yield a comparable pixel size (Fig. 3).
Although negatively stained specimens are not as sensitive to radiation damage as unstained specimens, we were concerned that the multiple exposures of the same specimen area that was needed to evaluate the various recording conditions might influence the results. We therefore collected images on film twice; one image was taken as the first exposure of the specimen area and one as the last (fifth) exposure. The class averages obtained with the particles selected from the last images appeared to have slightly more contrast but to display less fine structure (compare Fig. 2e with a and Fig. 2j with f). The loss in fine structure may be the result of beam damage, whereas the slight increase in contrast may reflect beam-induced redistribution of the stain [19]. The increase in contrast in the class averages is also reflected in the SNR analysis, which showed that the images taken last had higher SNRs than those taken first in an image set of the same specimen area (Fig. 3). The increase in the SNR with accumulated electron dose may not only be due to stain redistribution but may also reflect other effects, such as the burning off of impurities or increasing stability of the specimen holder over time. Regardless of the reason, the increase in the SNR between the first and last exposure recorded on film proves that the lower SNRs seen in images recorded on IPs (two to four exposures) does not result from the accumulated electron dose or the resulting increase in beam damage.
Images of the 20S proteasome have consistently higher SNRs than those of the Tf–TfR complex (Fig. 3). This difference may be the result of the different image sizes (the larger proteasomes were windowed into 96 × 96 pixel images, whereas the smaller Tf–TfR complexes were windowed into 64 × 64 pixel images) and/or a difference in the ratio between the particle and carbon film. Despite this difference, the SNRs were consistent with respect to the imaging conditions with the images on film having the highest SNRs, followed by the images recorded on IPs at 67 000× with 2 × 2 binning and the images recorded on IPs at 110 000× with 3 × 3 binning (the SNR of images recorded on IPs at 30 000× without binning could not be determined).
We determined that the best conditions to image negatively stained particles is a magnification of 67 000× combined with a binning of 2 × 2 pixels, but the SNR of these images is still not as good as that of images recorded on film. For samples with a low inherent contrast, such as single particles in vitrified ice, for which the image contrast has to be as high as possible, IPs do not appear to be a viable replacement for EM film. For samples with a high inherent contrast, however, a small decrease in the image contrast is tolerable. IPs can thus be used instead of EM film to record images of negatively stained specimens, especially since the negative stain limits the achievable resolution to ∼20 Å in any case. The comparable quality of the class averages obtained from images recorded on film and IPs corroborates this conclusion (compare Fig. 2c with a and Fig. 2h with f). Indeed, we have now calculated 2D averages and 3D reconstructions of several negatively stained molecules using data collected on imaging plates [20,21]. Most notably, a single-particle 3D reconstruction of human interferon α2 bound to the ectodomains of its two receptors ifnar1 and ifnar2, a complex only ∼100 kDa in size, resolved all the individual domains and made it possible to interpret the density map based on known atomic structures of the subunits [22]. These results confirm that images recorded on IPs are suitable for single-particle EM studies of negatively stained specimens.
It is worth to note that the currently available CCD cameras produce images with an excellent SNR that is comparable to that of images recorded on photographic film, even at a resolution of half the Nyquist frequency [14]. Although we did not directly compare the quality of images recorded with IPs and a CCD camera, comparison of our data with data published by Booth and co-workers [14] indicates that images recorded with a CCD camera have a better SNR than images recorded on IPs. The total number of pixels of a 4K × 4K CCD image is ∼60% of those in an IP image, which means that two 4K × 4K CCD images will contain the same number of particles as one IP image.
Use of IPs for collecting electron diffraction patterns
Our analysis of electron diffraction patterns collected on IPs and with a 2K × 2K CCD camera shows that the quality of the two data sets is very similar (Table 2). We did not optimize the read-out settings for electron diffraction patterns recorded on IPs. We read the IPs out with laser power and gain settings of 30% and 10 000, respectively, but we used the two-channel mode. In this mode, a second channel reads out another image with a 33-fold higher gain. The two 16-bit images are then automatically combined into a single, high-dynamic-range 32-bit image, ensuring that spots ranging from very weak to very strong intensity are properly read out.
The number of pixels available on the detector directly influences the resolution that can be accurately measured during data collection and processing. The maximum recorded resolution can be increased by decreasing the camera length of the microscope. The size of the array, however, is fixed resulting in decreased spacing between adjacent reflections. Proper spacing between reflections is necessary to accurately measure their intensities, particularly when they are smeared as is often the case in diffraction patterns of highly tilted samples. 2D crystals of AQP0 have a unit cell of a = b = 65.5 Å, which we have found allows accurate data collection to ∼3 Å using a 2K × 2K CCD camera [11] (Fig. 4c). Diffraction patterns of AQP0 2D crystals collected on IPs with similar spacing between reflections allowed data collection to better than 2.5 Å due to the larger number of pixels (Fig. 4d). This batch of 2D crystals did not appreciably diffract beyond 3 Å, so the size of the detector was not an impediment to data collection. In the case of crystals that diffract to higher resolution, such as previous batches of AQP0 2D crystals [18] or those that have a large unit cell (>100 Å), a 2K × 2K CCD camera would be insufficient for data collection. In this case, IPs could be helpful if a CCD camera with a larger chip is not available.
In contrast to CCD cameras, IPs do not require a beam stop to be used for the recording of diffraction patterns, which blocks out the low-resolution reflections and a band of reflections through the entire resolution range (compare Fig. 4a with c). Furthermore, the high linearity of the IPs should make it possible to also precisely read out the very high intensities of the very low resolution reflections. We therefore expected to measure more reflections from diffraction patterns recorded on IPs than from those recorded with the CCD camera. We found, however, that the opposite was true and that we measured about 19% fewer reflections from diffraction patterns recorded on IPs than from those recorded with the CCD camera (Table 2). One reason is that we were not able to reliably measure the intensities of the very low resolution reflections due to the very asymmetric background at low resolution caused by the inelastic scattering. This issue resulted in large differences in the intensities between Friedel pairs. The very low resolution spots, although not blocked out by the beam stop, thus had to be excluded from further processing. In addition, the CCD camera was normalized prior to data acquisition, while each of the individual IPs might have slight local differences that are not removed prior to data acquisition. Such local differences would result in differences in the intensities of Friedel pairs, which would then be removed from further processing.
Concluding remarks
Considering the convenience of collecting images with a CCD camera, a 4K × 4K CCD camera may be the best choice to collect images of negatively stained specimens for single-particle analysis. Nevertheless, using IPs is more efficient than using a 2K × 2K CCD camera, and, while delivering images of comparable quality, IPs are less expensive and more environmentally friendly than photographic film. IPs can also be used to record electron diffraction patterns, but fewer reflections can be measured from each pattern compared to patterns recorded with a CCD camera and since IPs have to be read out, CCD cameras are more convenient for data collection. CCD cameras thus remain the ideal recording medium for electron diffraction patterns.
Because the relative performance of film and IPs varies with different acceleration voltages and electron doses [7,23], we should point out that our results only apply to the recording conditions described in this manuscript.
Funding
National Institutes of Health (RO1 EY015107, RO1 GM082927 to T.W., PO1 GM62580 to S.C. Harrison); Giovanni Armenise Harvard Center for Structural Biology.
Supplementary data
Supplementary data is available at http://jmicro.oxfordjournals.org.
Acknowledgments
T.W. is an investigator of the Howard Hughes Medical Institute.
Reference
- 1.Sonoda M, Takano M, Miyahara J, Kato H. Computed radiography utilizing scanning laser stimulated luminescence. Radiology. 1983;148(3):833–838. doi: 10.1148/radiology.148.3.6878707. [DOI] [PubMed] [Google Scholar]
- 2.Amemiya Y, Miyahara J. Imaging plate illuminates many fields. Nature. 1988;336(6194):89–90. doi: 10.1038/336089a0. [DOI] [PubMed] [Google Scholar]
- 3.Amemiya Y, Wakabayashi K, Tanaka H, Ueno Y, Miyahara J. Laser-stimulated luminescence used to measure x-ray diffraction of a contracting striated muscle. Science. 1987;237(4811):164–168. doi: 10.1126/science.3496662. [DOI] [PubMed] [Google Scholar]
- 4.Ichihara S, Hayakawa S, Saga S, Hoshino M, Sakuma S, Ikeda M, Yamaguchi H, Hanaichi T, Kamiya Y. Usefulness of a scanning laser stimulated luminescence (SLSL) system for electron microscopy—a new image recording system. J. Electron Microsc. (Tokyo) 1984;33(3):255–257. [PubMed] [Google Scholar]
- 5.Mori N, Oikawa T, Katoh T, Miyahara J, Harada Y. Application of the ‘imaging plate’ to TEM image recording. Ultramicroscopy. 1988;25(3):195–201. doi: 10.1016/0304-3991(88)90014-9. [DOI] [PubMed] [Google Scholar]
- 6.Bele P, Ochs P, Angert I, Schroder R R. New high-DQE imaging plate scanner using the reflected readout laser signal for noise corrections. Microsc. Res. Tech. 2000;49:281–291. doi: 10.1002/(SICI)1097-0029(20000501)49:3<281::AID-JEMT6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 7.Zuo J M. Electron detection characteristics of a slow-scan CCD camera, imaging plates and film, and electron image restoration. Microsc. Res. Tech. 2000;49:245–268. doi: 10.1002/(SICI)1097-0029(20000501)49:3<245::AID-JEMT4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 8.Cheng Y, Zak O, Aisen P, Harrison S C, Walz T. Structure of the human transferrin receptor-transferrin complex. Cell. 2004;116(4):565–576. doi: 10.1016/s0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- 9.Rabl J, Smith D M, Yu Y, Chang S C, Goldberg A L, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell. 2008;30(3):360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification—powerful tools in modern electron microscopy. Biol. Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonen T, Sliz P, Kistler J, Cheng Y, Walz T. Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature. 2004;429(6988):193–197. doi: 10.1038/nature02503. [DOI] [PubMed] [Google Scholar]
- 12.Ludtke S J, Baldwin P R, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 13.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 14.Booth C R, Jakana J, Chiu W. Assessing the capabilities of a 4k×4k CCD camera for electron cryo-microscopy at 300kV. J. Struct. Biol. 2006;156(3):556–563. doi: 10.1016/j.jsb.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 15.Saad A, Ludtke S J, Jakana J, Rixon F J, Tsuruta H, Chiu W. Fourier amplitude decay of electron cryomicroscopic images of single particles and effects on structure determination. J. Struct. Biol. 2001;133(1):32–42. doi: 10.1006/jsbi.2001.4330. [DOI] [PubMed] [Google Scholar]
- 16.Hirai T, Murata K, Mitsuoka K, Kimura Y, Fujiyoshi Y. Trehalose embedding technique for high-resolution electron crystallography: application to structural study on bacteriorhodopsin. J. Electron Microsc. (Tokyo) 1999;48(5):653–658. doi: 10.1093/oxfordjournals.jmicro.a023731. [DOI] [PubMed] [Google Scholar]
- 17.Baldwin J, Henderson R. Measurement and evaluation of electron diffraction patterns from two-dimensional crystals. Ultramicroscopy. 1984;14(4):319–335. [Google Scholar]
- 18.Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison S C, Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438(7068):633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unwin P N. Electron microscopy of the stacked disk aggregate of tobacco mosaic virus protein: II. The influence of electron irradiation of the stain distribution. J. Mol. Biol. 1974;87(4):657–670. doi: 10.1016/0022-2836(74)90076-x. [DOI] [PubMed] [Google Scholar]
- 20.Ruch C, Skiniotis G, Steinmetz M O, Walz T, Ballmer-Hofer K. Structure of a VEGF-VEGF receptor complex determined by electron microscopy. Nat. Struct. Mol. Biol. 2007;14(3):249–250. doi: 10.1038/nsmb1202. [DOI] [PubMed] [Google Scholar]
- 21.Wu W, Sinha D, Shikov S, Yip C K, Walz T, Billings P C, Lear J D, Walsh P N. Factor XI homodimer structure is essential for normal proteolytic activation by factor XIIa, thrombin, and factor XIa. J. Biol. Chem. 2008;283(27):18655–18664. doi: 10.1074/jbc.M802275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Strunk J J, Lamken P, Piehler J, Walz T. The EM structure of a type I interferon-receptor complex reveals a novel mechanism for cytokine signaling. J. Mol. Biol. 2008;377(3):715–724. doi: 10.1016/j.jmb.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mori N, Oikawa T, Harada Y, Miyahara J. Development of the imaging plate for the transmission electron microscope and its characteristics. J. Electron Microsc. 1990;39:433–436. [Google Scholar]