Abstract
Spatial regulation of microtubule (MT) dynamics contributes to cell polarity and cell division. MT rescue, in which a MT stops shrinking and reinitiates growth, is the least understood aspect of MT dynamics. Cytoplasmic Linker Associated Proteins (CLASPs) are a conserved class of MT-associated proteins that contribute to MT stabilization and rescue in vivo. We show here that the Schizosaccharomyces pombe CLASP, Cls1p, is a homodimer that binds an αβ tubulin heterodimer through conserved TOG-like domains. In vitro, CLASP increases MT rescue frequency, decreases MT catastrophe frequency and moderately decreases MT disassembly rate. CLASP binds stably to the MT lattice, recruits tubulin and locally promotes rescues. Mutations in the CLASP TOG domains demonstrate that tubulin binding is critical for its rescue activity. We propose a mechanism for rescue in which CLASP-tubulin dimer complexes bind along the MT lattice and reverse MT depolymerization with their bound tubulin dimer.
Keywords: microtubule dynamics, mitosis, cell motility, TOG domain
Introduction
Microtubules (MTs) are dynamic polymers that exhibit two transitions: catastrophe, in which an assembling MT-plus end switches to rapid disassembly, and rescue, in which a plus end ceases disassembly and begins growing again (reviewed, Akhmanova and Steinmetz, 2008). In addition to intrinsic properties of tubulin polymerization, MT associated proteins (MAPs) participate in regulating MT dynamics in vivo. The mechanisms responsible for MT rescue are perhaps the least understood aspect of MT polymerization dynamics.
Cytoplasmic linker associated proteins (CLASPs) are members of a conserved class of MAPs that stabilize specific groups of MTs in yeast, plants and animals. CLASPs include S. cerevisiae Stu1 (Yin et al, 2002), S. pombe Cls1p (also called Peg1), Drosophila melangaster MAST/Orbit (reviewed by Bratman and Chang, 2008), Arabidopsis thaliana CLASP (Ambrose et al, 2007), and human CLASP1 and CLASP2 (Sousa et al, 2007). CLASPs have a broad range of functions in cell motility and mitosis and associate with the MT plus-ends and MT lattices (Mimori-Kiyosue et al, 2005; Kumar et al, 2009). CLASPs localize at kinetochores, the mitotic spindle midzone, centrosomes, the Golgi network and cell cortex (reviewed by Bratman and Chang, 2008; Kumar et al, 2009; Efimov et al, 2007). They contribute to the formation and maintenance of the mitotic spindle and regulate MT stability and growth at kinetochores and spindle midzone, (Pereira et al, 2006; Laycock et al, 2006; Maiato et al, 2003a; Maiato et al, 2005). In migrating fibroblasts, CLASPs are needed for a robust polarization of the MT cytoskeleton; their depletion causes an increase in the dynamics of anterior MTs and a decrease in rescue frequency (Drabek et al, 2006; Wittmann and Waterman-Storer, 2005; Mimori-Kiyosue et al, 2005). Inactivating mutations of Drosophila MAST/Orbit leads to defects in axonal growth cone motility (Lee et al, 2004).
CLASPs are targeted to different cellular processes through their interactions with other proteins. EB1 recruit a diverse group of +TIP proteins such as CLASPs to growing MT plus ends, by binding their Ser-X-Ile-Pro motifs (Honappa et al, 2009; Kumar et al, 2009; Galjart et al, 2005; Mimori-Kiyosue et al, 2005). In S. pombe, an interaction with Ase1 (PRC1 ortholog) localizes Cls1p to regions of anti-parallel MT bundling at the spindle midzone and in interphase MT bundles (Bratman and Chang, 2007). Interactions with the LL5β complex, GCC185 and CENP-E target human CLASP to the plasma membrane, Golgi network and kinetochores, respectively (Lansbergen et al, 2007; Efimov et al, 2007; Maffini et al, 2009). CLASP has also been shown to bind to MTs directly (Wittmann and Waterman-Storer, 2005; Mimori-Kiyosue et al, 2005).
Studies of S. pombe CLASP, Cls1p, indicate that it promotes MT rescue events (Bratman and Chang, 2007). Cls1p is not detectable at MT plus ends, but localizes to the MT lattice within MT bundles. Cls1p is essential for the frequent rescue events that maintain MTs in spindle midzone (Bratman and Chang, 2007; Grallert et al, 2006). In interphase cells, Cls1p localizes to dots near the middle of the MT bundles. Cls1p is required for all detectable rescue events, and cls1 mutant strains exhibit no defects in other MT kinetic parameters (Bratman and Chang, 2007). Overexpression of Cls1p causes it to accumulate all along the MT lattice and greatly increases rescue frequency (Bratman and Chang, 2007). Although Cls1p associates with CLIP-170 and EB1 (Grallert et al, 2006), its stabilizing effect on MTs is independent of these +TIPs and other MT associated proteins (Bratman and Chang, 2007), suggesting that CLASP regulates MTs directly.
CLASP orthologs share a common domain organization (Fig 1A). In S. pombe Cls1p, conserved N-terminal sequences are critical for function, and a fragment of Cls1p containing these N-terminal domains plus the serine/arginine-rich (S/R-rich) sequences, which follow them in the polypeptide chain, are sufficient when overexpressed for MT stabilization function and to rescue viability (Bratman and Chang, 2007). Likewise, the N-terminal domains are necessary for MT stabilization in motile fibroblasts and mitotic cells (Witmann and Waterman-Storer, 2005). It has been proposed that this N-terminal region has weak similarity to HEAT repeats characteristic of Tumor Overexpressed Gene (TOG) domains of MT polymerases, XMAP215 and its orthologs, which mediate tubulin dimer binding (Bratman and Chang, 2007; Al-Bassam et al, 2006, 2007; Slep and Vale, 2007). The C-terminal regions of CLASP have been implicated in binding the MT lattice (Wittmann and Waterman-Storer, 2005).
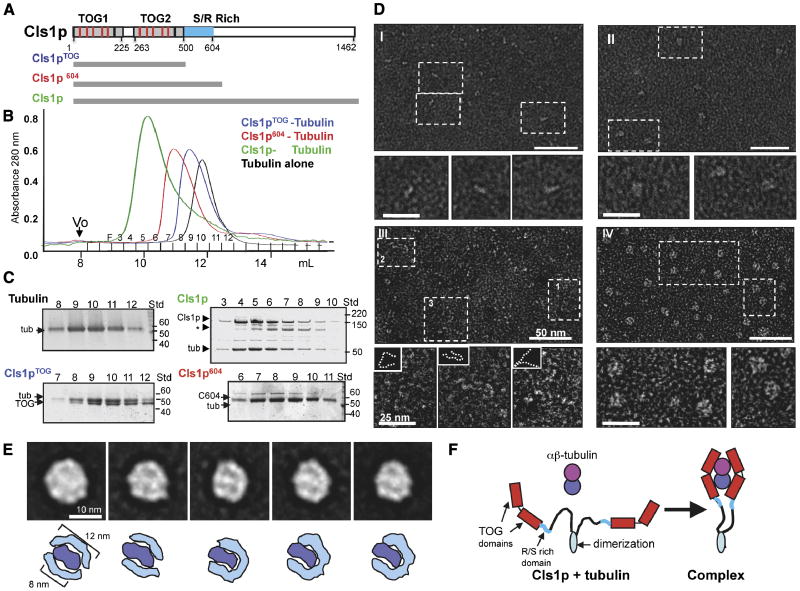
Figure 1. CLASP wraps around a tubulin dimer with two sets of TOG domains.
A) Domains of Cls1, as a typical CLASP protein. TOG-like and serine/arginine (S/R)-rich domains are in grey and blue, respectively. Studies with Cls1pTOG (residues 1-500), Cls1p604 (residues 1-604), and full length Cls1p (residues 1-1462) are described in this figure. CLASPs contain Two TOG-like domains each with five, tubulin-binding, intra-HEAT repeat turns (red lines in upper panel) identified by structure-based sequence alignments with XMAP215/Dis1 TOG domains (detailed in Fig S1A B).
B, C) Size exclusion chromatography of recombinant S. pombe Cls1p constructs. Intensity traces of Cls1pTOG (blue) and Cls1p604 (red) in complexes with soluble tubulin dimers indicate that Cls1p-tubulin complexes elute earlier than tubulin dimer alone (black). Cls1p-tubulin (green) elutes earlier than all other complexes, suggesting an extended conformation. SDS-PAGE of fractions from size exclusion chromatography (Panel C); Cls1pTOG and Cls1p604 co-elute with tubulin as complexes containing 1 Cls1p: 1 tubulin. The Cls1p604-tubulin complex elutes substantially earlier than Cls1pTOG-tubulin, indicating a higher Stokes radius (Supplementary Table S1). SDS-PAGE of fractions from Cls1p-tubulin complex peak shows that the peak saturates with a stoichiometry of 2 cls1p to 1 tubulin dimer, two-folds higher than the Cls1pTOG and Cls1p604 constructs. Asterisk (*) shows a very small amount of degraded inactive Cls1p. Supplementary data (Supplementary Table S1 Fig. S2) show that Cls1p is a dimer that binds a single tubulin dimer tightly, while Cls1pTOG and Cls1p604 are monomers that dissociate quickly from tubulin.
D) Negative stain electron microscopy of Cls1p and Cls1pTOG in the presence and absence of tubulin dimer. Panel I: Cls1pTOG; higher magnification images below panel I, show that the elongated Cls1pTOG molecules are ∼10 nm in length. In panel II, images of purified Cls1pTOG-tubulin dimer complexes immediately after size exclusion chromatography elution. Cls1pTOG appears to bind a tubulin dimer along its length. Panel III: Full-length Cls1p appears to be extended and contain flexible linkers. Panel IV: Full-length Cls1p-tubulin complexes are compact particles of uniform globular shape. Line traces of the lower magnification images in panel III are shown as broken lines as insets above each higher magnification image below panel III.
E) Upper panel: Images of five representative class averages from a reference-free classification of 2700 full length Cls1p-tubulin single particle images. The full image classification (Fig S2C) shows that Cls1p-tubulin complexes are homogenous with defined substructure. In each class average, two thin rim densities, about 12 nm in length encircle an elongated 8 nm density at the center of the particle. Lower panel: Interpretive drawing showing the outer rim densities as two sets of TOG domains (cyan) and the central density as a single tubulin dimer (blue).
F) Model for the CLASP conformational change accompanying the binding of a tubulin heterodimer. The C-terminal domains (900 residues) of Cls1p are flexibly linked with respect to the tubulin complex.
We describe here a mechanistic and structural analysis of S. pombe Cls1p. We show that the N-terminal region of Cls1p indeed contains two active Tumor Overexpressed Gene (TOG) domains that are similar to XMAP215 TOG domains. A Cls1p dimer binds a single αβ-tubulin heterodimer through two sets of TOG domains, and to the MT lattice through a serine arginine-rich domain. Despite this similarity, Cls1p behaves very differently from XMAP215, and has quite different effects on MT dynamics. In vitro, purified recombinant Cls1p is sufficient to promote MT rescue and to suppress MT catastrophe. The TOG-tubulin interaction is critical for rescue activity in vitro and in vivo. We propose that Cls1p promotes MT rescue by delivering tubulin dimers to the plus end. These studies provide a mechanistic model for MT rescue events in cells.
Results
CLASP binds a soluble tubulin dimer with two sets of TOG domains
Sequence alignment based upon the structures of XMAP215/Stu2 TOG domains suggests that CLASPs from various organisms contain two consecutive N-terminal TOG domains (TOG1 and TOG2), in which the amino acid residues in the predicted tubulin binding turns are similar to the corresponding residues in the XMAP215/Dis1 TOG domains (Fig S1). A region toward the C-terminus of human CLASP has also been proposed to be TOG-like (Slep and Vale, 2007; Slep, 2009), but it is not conserved in other organisms and deviates substantially from TOG consensus in the length and composition of the putative tubulin binding loops.
We tested whether the N-terminal TOG-like domains of CLASP bind tubulin. We expressed and purified full-length S. pombe Cls1p (residues 1-1462) and two shorter fragments: Cls1pTOG (residues 1-500), containing just the two TOG domains, and Cls1p604 (residues 1-604), including the TOG domains and the S/R-rich region (Fig 1A). Size-exclusion chromatography showed that full length Cls1p does indeed bind soluble tubulin dimer to form a stable complex (Fig. 1B and C; Supplementary Table S1); the Cls1pTOG or Cls1p604 fragments are sufficient to bind tubulin, but their tubulin complexes are unstable.
Size exclusion chromatography showed that full length Cls1p has a large apparent molecular mass (Fig S2A; Supplementary Table S1). By adding increasing amounts of Cls1p and determining the amount required to shift a constant amount of tubulin dimer into the complex peak, we found that the Cls1p-tubulin complex has a stoichiometry of two Cls1p molecules for each αβ-tubulin dimer (Fig S2A). Sedimentation equilibrium ultracentrifugation showed that Cls1p and Cls1p-tubulin complex have masses of 267±15 kDa and 370±25 kDa, respectively (Supplementary Table S1, Fig S2B). These results indicate that that Cls1p is homodimer and that the Cls1p-tubulin complex contains a Cls1p dimer bound with a single tubulin dimer.
Cls1p604 or Cls1pTOG are monomeric, as measured by sedimentation equilibrium ultracentrifugation, suggesting that sequences C-terminal to the S/R rich domain determine dimerization (Supplementary Table S1; Gallert et al 2006). These fragments dissociated from soluble tubulin dimers shortly after size exclusion chromatography, indicating they have lower affinity for tubulin than full-length Cls1p (Supplementary Table S1). Weak binding of one set of TOG domains to tubulin dimer in a Cls1pTOG monomer and strong binding of full-length Cls1p dimer are also properties of Stu2p— the S. cerevisiae XMAP215 ortholog (Al-Bassam et al, 2006). They suggest that dimerization enhances tubulin dimer binding.
We next studied Cls1p and shorter fragments and their tubulin complexes by electron microscopy. Images of negatively stained Cls1pTOG fragment showed that the Cls1pTOG domains form an extended structure, about 10 nm long and 2 nm wide. Images of Cls1pTOG-tubulin complexes showed more isometric particles, about 8 nm in diameter; these may represent a 1:1 association of the Cls1pTOG fragment with αβ-tubulin dimers, which are 8 nm in length (Fig 1D, panel II, data not shown). In contrast, full length Cls1p appeared as an extended structure with two thin “tentacles”, each shaped like Cls1pTOG (Fig 1D, panel I). Images of purified Cls1p-tubulin complex showed uniform globular complexes, 12 nm in diameter (Fig 1D, panel II). Reference-free classification of 2700 Cls1p-tubulin particle images in SPIDER showed that these particles are extremely homogenous with relatively small differences among 45 classes (Fig 1E, full classification is shown in Fig. S2C). Class averages containing the highest numbers of particles showed the Cls1p-tubulin complex to be a regular isometric particle, 12 nm in diameter, with two thin, elongated densities surrounding a ∼8 nm density at the particle center (Fig 1E, S2C). One interpretation is that two sets of TOG domains (each about 12 nm in length) encircle a tubulin dimer (8 nm long) in the complex, forming a particle that is about 30-34 nm in circumference (Fig 1F; Al-Bassam et al, 2006). These images of the Cls1p TOG domains, full-length proteins and the tubulin-complex are all very similar to those of those seen with XMAP215 and Stu2p (Brouhard et al, 2008; Al-Bassam et al, 2006). Thus, at a structural level, the CLASP and XMAP215/Stu2 TOG domain have a common mode of tubulin binding.
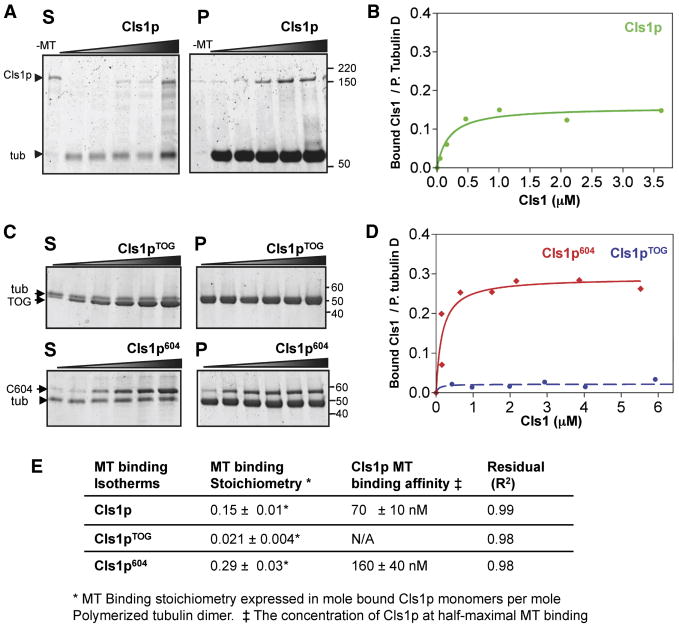
SR-rich domains contribute to high-affinity binding to the MT lattice
We tested in co-sedimentation experiments whether Cls1p and its fragments bind MTs. Full length Cls1p pelleted with GMPCPP stabilized MTs, with half maximal binding at 70 nM Cls1p (Fig 2C). The binding saturated at one Cls1p monomer per ∼6 polymerized tubulin dimers. This behavior is different from that of XMAP215, which exhibits only low affinity, non-saturating MT binding (Fig S2B). Cls1p604 also bound MTs with high affinity, whereas Cls1pTOG exhibited no significant MT binding and remained in the supernatant (Figs. 3A, 2D). Cls1p604 binding saturated at 1 Cls1 molecule per ∼3 polymerized tubulin dimers of the MT (Fig. 2B). We conclude that the S/R region, which is present in Cls1p604 but not Cls1pTOG, mediates the interaction with the MT lattice.
Figure 2. CLASP S/R rich domains bind MT lattices with high affinity.
A) Cls1p co-sedimentation with GMPCPP-stabilized MTs. SDS-PAGE of separated pellet (P) and supernatant (S) at low concentration, Cls1p is fully MT-bound (pellet), whereas at higher concentrations excess Cls1p is unbound (supernatant). Cls1p does not sediment in the absence of MTs (-MT lanes). Tubulin dimer does not affect Cls1p binding to GMPCPP-MTs (Fig S1A). Unlike Cls1p, XMAP215 does not bind GMPCPP-MTs with high affinity (Fig S2B).
B) Binding isotherm of Cls1p and GMPCPP-stabilized MTs determined from data shown in A (see Methods).
C) MT co-sedimentation analysis of Cls1pTOG, Cls1p604 constructs. Lower panel: Cls1p604 (containing S/R-rich domain) binds and sediments with GMPCPP-MTs with high affinity, while the top panel shows that Cls1pTOG does not sediment with MTs.
D) MT binding isotherms for Cls1pTOG and Cls1p604 determined from data shown C.
E) MT binding affinities and MT binding stoichiometries, in moles of Cls1p monomer per polymerized tubulin dimer, for full length Cls1p, Cls1pTOG and Cls1p604.
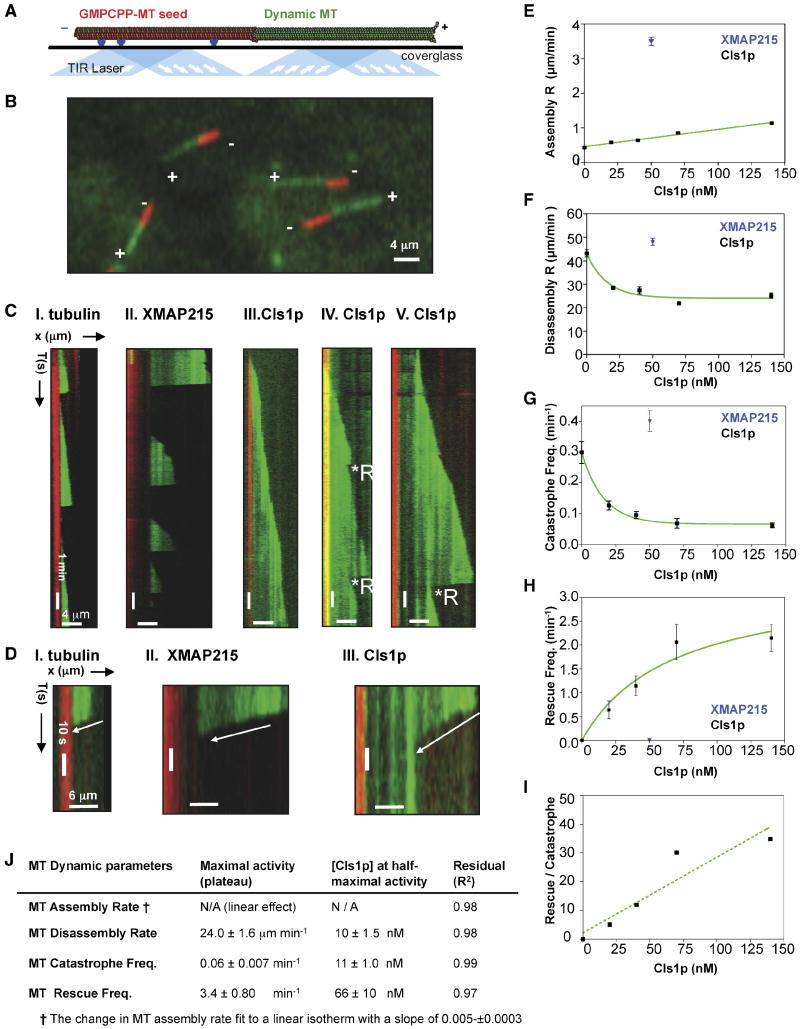
Figure 3. Effect of CLASP on MT dynamics parameters.
A) Scheme for total internal refection fluorescence (TIRF) microscopy used to study MT dynamics. Surface-attached anti-biotin antibodies (blue) bind biotin-labeled GMPCPP-stabilized MT seeds (red). The seeds near the surface nucleate AlexaFluor-488-labeled (green) dynamic MTs from 6 μM soluble tubulin dimers in the presence of GTP. TIRF illumination by 564 nm and 488 nm lasers. Image adapted from Brouhard et al (2008).
B) TIRF image of assembling MTs. Seeds (red) initiate assembly of dynamic MTs (green) at plus ends (+) only.
- I. 6 μM tubulin: dynamic MTs show slow assembly, rapid disassembly, and frequent catastrophes.
- II. 50 nM XMAP215 + 6 μM tubulin: dynamic MTs show rapid MT assembly and frequent catastrophes.
- III-V. 40 nM Cls1p + 6 μM tubulin: Panel III shows a relatively slow assembly MT rate and no catastrophes. Panel IV shows a dynamic MT assembling slowly with two catastrophes, each followed by a rescue. Panel V shows a dynamic MT assembling slowly and a catastrophe that leads to extended disassembly, which is followed by a rescue.
D) Close-up kymographs in C show MT disassembly at higher time resolution.
E) Effect of Cls1p concentration on MT assembly rate. Cls1p (black) accelerates MT assembly slightly, while 50 nM XMAP215 (blue) increases the assembly rate by ten-fold (Supplementary Table S2; Brouhard et al, 2008). Each point (Supplementary Table S2) represents the mean of Gaussian distribution fit of a large number of assembly events measured for each Cls1p concentration (distributions are shown in Fig S4A).
F) Effect of Cls1p concentration on MT disassembly rates. Each point (Supplementary Table I) represents the mean of Gaussian distribution fits for a large number of events at different Cls1p concentrations (distributions are shown in Fig S4B).
G) Effect of Cls1p concentration on MT catastrophe frequency (Supplementary Table S2). Each point (Supplementary Table S2) represents the mean from a Gaussian distribution to a large number of catastrophe frequencies measured for each Cls1p concentration (distributions are shown in Fig S4C).
H) Effect of Cls1p concentration on the MT rescue frequency. Each point (Supplementary Table S2) is the mean rescue frequency measured for each Cls1p concentration shown in Fig S4D.
I) Effect of Cls1p concentrations on the ratio of MT rescue to MT catastrophe. At 140 nM Cls1p, on average every MT catastrophe (one event per 15.6 minutes of MT assembly) was reversed by a rescue (one event per 45 seconds of MT disassembly).
J) Table fitted Cls1p concentration and maximal effects summarizing data from panels E-I.
To determine whether TOG-domain binding of tubulin dimer affects Cls1p association with MTs, we compared MT binding in the absence and presence of 50 nM Alexa-488-labeled tubulin dimers. The addition of soluble tubulin dimer did not influence the affinity of Cls1p for MTs (Fig S3A). These results indicate that Cls1p has two separable functions: binding to tubulin dimer and binding to the MT lattice.
CLASP promotes MT rescue and suppress MT disassembly and catastrophe
We analyzed the effects of Cls1p on the dynamics of individual MTs by total internal reflection fluorescence (TIRF) microscopy using a modified version of methods developed for work on XMAP215 (see Methods). Texas-red labeled MT seeds were assembled with the non-hydrolysable GTP analog, GMPPCP (Fig. 3A). Addition of a mixture of Alexa-fluor-488 labeled and unlabeled tubulin allowed us to visualize the dynamic behavior of MTs growing from the plus ends of these stable MT seeds (Fig 3B). Dynamic MTs in vitro undergo stochastic “catastrophes”, when the MT switches from assembly to disassembly, and “rescues”, when assembly reinitiates. At 6 μM tubulin concentration (in the absence of XMAP215 or Cls1p), assembly is relatively slow (0.43±0.03 μm/min), catastrophes are frequent (0.30±0.05 catastrophes/min), and MT rescues are not observed. As Brouhard et al (2008) previously observed, addition of 50 nM XMAP215 increased the average rate of MT assembly 10-fold and increased MT length distributions, but did not affect catastrophe frequency, disassembly rate, or rescue frequency (Supplementary Table S2; Fig S4).
Addition of 40 nM Cls1p led to many rescue events (as defined by the re-initiation of assembly before disassembly has reached the seed -- Fig. 3C, IV-V) and a decrease the apparent frequency of catastrophes (Fig. 3C, III). Some dynamic MTs assembled continuously, with no evident catastrophes in the time frame (20 min) of the experiment (Fig. 3C, III), while other MTs underwent occasional catastrophe followed promptly by a rescue (Fig. 3C, IV-V). To determine dose-dependent activities of Cls1p, we measured MT dynamic parameters at a series of Cls1p concentrations (Supplementary Table S2 and Fig S4; 20 nM to 140 nM). As Cls1p concentration increased, the catastrophe frequency declined by six-fold and the rescue frequency increased, reaching a plateau of 0.06 catastrophes/min and 2.15 rescues/min. The average total assembly time increased slightly with increasing the Cls1p concentration up to 13.5 min, almost 40% higher than that with 6 μM tubulin (Supplementary Table S2). Thus, at 140 nM Cls1p, a single catastrophe during a 20 minutes period was, on average, reversed by a rescue, which occurred at least once per 30 seconds of disassembly (Fig. 3I). The disassembly rate decreased with increasing Cls1p concentration, from 43 μm/min to 25 μm/min; the average disassembly time increased up two-fold with increasing Cls1p concentration, due to the decrease in disassembly rate (Supplementary Table S2). The assembly rate increased linearly up to two fold at 140 nM Cls1p. The linear increase in the assembly rate is very different from the increase observed with XMAP215, which accelerates assembly at the lower concentrations by more than ten-fold (see above; Brouhard et al, 2008). The average dynamic MT length increased with increasing Cls1p concentration (Supplementary Table S2, Fig S4E). At the highest Cls1p concentrations (70 nM and 140 nM), we observed a bimodal distribution for the dynamic MT lengths with two classes: 4.5-5.0 μm and 10-11 μm (Fig S4F, Supplementary Table S2). At 140 nM Cls1p, however, many MTs in the latter group were even longer (Fig S4F).
We also tested the activity of the monomeric Cls1p604, with a single set of TOG domains and an S/R rich sequence. Addition of 200 nM Cls1p604 produced no observable rescue and a threefold higher catastrophe frequency than with full-length Cls1p (Supplementary Table S2; Fig S4C). The average disassembly rate was 45 μm/min, similar to the disassembly rate for tubulin alone (Supplementary Table S2; Fig S4B). MTs were two-fold longer (Supplementary Table S2, Fig S4E). This Cls1p fragment's weakened effect on MT dynamics is consistent with its weakened grip on the tubulin dimer.
CLASP recruits tubulin dimers to the MT lattice
To visualize the behavior of Cls1p molecules, we expressed Cls1p with GFP fused to its C-terminus. This GFP fusion is functional in vivo, and can replace the endogenous Cls1p in the cell (Bratman and Chang, 2007). In vitro, the fused GFP does not affect free tubulin dimer binding and binds tubulin with a similar stoichiometry to untagged Cls1p (Supplementary Table S1; Fig S5A). Cls1p-GFP is a well-behaved dimer (300±16 kDa), and Cls1p-GFP tubulin complexes (400±12 kDa) contain a single tubulin dimer, just as do complexes with untagged Cls1p (Fig S5B). Furthermore, addition of GFP does not alter the effects of Cls1p on rates of MT catastrophe and rescue (see next section).
Using TIRF microscopy, we observed that Cls1p-GFP bound along MTs (Texas red labeled GMPCPP-MTs; Fig. 4, middle panel). When added at 50 nM, the distribution of Cls1p-GFP was non-uniform and punctate; the non-uniformity decreased as binding saturated at higher concentrations (data not shown). Time-lapse images showed that the pattern of bound Cls1p-GFP puncta did not change during the course of the experiment (Fig. 4A, lower panel), suggesting a stable association of Cls1p-GFP with the MT lattice.
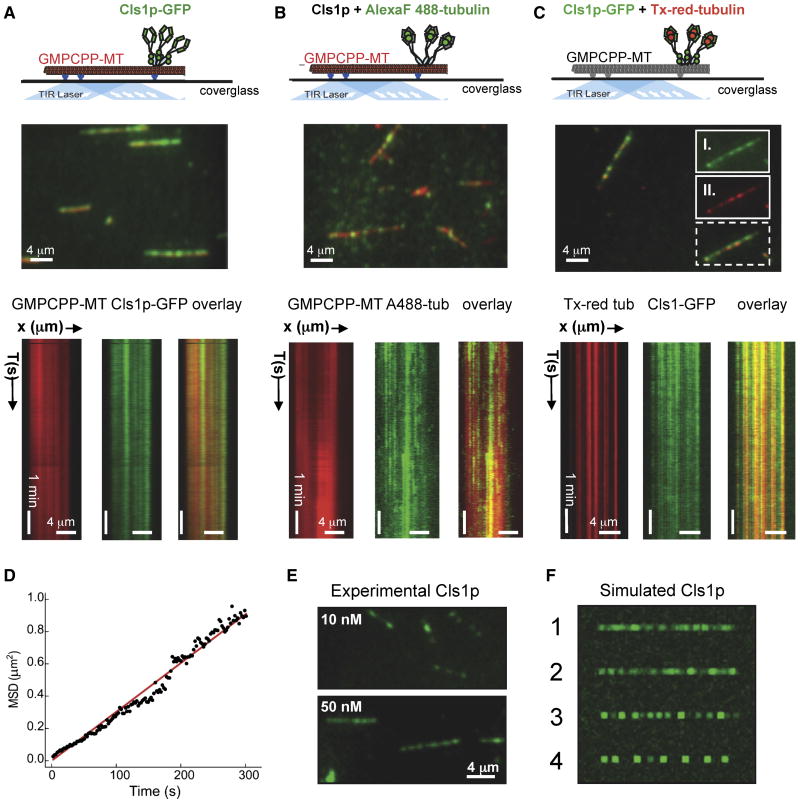
Figure 4. Cls1p-GFP molecules bind non-uniformly along GMPCPP-stabilized MTs and recruit soluble tubulin dimers.
A) Top panel, TIRF experiment scheme to detect Cls1p-GFP binding to Texas-red GMPCPP MTs (red). Middle panel, TIRF image of 50 nM Cls1p-GFP (green) binding non-uniformly along GMPCPP MTs (red). Lower panel, kymographs of Cls1p-GFP (green, left) on GMPCPP-MT (red, middle) in isolated channels and overlaid (right). Non-uniform patches of densely bound Cls1p-GFP remain stationary along GMPCPP MTs.
B) Top panel, TIRF experiment scheme to detect untagged Cls1p molecules (grey) recruiting Alexa-Fluor-488 labeled tubulin (green) while bound to GMPCPP MTs (red). Middle panel, TIRF image of Alexa-Fluor-488 labeled tubulin dimers (green) recruited to the GMPCPP MTs (red) by bound untagged-Cls1p. Lower panel Kymographs of Alexa-Fluor-488 labeled tubulin dimers (green, left) recruited by untagged Cls1p along GMPCPP-MTs (red, middle) in isolated channels and overlaid (right).
C) Top panel, Schematic of TIRF experiment to simultaneously detect recruitment of Cls1p-GFP (green) and Texas-red tubulin (red) along non-labeled GMPCPP MTs (grey). Middle panel, TIRF image of 50 nM Cls1p-GFP (green) and 50 nM Texas red tubulin (red) binding non-uniformly along GMPCPP MTs (red). Inset panels II and I show raw images of Cls1-GFP and Texas-red tubulin bound along GMPCPP MTs in the isolated channels above their overlaid image (broken lines). Lower panel, Kymographs of GMPCPP-MT (red) in isolated channels and overlaid. Non-uniform patches of densely bound Cls1p-GFP remain stationary along GMPCPP MTs. Images in top panels were adapted from Brouhard et al (2008).
D) Tracking of Cls1p puncta along dynamic MTs to determine the average diffusion coefficient (see supplementary materials and methods).
E) Experimental images of 10 nM (upper panel) and 50 nM (lower panel) of Cls1p-GFP puncta bound along GMPCPP-MTs.
F) Simulation of Cls1p-GFP binding along linear MT lattices comparing different degrees of self-association. Panel 1: a random association with MT lattice without diffusion. Panel 2: weak self-association among molecules while binding to the MT lattice binding. Panel 3: shows moderate self-association among Cls1p-GFP molecules. Panel 4; a high degree of self-association. Note that the degree of speckling and sharpness of puncta in experimental Cls1p-GFP images is similar to simulations in either panels 1 or 2, and different from simulations in panels 3 and 4
To determine whether Cls1p molecules can bind soluble tubulin dimers while attached to MT lattices, we added untagged Cls1p and Alexa-Fluor 488-labeled tubulin dimers to MTs (Fig 4B, upper panel; see methods). Alexa-Fluor-488-tubulin dimers were detected along GMPCPP MTs (Fig 4B, middle panel). This binding depended on addition of Cls1p. Bulk MT co-sedimentation experiments independently confirmed this finding (Fig S3A, panel IV). Like Cls1p, the distribution of Alexa-Fluor-488-tubulin dimers along GMPCPP-MTs was non-uniform, and there was little or no longitudinal diffusion (data not shown). The pattern of the tubulin dimers was slightly more dynamic than that of Cls1p-GFP, however, suggesting that tubulin dimers occasionally disassociated from Cls1p on the MT, with a mean residence time of 3-4 min. We further confirmed that Texas-red labeled tubulin dimers and Cls1-GFP directly co-localized along non-labeled GMCPP-MTs (Fig 4C). Thus, Cls1p dimers can bind tightly along MT lattices without diffusion, producing a non-uniform, punctate distribution, while retaining soluble tubulin heterodimers through TOG-domain interactions, with a slow exchange rate.
To determine the nature of Cls1p-GFP binding along lattices of GMPCPP-MTs, we tracked Cls1p-GFP puncta formed at a low Cls1p concentration (10 nM) and measured their diffusion rate. Cls1p-GFP puncta have a very low diffusion coefficient (D= 0.0015 μm2/s) along GMPCPP-MT lattices (see Fig 4D), significantly (135-fold) lower than the diffusion coefficient for XMAP215 (D = 0.2 μm2/s). This difference is consistent with the difference in their MT affinities derived from bulk MT-co sedimentation (Fig 2D and S3B). We conclude that the Cls1p binds the MT lattice tightly, while XMAP215 has weaker, largely electrostatic interactions that allow it to diffuse along the MT.
We asked whether the non-uniform distribution of Cls1p indicates that Cls1p molecules cluster together on the MT lattice through interactions among Cls1p molecules, or could simply reflect random binding along the MTs. We performed a computational simulation of Cls1p binding, using a “model convolution” approach (Gardner et al., 2010). The simulation generated artificial images of Cls1p-GFP on MTs, which were compared with experimental distributions (Fig 4E). Under conditions of a random distribution, Cls1p molecules produced images with non-uniform patches reminiscent of fluorescence speckling of Cls1p-GFP along MT lattices (Fig 4F, panel 1). A simulation that explicitly introduced weak oligomerization or clustering (see Methods) also produced images consistent with our data (Fig 4F, panel 2), but increasing the propensity of Cls1p to oligomerize caused the artificial Cls1p images to diverge from the experimental data (Fig 4F, panels 3 and 4). Comparing these artificial images with experimental data, we conclude that the non-uniform distribution of Cls1p-GFP puncta seen experimentally probably arises from the Poisson statistics that underlie random high affinity binding along the MT lattice, but we cannot rule out a limited degree of self-association and clustering among Cls1p molecules.
MT rescues occur at sites of a high local concentration of CLASP molecules
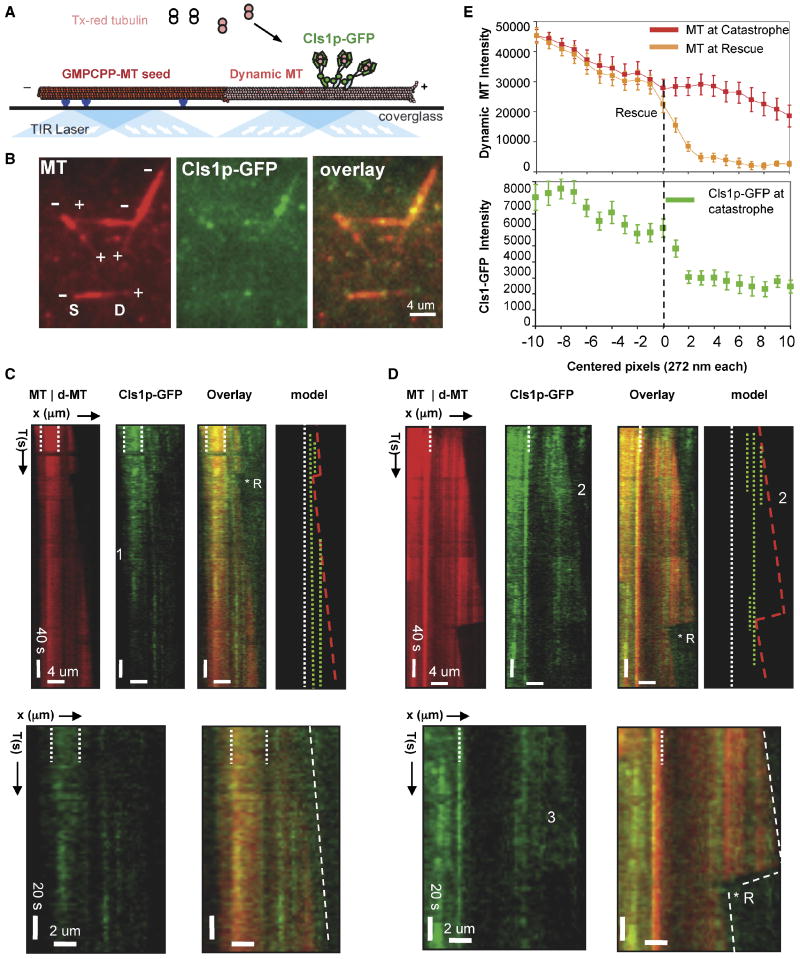
We devised a two-color TIRF experiment to determine the behavior of Cls1p on dynamic MTs, and to visualize Cls1p-mediated MT rescue events directly. The dynamic portion of the MT and the MT seeds were both labeled with Texas Red, but the difference in fluorescence intensities allowed us to identify dynamic transitions unambiguously (Fig 5B, S and D denote seed and dynamic MTs, respectively). We used a 70 nM mixture of Cls1p-GFP (10 nM) with untagged-Cls1p (60 nM) to decrease background; activities of this mixture were similar to those of 70 nM Cls1p (Supplementary Table S2; Fig S4, 2C-TIRF panels).
Figure 5. Sites of high CLASP concentration locally anticipate sites of MT rescues.
A) Schematic of TIRF microscopy assay used for imaging MT dynamics and for simultaneous imaging of dynamic MTs and Cls1p-GFP localization. Anti-biotin antibodies (blue) capture biotin- and Texas-red labeled GMPCPP polymerized MT seeds (red) near the silanized glass surface. Densely labeled Texas-red MT seeds nucleate dynamic MT assembly from 6 μM tubulin dimers, less densely labeled with Texas–red, in the presence of GTP and Cls1p-GFP (see methods). Image adapted from Brouhard et al (2008).
B) Raw TIRF image of dynamic MTs and Cls1p-GFP. Left panel: dynamic MTs growing at plus ends of MT seeds. Note, high fluorescence intensity of the MT seed compared to the dynamic MT. Middle panel: Cls1p-GFP non-uniformly bound along dynamic MTs (middle panel). Cls1p-GFP accumulates more densely along the seed. Right Panel: overlay of both images.
C) Cls1p-GFP binding along assembling dynamic MTs. Left panel: kymograph of dynamic MT (dMT) growing from a more densely labeled GMPCPP-MT seed (delimited by white broken lines). A single catastrophe occurred and was reversed by a rescue (*R). MT assembly then continued without further catastrophes. Second from left: Cls1p-GFP molecules bind more densely along the GMPCPP MT seed than along growing MT. Photobleaching decreased Cls1p-GFP fluorescence at the position marked by the numeral 1. Sites occupied at high Cls1p-GFP concentration remain at fixed positions without diffusion. Third from left: sites of Cls1p-GFP binding along a growing MT. Right: model kymograph showing the positions of high local concentration of Cls1p-GFP (broken green lines) along dynamic MTs (boundaries in red lines). Higher magnification kymographs show Cls1p-GFP molecules bound along an assembling MT. Note the absence of MT plus-end tracking. An additional example is shown in Fig S6A, IV.
D) Sites of high concentration of Cls1p-GFP molecules correlate with sites of MT rescue on a dynamic MT. Left panel: kymograph of dynamic MT (dMT) assembling from an intensely labeled GMPCPP-MT seed (MT, white broken lines). A catastrophe was followed by MT disassembly and a rescue (R*). Second panel from left: Cls1p-GFP (green) binds non-uniformly and without diffusion along the dynamic MT. Note that three Cls1p-GFP bands form, but two dissociate early during assembly (marked by the numeral 2). Third panel from left: sites of MT rescue correlate with positions of high Cls1p-GFP density (green) on a dynamic MT (red). Right: model kymograph showing the position of Cls1p-GFP dense bands (broken green) compared to the dynamic MT boundary (red) growing from the GMPCPP MT seed (broken white line). Higher magnification images show correlation of Cls1p “bands” with MT rescue. Additional examples are shown in Fig S6A, I-III. Note the loss in the diffuse Cls1p-GFP fluorescence (marked by #3) reproduces the MT disassembly rate. Fig S5C shows MT rescues were not observed in the absence of sites of high Cls1p-GFP concentration along dynamic MTs
E) Correlation of local Cls1p-GFP concentration on the MT with MT rescue. Average fluorescence intensity profile (see Methods) of a dynamic MT before catastrophe (red profile), dynamic MT at rescue (orange profile), and Cls1p-GFP along dynamic MT before catastrophe (green profile) for 20 different rescue events (more details in Fig S6B). Note that the Cls1p-GFP intensity peaks within 1 pixel (272 nm) of the site of rescue and the increase is three folds higher at that site than any pixel prior. The raw intensity profiles for Cls1p-GFP with MT rescue and statistical significance of Cls1-GFP intensity increase are shown in Fig. S6B.
We noted first that Cls1p-GFP bound more densely into non-uniform puncta along the MT seeds than along the dynamic MTs, even when the latter remained polymerized throughout the experiment (Fig 5C, D and Fig S6A). As the principal difference between the two MT forms is the nucleotide state of the polymerized tubulin (mostly GTP-like in the MT seed, and mostly GDP-like in the dynamic MT), it is possible that Cls1p has some preference for polymerized GTP tubulin in the MT lattice.
On the dynamic MTs, we observed that Cls1p-GFP bound with a combination of diffuse and punctate distributions along the lattice and near MT plus ends, but with a lower density than along the MT seeds (Fig 5C). The sites of MT rescues were anticipated by Cls1p-GFP puncta, which are the sites of high local concentration on the MT lattice (Fig. 5D, Fig S6A I-III). Average intensity profiles showed that the site of rescue coincided with a peak in the Cls1p-GFP fluorescence to within 1 pixel (270 nm) (Fig. 5E, lower panel; p value ≤ 0.01, Fig S6B, II). The average Cls1p-GFP intensity at the site of MT rescue was three fold higher than at sites that did not initiate rescue events. All MT rescues observed occurred at Cls1p puncta, and we did not observe rescues events at other sites. Thus, these results suggest that rescue requires a minimum local concentration of Cls1p along the MT lattice.
We did not observe Cls1p tracking with the growing MT plus end (Fig 5C, Fig S6A, III and IV). The presence of Cls1p-GFP puncta near growing plus ends may, however, explain the decrease in MT catastrophe. (Fig 5C, Fig S6A, IV). Catastrophes occurred with normal frequency at MT plus ends that did not accumulate Cls1-GFP puncta (Fig S6C). Rescue events occurring very near the position of catastrophe at plus ends cannot be spatially resolved by our imaging system (about 270 nm per pixel, equivalent to 34 tubulin dimers along a protofilament); such local rescues will appear as a decrease in the occurrence of MT catastrophes.
TOG-tubulin interaction is required for CLASP function
To test the function of tubulin binding, we generated mutations to eliminate CLASP tubulin binding without disrupting its structure. The alignment of TOG domains allows us to predict the tubulin-binding surface of CLASP (Fig. S1B, Al-Bassam et al, 2007). We mutated to alanine-conserved residues in two of the intra-HEAT repeat turns (W293A in T1 and K379A and K380A in T3: Fig. 6A). Multiple assays showed that this TOG domain mutant, designated Cls1pT1T3, is deficient specifically in tubulin dimer binding. Its affinity for tubulin was too low for co-elution in size-exclusion chromatography (Fig. 6B, Fig. S7A). MT co-sedimentation experiments showed that the Cls1pT1T3 bound MT lattices to the same extent as wt Cls1p, and the binding isotherms suggested a similar affinity and stoichiometry (Fig S7B). TIRF experiments showed that Cls1pT1T3 did not recruit AlexaF-488-labeled tubulin dimer to the lattices of GMPCPP-MTs (Fig S7B and Fig S4C).
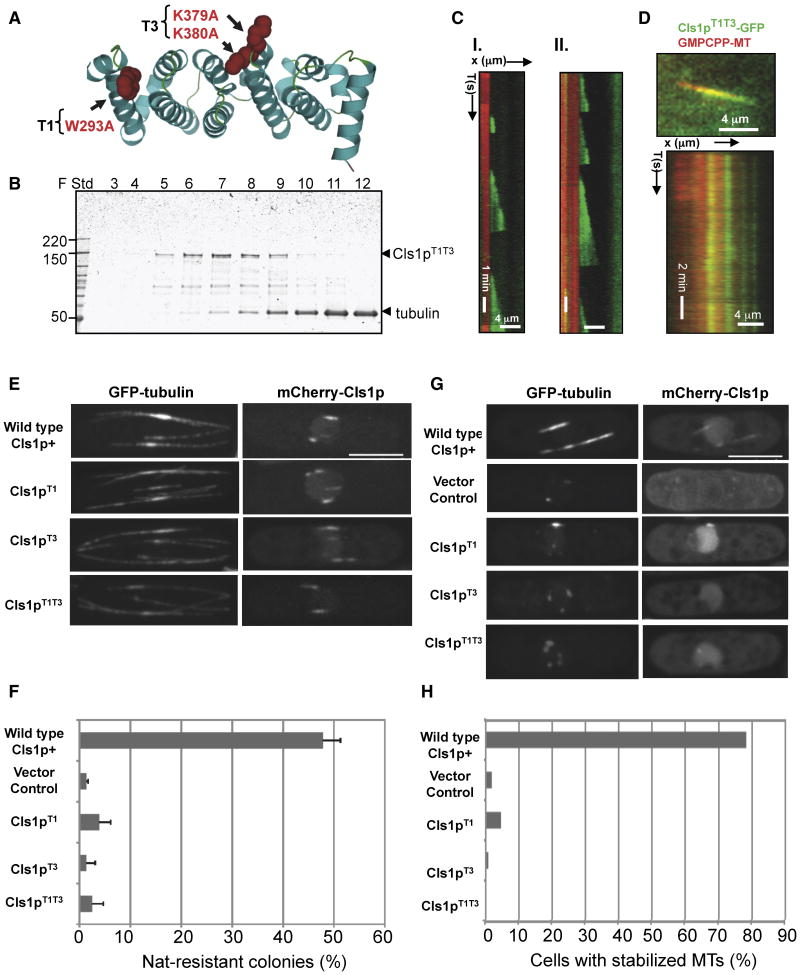
Figure 6. Defects of Cls1p mutants with inactivated TOG-tubulin interface.
A) Ribbon model of a TOG domain (PDB ID 2OF3; Al-Bassam et al, 2007) showing W293 and K379/K380 (red space filling) in the intra-HEAT turns (T1 and T3) mutated in Cls1pT1T3 to disrupt tubulin dimer binding
B) SDS-PAGE of size exclusion chromatography fractions of Cls1pT1T3 with tubulin dimer, as in the experiment with Cls1p in Fig 1A. The Cls1pT1T3 mutant does not co elute with tubulin dimer (tub); absorbance profile is shown in Fig S7A.
C) Kymographs of dynamic MTs assembled in the presence of 200 nM recombinant Cls1pT1T3 as described in Fig 3B; note the frequent catastrophes and loss of rescues. Dynamic MT parameters (Supplementary Table S2), determined based on distributions, are shown in Fig S6D.
D) Cls1p T1T3-GFP binding along GMPCPP MTs. Top panel: Raw image of Cls1T1T3-GFP binding non-uniformly along Texas-red GMPCPP MT (red) similar to Cls1p-GFP described in Fig 4E. Lower panel: kymograph of the Cls1pT1T3-GFP (green) on GMPCPP-MT (red), showing that Cls1T1T3-GFP distribution remained stationary throughout the experiment. Bulk MT co-sedimentation analysis shows that Cls1T1T3 binds MT with an affinity similar to wt Cls1p (Fig S7B).
E) Localization of cls1p mutant proteins in vivo. Full length Cls1pT1, Cls1pT3 and Cls1pT1T3 were expressed as mCherry fusion proteins driven by the thiamine-repressible nmt1* promoter in S. pombe cells expressing GFP-tubulin. Cells were grown in thiamine for relatively low levels of expression. Note that the localization of cls1p to regions of MT overlaps on MT bundles is not perturbed in the cls1 TOG mutants. Maximum projection confocal images of representative cells are shown.
F) Ability of the cls1 TOG mutant proteins to rescue viability of cls1 null cells. Diploid cls1+/cls1Δ cells carrying wild type or mutant cls1 plasmids were sporulated; resultant spores with plasmids were assayed for viability by measuring percentage of viable haploid Nat-resistant colonies carrying the with cls1Δ allele (see Methods). Full rescue of viability in cls1Δ is predicted to be 50% in this assay. Note that Cls1p T1, Cls1pT3, Cls1pT1T3 mutants are defective in rescuing the cls1p function in haploid cells, similar to vector control (detailed in Supplementary Table S3).
G, H) Cls1-TOG mutants are defective in MT stabilization activity. Panel G: Cells carrying m-Cherry Cls1pT1, Cls1pT3 and Cls1pT1T3 were induced for higher expression by growing in media lacking thiamine in wild type cells expressing GFP-tubulin, and then assayed for stabilization by treatment with MT destabilizing drug, methyl-benzidazole-carbamate (MBC). Maximum projections of confocal images of representative cells are shown. Note that MT bundles are stabilized in cells overexpressing wild type mCherry-Cls1p, which binds along the MT lattice, whereas MTs in cells overexpressing the Cls1pT1, Cls1pT3, Cls1pT1T3 or vector controls are not stabilized and shrink to peri-nuclear dots. (Scale bar, 5 μm). Panel H: percentage of cells with stabilized cytoplasmic MTs 10 minutes after MBC treatment. MTs were with MBC. We considered a MT bundle as stabilized if it was ≥ 2 μm in length (detailed in Supplementary Table S3).
We then examined the effect of these TOG mutations on MT dynamics in vitro. Addition of 200 nM Cls1pT1T3 had no effect on the frequencies of rescue or catastrophe (Supplementary Table S2; Figs. 6B and S7B). In contrast, 200 nM Cls1pT1T3 still had effects on MT assembly and disassembly (Supplementary Table S2). A Cls1pT1T3-GFP fusion protein also bound non-uniformly along GMPCPP-MTs, and formed puncta that were indistinguishable from those seen with Cls1p-GFP (Fig 6E). MT lattice binding and tubulin dimer binding are thus separable activities of Cls1p. These data suggest that regulation of rescue and catastrophe require tubulin dimer binding by the TOG domains, while the effects on MT disassembly and assembly are TOG-domain independent and thus may be caused by MT lattice binding activity.
We also examined the effects of the tubulin-binding mutations in vivo, by expressing these mutant proteins in S. pombe cells. We analyzed their localization and MT stabilizing activity using methods described previously (Supplementary Table S3; Bratman and Chang, 2007). Cls1 TOG-domain mutants (Cls1T1, Cls1T3, and Cls1T1T3) localized normally to regions of overlap within MT bundles (Fig. 6E). We determined if the TOG mutants could rescue the viability of cls1 null cells using a spore viability assay. Wild-type Cls1-expressing plasmids displayed nearly full rescue of Cls1 function, while the vector control and each of the cls1 TOG mutant genes showed little significant rescue (Fig 6F). To determine if TOG function is needed for MT stabilization, we induced high expression of the cls1 mutant genes. Overexpression of mCherry-Cls1 stabilized MT bundles and bound along the MT-lattices, so that they were resistant to the MT disrupting drug, methyl-benzidazole-carbamate (MBC; Bratman and Chang, 2007). In contrast, in each of the strains overexpressing the cls1 TOG mutants or in the vector control, MT bundles were not stabilized and shrank to single perinuclear dots (Fig. 6G, H). A Cls1T5 mutant (T5 mutation: R465A) showed similar but less severe defects (Supplementary Tables S2). Thus, binding of Cls1p to tubulin dimer is essential for its function and for MT stabilization activity both in vitro and in vivo.
Discussion
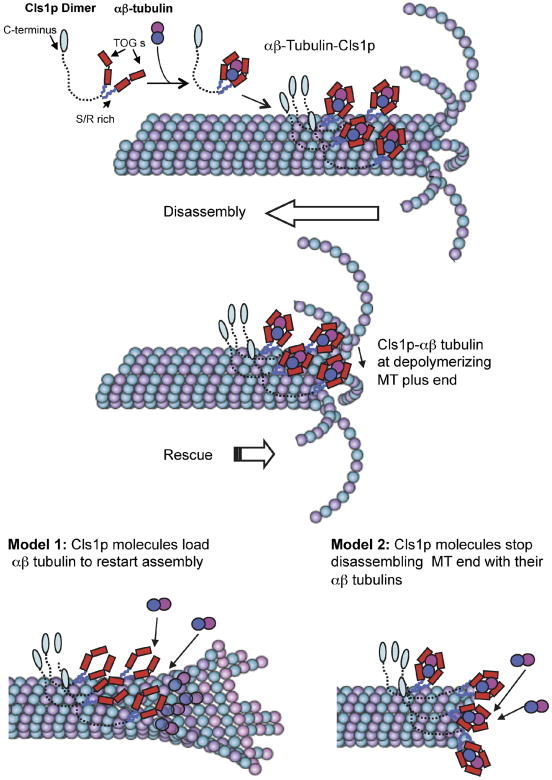
The experiments described here show that S. pombe CLASP, Cls1p, promotes MT rescue and suppresses catastrophe by recruiting tubulin dimer to the MT lattice. Consistent with previous studies indicating that the Cls1p is required for MT rescue in vivo (Bratman and Chang, 2007), we show that purified Cls1p alone is sufficient to mediate these activities without other proteins in vitro. Cls1p is a homodimer that captures a soluble αβ-tubulin heterodimer, encircling the tubulin with its two sets of tandem TOG domains; it also binds tightly to the MT lattice (Fig. 7). The binding of tubulin dimer is required for Cls1p to regulate rescue and catastrophe, but is not required for its effects on MT polymerization and depolymerization. These findings suggest the following model for rescue (Fig. 7). A group of Cls1p molecules clutching tubulin dimers bind on the MT lattice and await a MT depolymerization event. When a plus end shrinks to this site, these Cls1p-tubulin complexes halt depolymerization and restart growth (Fig. 7). In vitro studies with pure tubulin show that rescue depends on tubulin concentration; rescues are very rare at physiological concentrations (6 μM tubulin dimer), but are more frequent at much higher concentrations (Walker et al, 1988). Thus Cls1p could promote rescue by increasing the local tubulin dimer concentration. We envision two ways in which Cls1p could promote MT rescue (Fig. 7). In one model, Cls1 molecules deliver a GTP tubulin dimer into MT plus ends to restart MT assembly. The tubulin is released and incorporated into the MT. In this model, Cls1p acts like a polymerase to promote growth. In another (not mutually exclusive) model, local clusters of Cls1p-tubulin halt protofilament disassembly by transiently stabilizing the MT plus end, presenting the bound tubulin, without necessarily releasing it onto the MT plus end. Cls1p may suppress catastrophe by a related mechanism, promoting local rescue events so close to the plus end that the disassembly phase remains undetected.
Figure 7. Mechanism of CLASP in promoting MT rescues.
The S. pombe CLASP, Cls1p, is a dimer that binds a single αβ-tubulin dimer through two sets of TOG domains and binds to the MT lattice through two S/R rich domains. The C-terminal domain, shown extended, may bind other molecules. Cls1p dimers loaded with αβ-tubulin dimer bind non-uniformly along the MT lattice. When the disassembling end reaches a position of high local Cls1p density, Cls1p promotes rescue by halting disassembly and stimulating re-growth. Two models may explain Cls1p activity: model 1 shows Cls1p molecules loading their bound tubulin to MT plus end to restart assembly (left). Model 2 shows Cls1p molecules using their bound tubulin to halt protofilament and pause MT plus end disassembly. Dynamic MT Images were adapted from Akhmanova and Steinmetz (2008).
The combined in vitro and in vivo evidence makes CLASP a compelling candidate for a rescue factor. Our data show that CLASP promotes rescues at essentially physiological tubulin concentrations. In S. pombe, genetic studies show that Cls1p is responsible for all measurable rescue events in these cells and does not affect any other MT parameters (Bratman and Chang, 2007). Loss of CLASP in motile fibroblasts leads to a decrease of MT rescue frequency (Mimori-Kiyosue et al, 2005). Although activities of other CLASPs have not yet been examined in vitro, we propose that CLASPs have a general function in promoting MT rescue, consistent with their various biological roles. Near MT minus ends, such as at the centrosome and sites of MT nucleation at the Golgi, CLASP may wait on the MT lattice and prevent complete depolymerization, stabilizing a MT stub that is competent to grow again (Efimov et al, 2007; Bratman and Chang, 2007). At the kinetochore and at the leading edge of migrating cells, CLASP is needed for sustained MT assembly and suppression of catastrophes (Maiato et al, 2003b; Mimori-Kiyosue et al, 2005). We suggest that if CLASP is targeted to the MT plus end, it may suppress catastrophe by promoting local rescue events at plus ends so that MT disassembly is not detected.
Our work shows that CLASP is structurally related to XMAP215-- another TOG-domain protein. The two sets of TOG domains in a CLASP dimer wrap around tubulin just as do the two sets of tandem TOG domains in a Stu2p dimer or those within an XMAP215 monomer (Al-Bassam et al, 2006; Brouhard et al, 2008). These proteins contact tubulin dimer through a conserved surface located in the turns between successive helices of a HEAT repeat (Al-Bassam et al 2007; Slep and Vale, 2007). TOG domains bind free tubulin dimers and not tubulins within a MT, presumably because sites of interaction on the tubulin are occluded when the tubulin is incorporated into the lattice.
Despite their structural similarity, CLASPs and XMAP215/Dis1 proteins have quite different effects on MT dynamics. One evident difference between these proteins is how they interact with the MT lattice. XMAP215 proteins diffuse along the MT lattice and bind selectively at MT plus ends, where they accelerate both assembly and disassembly (Brouhard et al, 2008). In contrast, we detect no accumulation of Cls1p at MT plus ends either in vitro or in vivo. Instead, Cls1p molecules bind with high affinity to the MT lattice and exhibit little movement. It will be interesting to see whether other differences between the two proteins, for instance within the TOG domain, also contribute to their differences in activity.
Several other MAPs including MAP4, MAP2, EB1 and kinesin-8 have also been shown increase MT rescue frequencies in vitro (Pryer et al, 1992; Ookata et al 1995; Ichihara et al, 2001; Gupta et al, 2006; Katsuki et al, 2009), but in general, the in vivo significance of their rescue activities are not yet clear. Conventional MAPs, like MAP2, may promote rescue by binding and stabilizing the MT lattice (Ichihara et al, 2001), while the kinesin-8 Kif18A binds tightly to the MT plus end upon reaching the end of its motile cycle and thereby decreases MT dynamic transitions (Du et al, 2010). Our work shows that Cls1p promotes MT rescue by a mechanism distinct from those of other factors. Some recent work suggests that many MT rescue events in mammalian cultured cells occur at sites on the MT lattice that also stain with an antibody specific for the GTP form of tubulin (Dimitrov et al, 2008). It will be important to test if MAPs such as CLASP are located at these sites. We observed in our in vitro experiments that CLASP binds more readily to a GMPCPP-MT seeds than it does to the dynamic parts of the MT, suggesting that CLASP recognizes structural features of the MT lattice that reflect the nucleotide state of the polymerized tubulin. It is also possible that the GTP-tubulins associated with the MT lattice in vivo are actually GTP-tubulin dimers bound to the lattice by CLASP.
Materials and Methods
S. pombe Cls1p constructs, protein expression and purification
cDNA encoding full-length S. pombe Cls1p, Cls1pT1T3 (residues 1-1462 with or without C-terminal GFP), Cls1pTOG (residues 1-500) and Cls1p604 (residues 1-604) was cloned into a modified baculovirus expression vector. Proteins were expressed in Hi5 insect cells and purified using standard approaches, as described in detail in the supplementary materials and methods.
Biophysical analyses of Cls1p constructs and their complexes with tubulin dimer
Tubulin dimers were purified from bovine brains using the standard approaches (Mitchison and Kirschner, 1984). For size exclusion chromatography (SEC), Cls1p constructs were allowed to form complexes with various amounts of tubulin dimer for 5 minutes, and were loaded onto a 10/5 Superdex 200 column pre-equilibrated at 4°C with 25 mM HEPES, 80 mM KCl, 1 mM EGTA, pH 7.0, and eluted in 0.5 ml fractions, which were evaluated by SDS-PAGE. Apparent and measured molecular masses for Cls1p and constructs and tubulin complexes were determined by size-exclusion column and by sedimentation equilibrium analytical ultracentrifugation, respectively, as described in the supplementary materials and methods.
MT co sedimentation
Tubulin at 1.8 mg/ml was polymerized into MTs in the presence GMPPCPP for 2 hours. Various Cls1p constructs and XMAP215 were added in increasing amounts to a constant concentration (8.1 μM) of GMCPPP-stabilized MTs and co-sedimented by ultracentrifugation. The contents of Cls1p and its constructs in the supernatants and pellet were determined by SDS-PAGE. Gels were scanned and quantified by densitometry of the Cls1p-protein bands. Binding curves and dissociation constants for Cls1p and other constructs were fit using Prizm software. We detect Alexa-F-488 fluorescence at 488 nm in SDS-PAGE using a Typhoon™ imager.
Electron microscopy of Cls1p-tubulin complexes
Early size exclusion chromatography fractions for Cls1p and Cls1pTOG alone or in complex with tubulin dimer were diluted to 0.1 mg/ml, and incubated for 2 minutes on glow discharged continuous carbon support on 400 copper mesh grids. Grids were washed quickly with gel filtration buffer, and then stained with 0.5 % uranyl formate and dried. Samples were imaged at 52,000 × magnification using a Techni-12 electron microscope (Philips) operated at 120 kV using a low dose protocol. EM images were recorded on a 2K × 2K Gatan CDD (model 894) using Digital Micrograph software (Gatan) or image plates (Datibis Corp). Cls1-tubulin image classification and alignment were carried out with the program SPIDER (Shaikh et al, 2008), described in detail in the supplementary materials and methods.
TIRF microscopy studies of dynamic or stable MTs with Cls1p
We used a modified version of the approach described by Brouhard et al (2008). Tubulin dimers were labeled with succinimidyl ester or tetraflourophenyl esters of biotin, Alexa-Fluor-488 or Texas-Red esters (Invitrogen) and recycled as described by Mitchison and Kirschner (1984). MT seeds were polymerized with 2 mM GMPCPP from a 1.8 mg/ml mixture of Texas red, biotin labeled and unlabelled tubulin dimers in a 4:2:1 ratio at 37 °C for 2 hours. Glass coverslips were cleaned and silanized as described (Brouhard et al, 2008). Flow cells were constructed (Brouhard et al, 2008) and treated as described below and in the Supplementary materials and methods. Fully sealed flow cells were mounted onto a Nikon microscope with a 60× Nikon TIRF lens, warmed to 35 °C by a temperature collar. MTs were imaged in the evanescent wave by 488 and 568 nm laser excitation. Dual emission data were collected in 2.1 sec intervals for 18.4 min (1125 s) with a side entry iXon EM CCD (Andor Sciences) with each channel projected onto split fields. Details for dynamic and stabilized MT imaging is described in supplementary materials and methods.
Image analysis and measurement of MT dynamics parameters
Image stacks were analyzed using EMBL-ImageJ software (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997-2009). Raw image stacks were bleach corrected by normalization of total image intensity to the average intensity of the first image and scaling up the value of bleached images accordingly. Kymographs were constructed for dynamic MTs growing from each MT seed to study the transitions and rates of assembly and disassembly. MT assembly and disassembly rates were measured as slopes in kymographs of individual dynamic MTs. Frequencies of catastrophe and rescue events were measured for individual dynamic MTs formed from each seed by dividing the number of events by the duration of assembly (for catastrophe) and disassembly (for rescue). A rescue event was identified as a reversal of measureable disassembly (at least 10 pixels) to assembly. MT pauses were extremely rare and were not counted as rescue events. Further details are described in the supplementary materials and methods
Cls1p TOG mutants in S. pombe
A Stratagene Site-directed mutagenesis kit was used to introduce mutations into pREP42x nmt*-mCherry-Cls1 (1-1462; Bratman and Chang, 2007). For induction of Cls1p expression from the nmt1* promoter, cells were grown for 14-16 hrs at 30°C in media lacking thiamine. Assays for Cls1p rescue of viability, MT stabilization and Cls1p localization are as described in Bratman and Chang, 2007. Maximum intensity projections of spinning disc confocal images are shown.
Supplementary Material
Acknowledgments
We thank Anthony Hyman and Jonathan Howard and their laboratories (Max Planck Institute-CBG, Dresden) for hosting J.A.B to gain expertise in TIRF microscopy prior to this work. We thank Scott Bratman and members of Harrison and Chang laboratories for suggestions and advice throughout this work. J.A.B and F.C. acknowledge support from the National Institutes of Health (Pathway to Independence GMO8429 for J.A.B) and (GM069670 for F.C.), respectively. S.C.H is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, Galjart N. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. Epub 2008 Mar 2005. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, Larsen NA, Hyman AA, Harrison SC. Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure. 2007;15:355–362. doi: 10.1016/j.str.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, van Breugel M, Harrison SC, Hyman A. Stu2p binds tubulin and undergoes an open-to-closed conformational change. J Cell Biol. 2006;172:1009–1022. doi: 10.1083/jcb.200511010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose JC, Shoji T, Kotzer AM, Pighin JA, Wasteneys GO. The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell. 2007;19:2763–2775. doi: 10.1105/tpc.107.053777. Epub 2007 Sep 2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose JC, Wasteneys GO. CLASP modulates microtubule-cortex interaction during self-organization of acentrosomal microtubules. Mol Biol Cell. 2008;19:4730–4737. doi: 10.1091/mbc.E08-06-0665. Epub 2008 Aug 4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Laan L, Schek H, Munteanu EL, Sandblad L, Dogterom M, Brunner D, Surrey T. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. doi: 10.1038/nature06386. Epub 2007 Dec 1102. [DOI] [PubMed] [Google Scholar]
- Bratman SV, Chang F. Stabilization of overlapping microtubules by fission yeast CLASP. Dev Cell. 2007;13:812–827. doi: 10.1016/j.devcel.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratman SV, Chang F. Mechanisms for maintaining microtubule bundles. Trends Cell Biol. 2008;18:580–586. doi: 10.1016/j.tcb.2008.09.004. Epub 2008 Oct 2023. [DOI] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov A, Quesnoit M, Moutel S, Cantaloube I, Pous C, Perez F. Detection of GTP-tubulin conformation in vivo reveals a role for GTP remnants in microtubule rescues. Science. 2008;322:1353–1356. doi: 10.1126/science.1165401. Epub 2008 Oct 1316. [DOI] [PubMed] [Google Scholar]
- Drabek K, van Ham M, Stepanova T, Draegestein K, van Horssen R, Sayas CL, Akhmanova A, Ten Hagen T, Smits R, Fodde R, et al. Role of CLASP2 in microtubule stabilization and the regulation of persistent motility. Curr Biol. 2006;16:2259–2264. doi: 10.1016/j.cub.2006.09.065. [DOI] [PubMed] [Google Scholar]
- Du Y, English CA, Ohi R. The kinesin-8 Kif18A dampens microtubule plus end dynamics. Curr Biol. 20:374–380. doi: 10.1016/j.cub.2009.12.049. Epub 2010 Feb. [DOI] [PubMed] [Google Scholar]
- Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Beuter C, Craven RA, Bagley S, Wilks D, Fleig U, Hagan IM. S. pombe CLASP needs dynein, not EB1 or CLIP170, to induce microtubule instability and slows polymerization rates at cell tips in a dynein-dependent manner. Genes Dev. 2006;20:2421–2436. doi: 10.1101/gad.381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galjart N, Grosveld F, Vorobjev I, Tsukita S, Akhmanova A. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Odde DJ. Stochastic simulation and graphic visualization of mitotic processes. Methods. 2010:22. doi: 10.1016/j.ymeth.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. Epub 2006 Aug 2013. [DOI] [PubMed] [Google Scholar]
- Ichihara K, Kitazawa H, Iguchi Y, Hotani H, Itoh TJ. Visualization of the stop of microtubule depolymerization that occurs at the high-density region of microtubule-associated protein 2 (MAP2) J Mol Biol. 2001;312:107–118. doi: 10.1006/jmbi.2001.4934. [DOI] [PubMed] [Google Scholar]
- Katsuki M, Drummond DR, Osei M, Cross RA. Mal3 masks catastrophe events in Schizosaccharomyces pombe microtubules by inhibiting shrinkage and promoting rescue. J Biol Chem. 2009;284:29246–29250. doi: 10.1074/jbc.C109.052159. Epub 22009 Sep 29249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Lyle KS, Gierke S, Matov A, Danuser G, Wittmann T. GSK3beta phosphorylation modulates CLASP-microtubule association and lamella microtubule attachment. J Cell Biol. 2009;184:895–908. doi: 10.1083/jcb.200901042. Epub 2009 Mar 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G, Grigoriev I, Mimori-Kiyosue Y, Ohtsuka T, Higa S, Kitajima I, Demmers J, Galjart N, Houtsmuller AB, Grosveld F, Akhmanova A. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev Cell. 2006;11:21–32. doi: 10.1016/j.devcel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Laycock JE, Savoian MS, Glover DM. Antagonistic activities of Klp10A and Orbit regulate spindle length, bipolarity and function in vivo. J Cell Sci. 2006;119:2354–2361. doi: 10.1242/jcs.02957. [DOI] [PubMed] [Google Scholar]
- Lee H, Engel U, Rusch J, Scherrer S, Sheard K, Van Vactor D. The microtubule plus end tracking protein Orbit/MAST/CLASP acts downstream of the tyrosine kinase Abl in mediating axon guidance. Neuron. 2004;42:913–926. doi: 10.1016/j.neuron.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Maffini S, Maia AR, Manning AL, Maliga Z, Pereira AL, Junqueira M, Shevchenko A, Hyman A, Yates JR, 3rd, Galjart N, et al. Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr Biol. 2009;19:1566–1572. doi: 10.1016/j.cub.2009.07.059. Epub 2009 Sep 1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Fairley EA, Rieder CL, Swedlow JR, Sunkel CE, Earnshaw WC. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 2003;113:891–904. doi: 10.1016/s0092-8674(03)00465-3. [DOI] [PubMed] [Google Scholar]
- Maiato H, Rieder CL, Earnshaw WC, Sunkel CE. How do kinetochores CLASP dynamic microtubules? Cell Cycle. 2003;2:511–514. doi: 10.4161/cc.2.6.576. [DOI] [PubMed] [Google Scholar]
- Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. Epub 2004 Dec 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, Akhmanova A. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Ookata K, Hisanaga S, Bulinski JC, Murofushi H, Aizawa H, Itoh TJ, Hotani H, Okumura E, Tachibana K, Kishimoto T. Cyclin B interaction with microtubule-associated protein 4 (MAP4) targets p34cdc2 kinase to microtubules and is a potential regulator of M-phase microtubule dynamics. J Cell Biol. 1995;128:849–862. doi: 10.1083/jcb.128.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira AL, Pereira AJ, Maia AR, Drabek K, Sayas CL, Hergert PJ, Lince-Faria M, Matos I, Duque C, Stepanova T, et al. Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol Biol Cell. 2006;17:4526–4542. doi: 10.1091/mbc.E06-07-0579. Epub 2006 Aug 4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer NK, Walker RA, Skeen VP, Bourns BD, Soboeiro MF, Salmon ED. Brain microtubule-associated proteins modulate microtubule dynamic instability in vitro. Real-time observations using video microscopy. J Cell Sci. 1992;103:965–976. doi: 10.1242/jcs.103.4.965. [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep KC. The role of TOG domains in microtubule plus end dynamics. Biochem Soc Trans. 2009;37:1002–1006. doi: 10.1042/BST0371002. [DOI] [PubMed] [Google Scholar]
- Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A, Reis R, Sampaio P, Sunkel CE. The Drosophila CLASP homologue, Mast/Orbit regulates the dynamic behaviour of interphase microtubules by promoting the pause state. Cell Motil Cytoskeleton. 2007;64:605–620. doi: 10.1002/cm.20208. [DOI] [PubMed] [Google Scholar]
- Walker RA, O'Brien ET, Pryer NK, Soboeiro MF, Voter WA, Erickson HP, Salmon ED. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Waterman-Storer CM. Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3beta in migrating epithelial cells. J Cell Biol. 2005;169:929–939. doi: 10.1083/jcb.200412114. Epub 2005 Jun 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, You L, Pasqualone D, Kopski KM, Huffaker TC. Stu1p is physically associated with beta-tubulin and is required for structural integrity of the mitotic spindle. Mol Biol Cell. 2002;13:1881–1892. doi: 10.1091/mbc.01-09-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.