Abstract
Stromal processing peptidase (SPP) is a metalloendopeptidase located in the stroma of chloroplasts, and it is responsible for the cleavage of transit peptides from preproteins upon their import into the organelle. Two independent mutant Arabidopsis lines with T-DNA insertions in the SPP gene were analysed (spp-1 and spp-2). For both lines, no homozygous mutant plants could be detected, and the segregating progeny of spp heterozygotes contained heterozygous and wild-type plants in a ratio of 2∶1. The siliques of heterozygous spp-1 and spp-2 plants contained many aborted seeds, at a frequency of ∼25%, suggesting embryo lethality. By contrast, transmission of the spp mutations through the male and female gametes was found to be normal, and so gametophytic effects could be ruled out. To further elucidate the timing of the developmental arrest, mutant and wild-type seeds were cleared and analysed by Nomarski microscopy. A significant proportion (∼25%) of the seeds in mutant siliques exhibited delayed embryogenesis compared to those in wild type. Moreover, the mutant embryos never progressed normally beyond the 16-cell stage, with cell divisions not completing properly thereafter. Heterozygous spp mutant plants were phenotypically indistinguishable from the wild type, indicating that the spp knockout mutations are completely recessive and suggesting that one copy of the SPP gene is able to produce sufficient SPP protein for normal development under standard growth conditions.
Introduction
The chloroplast is a unique plant cell compartment which harbours many essential processes such as photosynthesis, starch metabolism, and the biosynthesis of lipids and secondary metabolites [1], [2]. Like all plastids, chloroplasts are derived from an ancient free-living cyanobacterial ancestor that was incorporated into early eukaryotic cells through endosymbiosis [3]. As a result of this evolutionary origin, modern chloroplasts contain DNA and are able to synthesize roughly one hundred of their own proteins [4]. Nonetheless, the bulk of the ∼3000 different proteins in chloroplasts are encoded in the nuclear genome and must be imported post-translationally from the cytosol [5], [6].
Soon after the emergence of the signal hypothesis to account for the translocation of ER proteins, it was suggested that nucleus-encoded chloroplast proteins are similarly synthesized with a targeting tag that directs them to the organelle [7], [8]. This tag is an N-terminal extension of the protein called a transit peptide, and it is cleaved off after organellar import, producing a smaller, mature form of the chloroplast protein [9]. Chloroplast transit peptides vary greatly in length and amino acid sequence, and while secondary structural features have been reported in some cases the general significance of such observations remains uncertain [10], [11]. Thus, it is not fully understood how different preproteins are all targeted quite specifically to the same organelle. Transit peptides do contain slightly more hydroxylated residues and fewer acidic residues than average, giving them a net positive charge, and it has been suggested that a lack of a secondary structure might be necessary for their targeting properties [12].
The transit peptide is recognized by receptor components at the chloroplast surface, and subsequently the preprotein is guided through pores in the outer and inner envelope membranes. The multiprotein assemblies responsible for these recognition and translocation events are the TOC and TIC complexes (translocon at the outer/inner envelope membrane of chloroplasts) [13], [14], [15], [16], [17]. Upon reaching the stromal side of the envelope, the transit peptide is removed by the stromal processing peptidase (SPP), a metalloendopeptidase of the M16 family (members of which include subunit β of the mitochondrial processing peptidase, MPP, and Escherichia coli pitrilysin) which has a high specificity for chloroplast transit peptides [18], [19], [20]. The SPP enzyme recognizes a stretch of basic residues with weak sequence or physicochemical conservation at the C-terminus of the transit peptide [21], [22], [23]. Following recognition, it cleaves the transit peptide from the mature sequence using the catalytic activity of its zinc-binding domain, and subsequently proteolyses the C-terminal binding site of the transit peptide which facilitates release of the peptide fragments so that they may be degraded by the presequence protease, PreP [22], [24], [25]. Homologues of SPP exist in red and green algae as well as in the malaria parasite, Plasmodium falciparum, suggesting that the protein's function is well conserved amongst plastid-containing organisms [20]. An ancestral activity was probably inherited with the original endosymbiont, as SPP-related sequences even exist in cyanobacteria.
Antisense mediated down-regulation of SPP gene expression in Arabidopsis or tobacco plants resulted in chlorotic, albino or even a seedling-lethal phenotypes, indicating that the SPP enzyme plays an important role in chloroplast biogenesis [26], [27]. Indeed, the antisense lines displayed reduced numbers of chloroplasts per cell, and those organelles that were present were structurally abnormal. Both in vitro import experiments (using isolated chloroplasts and radiolabelled preproteins) and an in vivo targeting assay (involving expression of a transit peptide fusion to green fluorescent protein) revealed defects in chloroplast protein import in the antisense lines [26], [27]. Such defects may reflect the fact that most components of the TOC-TIC import machinery are themselves synthesized as preproteins (with transit peptides that presumably must be removed before those components can begin to operate), or indicate that transit peptide cleavage is a fully-integrated step of the translocation mechanism. More recently, a hypomorphic spp allele was identified in a forward-genetic screen of ethyl methanesulfonate-mutagenized rice plants [28]. The relevant mutant lacks a conserved glutamate residue in a C-terminal M16 domain of SPP, and it exhibits chlorosis associated with small, underdeveloped chloroplasts as well as defective root development.
The aforementioned in vivo studies all analysed the consequences of reduced levels of SPP activity for plant development. To determine the consequences of complete loss of SPP protein, we identified and characterized T-DNA knockout mutants in the Arabidopsis background.
Results and Discussion
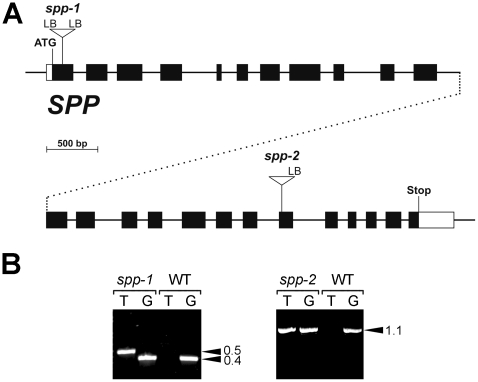
To further assess the importance of SPP during plant development, two different Arabidopsis lines with T-DNA insertions in the SPP gene (At5g42390) were obtained from the Nottingham Arabidopsis Stock Centre (NASC). These mutants were called spp-1 (SALK_087683) and spp-2 (SAIL_242_H11). Firstly, both T-DNA insertion sites were confirmed by genomic PCR, and by sequencing the T-DNA/gene junctions at one or both sides, as indicated (Figure 1A). In spp-1, the T-DNA disrupts the first exon of the SPP gene, whereas in spp-2 the insertion lies in the nineteenth exon (Figure 1A).
Figure 1. Molecular characterization of the SPP T-DNA insertion mutants.
(A) Schematic diagram of the SPP gene showing the location of each T-DNA insertion. Protein-coding exons are represented by black boxes and untranslated regions by white boxes; introns are represented by thin lines between the boxes. The gene model shown is based on the RIKEN full-length cDNA clone, RAFL07-08-B09; it is separated over two lines (with the division at an arbitrary point near the middle, after intron 11; dotted lines) to aid presentation. Positions of T-DNA insertions are indicated precisely, but the insertion sizes are not to scale. ATG, translation initiation codons; Stop, translation termination codon; p(A), polyadenylation site. (B) Mutant genotype analysis by PCR. Genomic DNA samples extracted from wild-type and mutant plants (spp-1 and spp-2) were analysed by PCR. Appropriate T-DNA- and SPP-gene-specific primers were employed in two different combinations: the first (T) comprised one T-DNA primer (LB) and one gene-specific primer (forward for spp-1; reverse for spp-2); the second (G) comprised two gene-specific primers flanking the T-DNA insertion site. The results shown for spp-1 and spp-2 are representative of those obtained for all antibiotic-resistant plants tested; amplification using both T and G indicated the presence of both mutant and wild-type alleles, respectively, and demonstrated that the plants were heterozygous. Amplicon sizes are indicated at right (in kb).
Next, we attempted to identify homozygous mutant lines in each case, by genotyping 30 antibiotic-resistant plants for each line in PCR reactions using gene- and T-DNA-specific primers. However, when we did this, we found that all of the 60 tested plants (30 for each allele) were hemizygous for the relevant T-DNA insertion (Figure 1B), suggesting that the homozygous genotypes are not viable. Consistent with this notion, when segregation of the antibiotic-resistance marker associated with each T-DNA insertion was analysed (by plating seeds from heterozygous plants on medium containing either kanamycin [spp-1] or phosphinothricin [spp-2]), significant deviations from standard Mendelian inheritance were observed: only two antibiotic-resistant plants were observed for every one antibiotic-sensitive plant, instead of the more normal 3∶1 segregation ratio (Table 1).
Table 1. Segregation of the T-DNA-borne antibiotic resistance markers in the spp mutant lines.
| Parental genotype | Plant number | Antibiotic | Resistant (R) | Sensitive (S) | R∶S ratio | P-valuea |
| +/spp-1 | 1 | Kanamycin | 87 | 37 | 2.35 | 0.41 |
| 2 | 82 | 44 | 1.86 | 0.71 | ||
| 3 | 99 | 42 | 2.36 | 0.37 | ||
| 4 | 69 | 46 | 1.50 | 0.13 | ||
| sum | 337 | 169 | 1.99 | 0.97 | ||
| +/spp-2 | 1 | Phosphinothricin | 74 | 38 | 1.95 | 0.89 |
| 2 | 105 | 43 | 2.44 | 0.27 | ||
| 3 | 78 | 43 | 1.81 | 0.61 | ||
| 4 | 100 | 45 | 2.22 | 0.56 | ||
| 5 | 83 | 40 | 2.08 | 0.85 | ||
| sum | 440 | 209 | 2.11 | 0.54 |
Goodness of fit of the observed ratios to 2∶1 was assessed by χ2 analysis. P-values are the probabilities that the observed ratios differ from 2∶1 due to random chance only.
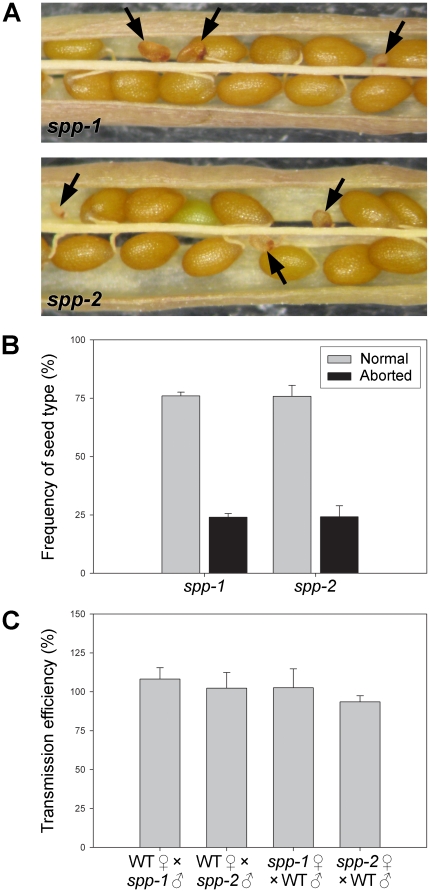
To investigate the possibility of embryo lethality, we carefully examined the seeds within ripe siliques of heterozygous spp-1 and spp-2 plants. Significant numbers of small, aborted seeds were observed in both mutant genotypes (Figure 2A), but not within wild-type siliques (data not shown). Amongst the fertilized seeds, abortions occurred with a frequency of almost exactly 25% (Figure 2B), strongly supporting the notion that the homozygous mutant genotypes were responsible for developmental arrest. Small numbers (∼3–5%) of what appeared to be failed ovules were also apparent in the spp siliques (data not shown), suggesting that there might be an additional effect of the mutations on gametophytic transmission [29], [30]. To assess this possibility, we conducted reciprocal crossing experiments, between both spp alleles and wild-type plants, analysing transmission of the mutations to the resulting F1 progenies by plating on selective media. However, the results revealed essentially normal transmission of both mutations through both male and female gametes (Figure 2C) [31]. This indicated that the spp-mediated block in development is exclusively post-fertilization, occurring during embryogenesis, and that the presumed failed ovules observed in ripe siliques (mentioned earlier) were perhaps very early seed abortions and/or the consequences of environmental stresses.
Figure 2. Embryo lethality of the spp-1 and spp-2 mutations.
(A) Appearance of aborted seeds within mature siliques of heterozygous spp-1 and spp-2 plants. Aborted seeds indicative of embryo lethality (see black arrows) are smaller in size than normal seeds, and have a darker, shrivelled appearance. (B) Frequencies of normal and aborted seeds within ripe siliques of spp-1 and spp-2 heterozygotes. The data shown are means (±SD) derived from analyses of six different siliques per genotype, each one from a different plant. Values shown refer to fertilized seeds only. (C) Reciprocal crossing analysis. Transmission of the spp-1 and spp-2 mutations through the male and female gametes was assessed by crossing heterozygotes of both mutants to wild type, in both directions, multiple times (28–50 crosses using 3–4 different plants were conducted per direction, per allele). Inheritance of the spp mutations in the F1 progeny was assessed by determining antibiotic resistance of the F1 plants (kanamycin for spp-1; phosphinothricin for spp-2). Transmission efficiencies were calculated for each plant as described previously [31], and then these values were used to derive the means shown (±SD).
It is well documented that the disruption of chloroplast functions can lead to a block in embryogenesis [32]. In fact, it has been estimated that a disproportionately large number (∼25–30%) of non-redundant, embryo-lethal mutations in Arabidopsis affect chloroplast proteins [33], [34]. Prominent amongst the chloroplast functions that lead to embryo arrest, when disrupted, are: plastid gene expression (including RNA and protein synthesis); non-photosynthetic metabolism (including amino acid, vitamin and nucleotide biosynthesis); and, protein modification, transport and degradation [32], [34]. Of the previous reports linking chloroplast function to embryogenesis, those pertaining to two core components of the chloroplast protein import machinery, atToc75-III and atTic110, are most relevant [35], [36], [37], [38]. The aborted seeds observed in the spp mutant siliques (Figure 2A) appeared to be somewhat larger than those in toc75-III [35], and smaller in size than those in tic110 siliques [37], suggesting that the spp mutations may affect embryo development at a stage intermediate between toc75-III and tic110.
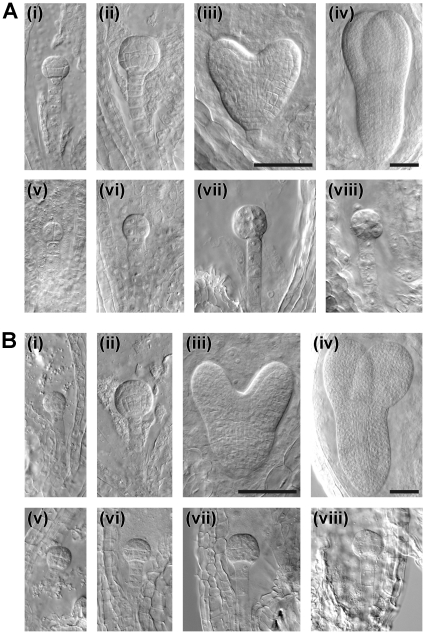
To precisely determine the stage of embryogenesis during which the spp mutations arrest growth, we conducted a detailed examination of developing embryos in both mutants as well as in the wild type, using Nomarski optics microscopy (Table 2). Figure 3 shows equivalent developmental series for normal (i–iv) and mutant (v–viii) embryos within immature siliques of self-pollinated spp-1 (Figure 3A) and spp-2 (Figure 3B) heterozygotes. Typically, when normal embryos were at the 16-cell stage (Figure 3, i panels), mutant embryos were delayed at the 2- to 8-cell stages (Figure 3, v panels). As the normal embryos progressed to the globular and heart stages (Figure 3, ii and iii panels), the mutants were retarded at the 8- to 16-cell stages (Figure 3, vi and vii panels). In fact, the mutant embryos were never seen to develop normally beyond the 16-cell stage, even when the other embryos had reached the torpedo stage (Figure 3, iv and viii panels). Cell boundaries in the most mature mutant embryos were frequently indistinct, and such embryos often had an irregular or swollen appearance. As the normal seeds reached the cotyledon stage, significant numbers of aborted seeds (with degenerated structures containing no discernable embryo) became apparent within the mutant siliques (Table 2). In contrast with the situation in spp siliques, where two distinct classes of embryos could be observed (normal and mutant; the latter corresponding to ∼25% of the total), embryos within individual wild-type siliques rarely spanned more than three consecutive developmental stages (Table 2).
Table 2. Distribution of embryo phenotypes in single siliques of wild type and spp mutant heterozygotes.a .
| Genotype | Silique numberb | 1-cell | 2-cell | 8-cell | 16-cell | Aborted seedc | EG | LG | EH | Heart | Torpedo | Cotyledon | Total scored | Fraction delayed or abnormal |
| WT | 2 | - | - | 9 | 22 | - | 5 | - | - | - | - | - | 36 | - |
| 4 | - | - | - | 4 | - | 11 | 23 | 6 | - | - | - | 44 | - | |
| 6 | - | - | - | - | - | 4 | 33 | 10 | - | - | - | 47 | - | |
| 8 | - | - | - | - | - | - | 20 | 18 | 9 | - | - | 47 | - | |
| 10 | - | - | - | - | - | - | - | 17 | 33 | - | - | 50 | - | |
| 12 | - | - | - | - | - | - | - | - | 37 | 18 | - | 55 | - | |
| 14 | - | - | - | - | - | - | - | - | 1 | 19 | 33 | 53 | - | |
| 16 | - | - | - | - | - | - | - | - | - | - | 53 | 53 | - | |
| spp-1 | 2 | 2 | 15 | 22 | 5 | - | - | - | - | - | - | - | 44 | - |
| 4 | - | 2 | 12 | 25 | - | 2 | - | - | - | - | - | 41 | - | |
| 6 | 1 | 3 | 7 | 17 | - | 17 | - | - | - | - | - | 45 | - | |
| 8 | - | - | 3 | 12 | - | 3 | 15 | 15 | - | - | - | 48 | 0.31 | |
| 10 | - | 2 | 2 | 8 | - | - | 5 | 4 | 21 | - | - | 42 | 0.29 | |
| 12 | - | - | - | 8 | 8 | - | - | - | 5 | 22 | 2 | 45 | 0.36 | |
| 14 | - | - | - | 6 | 6 | - | - | - | - | 3 | 26 | 41 | 0.29 | |
| spp-2 | 2 | - | 2 | 12 | 6 | - | 4 | - | - | - | - | - | 24 | - |
| 4 | - | - | 12 | 11 | - | 7 | 1 | 1 | - | - | - | 32 | - | |
| 6 | - | - | 4 | 6 | - | 12 | 5 | 3 | - | - | - | 30 | - | |
| 8 | - | - | 8 | 2 | - | 2 | 10 | 13 | - | - | - | 35 | - | |
| 10 | - | - | 2 | 5 | 1 | 9 | 5 | 6 | 10 | - | - | 38 | 0.21 | |
| 12 | - | - | 2 | 8 | 2 | - | - | - | 15 | 21 | - | 48 | 0.25 | |
| 14 | - | - | - | 2 | 7 | - | - | - | 2 | 30 | - | 41 | 0.22 | |
| 16 | - | - | - | 1 | 9 | - | - | - | - | 6 | 26 | 42 | 0.24 |
Embryo developmental stage names refer to the cell number or morphology of the embryo proper. Abbreviations are defined as follows: EG, early globular; LG, later globular; EH, early heart.
Siliques were numbered consecutively from the top of the inflorescence, such that the oldest siliques have the highest numbers.
Aborted seed were shrivelled with no visible embryo or endosperm development.
Figure 3. Analysis of embryo development in the spp mutants using Nomarski optics.
Equivalent developmental series for normal (i–iv) and mutant (v–viii) embryos within immature heterozygous siliques of spp-1 (A) and spp-2 (B). Normal embryos: i, 16-cell stage; ii, early globular stage; iii, heart stage; iv, torpedo stage. Corresponding mutant embryos from the same siliques: v, 2- to 8-cell stages; vi, 8- to 16-cell stages; vii and viii, arrested or abnormal 16-cell stages. Embryo developmental stage names refer to the cell number or morphology of the embryo proper. Images i–iii and v–viii are all at the same magnification (40× objective); images iv are at lower magnification (20× objective). Bars = 50 µm.
Thus, the spp mutations indeed arrest embryogenesis at a stage (the 16-cell stage) intermediate between those during which toc75-III (two-cell stage) and tic110 (globular stage) block growth [35], [36], [37], [38]. These differences in phenotype severity may partly reflect the differing expression patterns of the genes during embryogenesis: publicly-available microarray data show that atTOC75-III expression peaks at a much earlier stage in embryogenesis than that of either atTIC110 or SPP (Figure S1) [39], [40], [41]. The phenotypic differences may also be linked to the roles of the proteins, as has been discussed [34]. Toc75 is the channel component of the outer membrane through which the vast majority of chloroplast proteins gain entry, including many envelope proteins, while Tic110 is thought to play roles in channel formation and/or the coordination of chaperones at the inner membrane, and is likely to be utilized by a smaller subset of chloroplast-destined proteins [16], [17]. The proteins that are dependent upon SPP for proper targeting may be intermediate in number and/or importance. An alternative explanation is that the differing phenotypes are a reflection of differing stabilities of the proteins, and the extent to which they can persist at functional levels during embryogenesis following the segregative loss of a functional gene. Irrespective of the basis for these differences in severity, the data leave no doubt that all three proteins play crucial roles in protein transport, and that efficient protein transport into plastids is essential during embryogenesis.
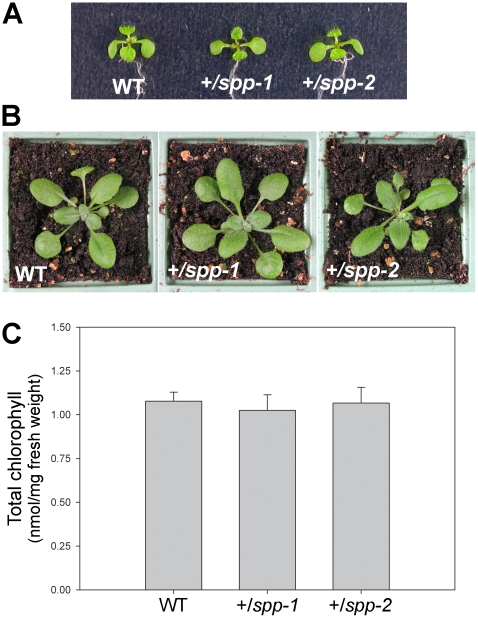
While toc75-III mutations are completely recessive [35], heterozygous tic110 plants are clearly chlorotic with quantifiable defects in chloroplast biogenesis [37]. In this respect, the spp mutations are more similar to toc75-III, as spp heterozygotes were visibly indistinguishable from wild type throughout development (Figure 4, A and B), and accumulated normal levels of chlorophyll pigment (Figure 4C). This indicates that a single copy of the SPP gene is able to produce sufficient quantities of the peptidase for normal growth under standard conditions. The phenotypic difference between tic110 heterozygotes and toc75-III or spp heterozygotes cannot easily be explained in terms of mRNA or protein levels, as quantitative RT-PCR and immunoblot experiments did not reveal more severe deficiencies in +/tic110 plants (Figure S2). As was discussed previously [42], the greater dosage dependency of the tic110 mutation may reflect the absence of excess expression capacity for atTic110 in the wild type.
Figure 4. Phenotypic analysis of the spp heterozygotes.
(A, B) Plants of the indicated genotypes were grown in vitro on selective medium (non-selective in the case of wild type) for 7 days, and then photographed (A). Additional similar plants were transferred to soil on day 7, and then allowed to grow further until they were 21 days old prior to photography (B). Representative plants are shown in each case. (C) Chlorophyll concentrations in 21-day-old plants grown as described above were determined using a SPAD-502 meter. Values shown are means (±SD) derived from six independent measurements per genotype, each one taken using a different plant. Units are nmol chlorophyll a+b per mg fresh weight of tissue.
Conclusion
While it was evident from earlier studies that SPP is an important chloroplast protein, the phenotype of homozygous knockout plants had not previously been reported. In this study, we employed Arabidopsis T-DNA insertion lines to demonstrate that SPP is indispensable in vivo. Homozygotes were absent from the progeny of plants carrying spp mutations, while a quarter of the seeds in the siliques of such plants were aborted, consistent with an embryo lethal phenotype. The mutant embryos exhibited delayed development, with cell divisions not terminating properly after the 16-cell stage. This work further emphasizes the importance of plastid functions during seed development, and extends the list of protein transport components known to be essential for plant viability.
Materials and Methods
Plant growth and chlorophyll analysis
All Arabidopsis thaliana plants were of the Columbia-0 ecotype. For in vitro growth, seeds were surface sterilized, sown on Murashige and Skoog agar medium containing 0.5% (w/v) sucrose in petri plates, cold treated at 4°C, and thereafter kept in a growth chamber, as described previously [43]. To select for the presence of T-DNA insertions, the following antibiotics were added to the medium: kanamycin monosulfate, 50 µg/ml (spp-1); phosphinothricin, 10 µg/ml (spp-2). All plants were grown under a long-day cycle (16 h light, 8 h dark).
Chlorophyll content was measured using a SPAD-502 meter following the manufacturer's instructions (Konica-Minolta, Osaka, Japan). Conversion from SPAD units to chlorophyll concentration values (nmol chlorophyll a+b per mg fresh weight) was done using a validated calibration curve [44].
Identification of the spp mutants
The spp-1 mutant was from the Salk Institute Genomic Analysis Laboratory (line number SALK_087683) [45], while the spp-2 mutant was from Syngenta (line number SAIL_242_H11) [46]. Mutant genotypes were assessed by PCR (Figure 1B). Genomic DNA was extracted from Arabidopsis plants using a published protocol [47] and PCR was conducted using standard procedures. The primers used were as follows: spp-1 forward, 5′-CTTCAAACCCTTTGCTACAAA-3′; spp-1 reverse, 5′-GACGATGGATTAAACCTAACT-3′; spp-1 T-DNA LB, 5′-GCGTGGACCGCTTGCTGCAACT-3′; spp-2 forward, 5′-AAACTGTGTATAGGTCTGGTT- 3′; spp-2 reverse, 5′-GGAGACGAGAGATGAGTATAGATAATGGGG-3′; spp-2 T-DNA LB, 5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′. Amplification products were resolved by agarose gel electrophoresis and stained with SYBR Safe (Invitrogen). The location of each T-DNA insertion was determined precisely (Figure 1A) by sequencing PCR products spanning both junctions (except in the case of spp-2, where only one junction was identified).
Seed and embryo analyses
The phenotypes of seeds in ripe siliques were determined by dissecting siliques held on double-sided sticky tape using a stereo microscope (Zeiss Stemi 2000). The analysis of cleared wild-type and spp mutant embryos using Nomarski optics was performed as described previously [35], [48], using a microscope equipped for differential interference contrast (Nikon Eclipse 80i).
Supporting Information
Expression patterns of essential chloroplast protein import apparatus genes during embryogenesis. Publicly-available Affymetrix microarray data corresponding to defined tissues and developmental stages of Arabidopsis embryogenesis [39], [40] were accessed using an electronic fluorescent pictograph (eFP) browser online [41]. Data for atTOC75-III (At3g46740), atTIC110 (At1g06950) and SPP (At5g42390) are shown. Values are means (±SE) derived from three or six independent measurements. SAM, shoot apical meristem.
(TIF)
Analyses of mRNA and protein expression in the
toc75-III
,
tic110
and
spp
heterozygotes. (A) Quantitative RT-PCR analysis of mRNA levels. Total-RNA samples were extracted from ∼10–30 whole seedlings of the indicated genotypes (and wild type) that had been grown in vitro for 14 days under standard conditions. RNA was isolated using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany), and was then treated with DNAse I (DNA-free; Ambion, Texas, USA). Quantitative RT-PCR was performed using an MJ Research Chromo4 Gradient Cycler (Bio-Rad, Hercules, CA, USA) and SYBR Green Jump Start Taq Ready Mix (Sigma, St. Louis, MO, USA) for high-throughput quantitative PCR. Relative quantification was determined according to published methods [49], [50]. Reactions were characterized by comparing threshold cycle (C
T) values; C
T is a unit-less value defined as the fractional cycle number at which the sample fluorescence signal passes a fixed threshold. The relative amount of transcript was calculated by subtracting the control gene (ACTIN 2; At3g18780) C
T-value from the gene-of-interest (atTOC75-III, atTIC110, SPP) C
T-value (ΔC
T). Then, ΔC
T values for the wild type were subtracted from those for the mutants to yield ΔΔC
T values, and these were used to estimate expression levels as 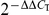 . Data shown are means (±SE) derived from four independent amplifications performed on three biological replicates. The ACTIN2 primers have been described previously [51]; the other primers used were: atTOC75-III sense 5′-TCG CAT CTC CAC TCA ATC-3′; atTOC75-III antisense, 5′-GTC TCT GTA TCT CGG TTA GG-3′; atTIC110 sense, 5′-CTC CTC AGG TGC CTT ATC AGA AG-3′; atTIC110 antisense, 5′-CGA GCA AGA GCA GCC GAG AAC-3′; SPP sense, 5′-AAG CTA GCC ATG ATT CTG CAA-3′; SPP antisense, 5′-CAT CAT GAG CAA CAG GAA GTT-3′. (B) Immunoblot analysis of protein levels. Total-protein was extracted from plant material samples equivalent to those employed in panel A, using previously-described procedures [51]. Three different protein amounts (80, 60 and 40 µg) of each genotype were analysed by immunoblotting, as described previously [51], using primary antibodies against atToc75-III, atTic110 and the plasma membrane H+-ATPase, PMA2 [51], [52], [53]; unfortunately, an SPP antibody was not available for this work. The secondary antibody was anti-rabbit IgG conjugated with horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and the detection reagent was ECL Plus (GE Healthcare, Chalfont St. Giles, UK). Chemiluminescence detection employed a Fujifilm LAS-4000 imager. Quantification of all images was performed using Aida software (Raytest, Straubenhardt, Germany). The atToc75-III and atTic110 data were normalized using equivalent PMA2 data. Values shown are means (±SE) derived from three independent measurements.
. Data shown are means (±SE) derived from four independent amplifications performed on three biological replicates. The ACTIN2 primers have been described previously [51]; the other primers used were: atTOC75-III sense 5′-TCG CAT CTC CAC TCA ATC-3′; atTOC75-III antisense, 5′-GTC TCT GTA TCT CGG TTA GG-3′; atTIC110 sense, 5′-CTC CTC AGG TGC CTT ATC AGA AG-3′; atTIC110 antisense, 5′-CGA GCA AGA GCA GCC GAG AAC-3′; SPP sense, 5′-AAG CTA GCC ATG ATT CTG CAA-3′; SPP antisense, 5′-CAT CAT GAG CAA CAG GAA GTT-3′. (B) Immunoblot analysis of protein levels. Total-protein was extracted from plant material samples equivalent to those employed in panel A, using previously-described procedures [51]. Three different protein amounts (80, 60 and 40 µg) of each genotype were analysed by immunoblotting, as described previously [51], using primary antibodies against atToc75-III, atTic110 and the plasma membrane H+-ATPase, PMA2 [51], [52], [53]; unfortunately, an SPP antibody was not available for this work. The secondary antibody was anti-rabbit IgG conjugated with horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and the detection reagent was ECL Plus (GE Healthcare, Chalfont St. Giles, UK). Chemiluminescence detection employed a Fujifilm LAS-4000 imager. Quantification of all images was performed using Aida software (Raytest, Straubenhardt, Germany). The atToc75-III and atTic110 data were normalized using equivalent PMA2 data. Values shown are means (±SE) derived from three independent measurements.
(TIF)
Acknowledgments
We are extremely grateful to Ramesh Patel for initial observations made using the spp-2 allele, and for general advice and technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a Gatsby Charitable Foundation Sainsbury PhD Studentship (http://www.gatsby.org.uk/) (to RT), and by Biotechnology and Biological Sciences Research Council (http://www.bbsrc.ac.uk/) grants BB/D016541/1, BB/F020325/1, and BB/H008039/1 (to PJ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Nelson N, Ben-Shem A. The complex architecture of oxygenic photosynthesis. Nat Rev Mol Cell Biol. 2004;5:971–982. doi: 10.1038/nrm1525. [DOI] [PubMed] [Google Scholar]
- 2.López-Juez E, Pyke KA. Plastids unleashed: their development and their integration in plant development. Int J Dev Biol. 2005;49:557–577. doi: 10.1387/ijdb.051997el. [DOI] [PubMed] [Google Scholar]
- 3.Reyes-Prieto A, Weber AP, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu Rev Genet. 2007;41:147–168. doi: 10.1146/annurev.genet.41.110306.130134. [DOI] [PubMed] [Google Scholar]
- 4.Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 5.Keegstra K, Cline K. Protein import and routing systems of chloroplasts. Plant Cell. 1999;11:557–570. doi: 10.1105/tpc.11.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leister D. Chloroplast research in the genomic age. Trends Genet. 2003;19:47–56. doi: 10.1016/s0168-9525(02)00003-3. [DOI] [PubMed] [Google Scholar]
- 7.Blobel G, Sabatini DD. Ribosome-membrane interactions in eukaryotic cells. In: Manson LA, editor. Biomembranes. New York: Plenum; 1971. pp. 193–195. [Google Scholar]
- 8.Blair GE, Ellis RJ. Protein synthesis in chloroplasts. I. Light-driven synthesis of the large subunit of fraction I protein by isolated pea chloroplasts. Biochim Biophys Acta. 1973;319:223–234. doi: 10.1016/0005-2787(73)90013-0. [DOI] [PubMed] [Google Scholar]
- 9.Bruce BD. Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol. 2000;10:440–447. doi: 10.1016/s0962-8924(00)01833-x. [DOI] [PubMed] [Google Scholar]
- 10.Krimm I, Gans P, Hernandez JF, Arlaud GJ, Lancelin JM. A coil-helix instead of a helix-coil motif can be induced in a chloroplast transit peptide from Chlamydomonas reinhardtii. Eur J Biochem. 1999;265:171–180. doi: 10.1046/j.1432-1327.1999.00701.x. [DOI] [PubMed] [Google Scholar]
- 11.Bruce BD. The paradox of plastid transit peptides: conservation of function despite divergence in primary structure. Biochim Biophys Acta. 2001;1541:2–21. doi: 10.1016/s0167-4889(01)00149-5. [DOI] [PubMed] [Google Scholar]
- 12.von Heijne G, Nishikawa K. Chloroplast transit peptides. The perfect random coil? FEBS Lett. 1991;278:1–3. doi: 10.1016/0014-5793(91)80069-f. [DOI] [PubMed] [Google Scholar]
- 13.Soll J, Schleiff E. Protein import into chloroplasts. Nat Rev Mol Cell Biol. 2004;5:198–208. doi: 10.1038/nrm1333. [DOI] [PubMed] [Google Scholar]
- 14.Kessler F, Schnell DJ. The function and diversity of plastid protein import pathways: a multilane GTPase highway into plastids. Traffic. 2006;7:248–257. doi: 10.1111/j.1600-0854.2005.00382.x. [DOI] [PubMed] [Google Scholar]
- 15.Inaba T, Schnell DJ. Protein trafficking to plastids: one theme, many variations. Biochem J. 2008;413:15–28. doi: 10.1042/BJ20080490. [DOI] [PubMed] [Google Scholar]
- 16.Jarvis P. Targeting of nucleus-encoded proteins to chloroplasts in plants (Tansley Review). New Phytol. 2008;179:257–285. doi: 10.1111/j.1469-8137.2008.02452.x. [DOI] [PubMed] [Google Scholar]
- 17.Li HM, Chiu CC. Protein transport into chloroplasts. Annu Rev Plant Biol. 2010;61:157–180. doi: 10.1146/annurev-arplant-042809-112222. [DOI] [PubMed] [Google Scholar]
- 18.VanderVere PS, Bennett TM, Oblong JE, Lamppa GK. A chloroplast processing enzyme involved in precursor maturation shares a zinc-binding motif with a recently recognized family of metalloendopeptidases. Proc Natl Acad Sci USA. 1995;92:7177–7181. doi: 10.1073/pnas.92.16.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richter S, Lamppa GK. A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc Natl Acad Sci USA. 1998;95:7463–7468. doi: 10.1073/pnas.95.13.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter S, Zhong R, Lamppa G. Function of the stromal processing peptidase in the chloroplast import pathway. Physiol Plant. 2005;123:362–368. [Google Scholar]
- 21.Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richter S, Lamppa GK. Determinants for removal and degradation of transit peptides of chloroplast precursor proteins. J Biol Chem. 2002;277:43888–43894. doi: 10.1074/jbc.M206020200. [DOI] [PubMed] [Google Scholar]
- 23.Rudhe C, Clifton R, Chew O, Zemam K, Richter S, et al. Processing of the dual targeted precursor protein of glutathione reductase in mitochondria and chloroplasts. J Mol Biol. 2004;343:639–647. doi: 10.1016/j.jmb.2004.08.053. [DOI] [PubMed] [Google Scholar]
- 24.Richter S, Lamppa GK. Structural properties of the chloroplast stromal processing peptidase required for its function in transit peptide removal. J Biol Chem. 2003;278:39497–39502. doi: 10.1074/jbc.M305729200. [DOI] [PubMed] [Google Scholar]
- 25.Moberg P, Stahl A, Bhushan S, Wright SJ, Eriksson A, et al. Characterization of a novel zinc metalloprotease involved in degrading targeting peptides in mitochondria and chloroplasts. Plant J. 2003;36:616–628. doi: 10.1046/j.1365-313x.2003.01904.x. [DOI] [PubMed] [Google Scholar]
- 26.Wan J, Bringloe D, Lamppa GK. Disruption of chloroplast biogenesis and plant development upon down-regulation of a chloroplast processing enzyme involved in the import pathway. Plant J. 1998;15:459–468. [Google Scholar]
- 27.Zhong R, Wan J, Jin R, Lamppa G. A pea antisense gene for the chloroplast stromal processing peptidase yields seedling lethals in Arabidopsis: survivors show defective GFP import in vivo. Plant J. 2003;34:802–812. doi: 10.1046/j.1365-313x.2003.01772.x. [DOI] [PubMed] [Google Scholar]
- 28.Yue R, Wang X, Chen J, Ma X, Zhang H, et al. A rice stromal processing peptidase regulates chloroplast and root development. Plant Cell Physiol. 2010;51:475–485. doi: 10.1093/pcp/pcq012. [DOI] [PubMed] [Google Scholar]
- 29.Kovacheva S, Bédard J, Wardle A, Patel R, Jarvis P. Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J. 2007;50:364–379. doi: 10.1111/j.1365-313X.2007.03060.x. [DOI] [PubMed] [Google Scholar]
- 30.Kasmati AR, Töpel M, Patel R, Murtaza G, Jarvis P. Molecular and genetic analyses of Tic20 homologues in Arabidopsis thaliana chloroplasts. Plant J. 2011 doi: 10.1111/j.1365-313X.2011.04551.x. in press. [DOI] [PubMed] [Google Scholar]
- 31.Howden R, Park SK, Moore JM, Orme J, Grossniklaus U, et al. Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics. 1998;149:621–631. doi: 10.1093/genetics/149.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D. Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis thaliana. Plant Physiol. 2011 doi: 10.1104/pp.110.168120. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, et al. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics. 2001;159:1751–1763. doi: 10.1093/genetics/159.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu SC, Belmonte MF, Harada JJ, Inoue K. Indispensable roles of plastids in Arabidopsis thaliana embryogenesis. Curr Genomics. 2010;11:338–349. doi: 10.2174/138920210791616716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baldwin A, Wardle A, Patel R, Dudley P, Park SK, et al. A molecular-genetic study of the Arabidopsis Toc75 gene family. Plant Physiol. 2005;138:715–733. doi: 10.1104/pp.105.063289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hust B, Gutensohn M. Deletion of core components of the plastid protein import machinery causes differential arrest of embryo development in Arabidopsis thaliana. Plant Biol (Stuttg) 2006;8:18–30. doi: 10.1055/s-2005-873044. [DOI] [PubMed] [Google Scholar]
- 37.Kovacheva S, Bédard J, Patel R, Dudley P, Twell D, et al. In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 2005;41:412–428. doi: 10.1111/j.1365-313X.2004.02307.x. [DOI] [PubMed] [Google Scholar]
- 38.Inaba T, Alvarez-Huerta M, Li M, Bauer J, Ewers C, et al. Arabidopsis Tic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell. 2005;17:1482–1496. doi: 10.1105/tpc.105.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casson S, Spencer M, Walker K, Lindsey K. Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. Plant J. 2005;42:111–123. doi: 10.1111/j.1365-313X.2005.02355.x. [DOI] [PubMed] [Google Scholar]
- 40.Spencer MW, Casson SA, Lindsey K. Transcriptional profiling of the Arabidopsis embryo. Plant Physiol. 2007;143:924–940. doi: 10.1104/pp.106.087668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, et al. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel R, Hsu S, Bédard J, Inoue K, Jarvis P. The Omp85-related chloroplast outer envelope protein OEP80 is essential for viability in Arabidopsis. Plant Physiol. 2008;148:235–245. doi: 10.1104/pp.108.122754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aronsson H, Jarvis P. A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 2002;529:215–220. doi: 10.1016/s0014-5793(02)03342-2. [DOI] [PubMed] [Google Scholar]
- 44.Ling Q, Huang W, Jarvis P. Use of a SPAD-502 meter to measure leaf chlorophyll concentration in Arabidopsis thaliana. Photosynth Res. 2011;107:209–214. doi: 10.1007/s11120-010-9606-0. [DOI] [PubMed] [Google Scholar]
- 45.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 46.Sessions A, Burke E, Presting G, Aux G, McElver J, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goubet F, Misrahi A, Park SK, Zhang Z, Twell D, et al. AtCSLA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol. 2003;131:547–557. doi: 10.1104/pp.014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allen T, Ingles PJ, Praekelt U, Smith H, Whitelam GC. Phytochrome-mediated agravitropism in Arabidopsis hypocotyls requires GIL1 and confers a fitness advantage. Plant J. 2006;46:641–648. doi: 10.1111/j.1365-313X.2006.02727.x. [DOI] [PubMed] [Google Scholar]
- 51.Kasmati AR, Töpel M, Patel R, Murtaza G, Jarvis P. Molecular and genetic analyses of Tic20 homologues in Arabidopsis thaliana chloroplasts. Plant J. 2011;66:877–889. doi: 10.1111/j.1365-313X.2011.04551.x. [DOI] [PubMed] [Google Scholar]
- 52.Aronsson H, Combe J, Patel R, Agne B, Martin M, et al. Nucleotide binding and dimerization at the chloroplast pre-protein import receptor, atToc33, are not essential in vivo but do increase import efficiency. Plant J. 2010;63:297–311. doi: 10.1111/j.1365-313X.2010.04242.x. [DOI] [PubMed] [Google Scholar]
- 53.Morsomme P, Dambly S, Maudoux O, Boutry M. Single point mutations distributed in 10 soluble and membrane regions of the Nicotiana plumbaginifolia plasma membrane PMA2 H+-ATPase activate the enzyme and modify the structure of the C-terminal region. J Biol Chem. 1998;273:34837–34842. doi: 10.1074/jbc.273.52.34837. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression patterns of essential chloroplast protein import apparatus genes during embryogenesis. Publicly-available Affymetrix microarray data corresponding to defined tissues and developmental stages of Arabidopsis embryogenesis [39], [40] were accessed using an electronic fluorescent pictograph (eFP) browser online [41]. Data for atTOC75-III (At3g46740), atTIC110 (At1g06950) and SPP (At5g42390) are shown. Values are means (±SE) derived from three or six independent measurements. SAM, shoot apical meristem.
(TIF)
Analyses of mRNA and protein expression in the
toc75-III
,
tic110
and
spp
heterozygotes. (A) Quantitative RT-PCR analysis of mRNA levels. Total-RNA samples were extracted from ∼10–30 whole seedlings of the indicated genotypes (and wild type) that had been grown in vitro for 14 days under standard conditions. RNA was isolated using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany), and was then treated with DNAse I (DNA-free; Ambion, Texas, USA). Quantitative RT-PCR was performed using an MJ Research Chromo4 Gradient Cycler (Bio-Rad, Hercules, CA, USA) and SYBR Green Jump Start Taq Ready Mix (Sigma, St. Louis, MO, USA) for high-throughput quantitative PCR. Relative quantification was determined according to published methods [49], [50]. Reactions were characterized by comparing threshold cycle (C
T) values; C
T is a unit-less value defined as the fractional cycle number at which the sample fluorescence signal passes a fixed threshold. The relative amount of transcript was calculated by subtracting the control gene (ACTIN 2; At3g18780) C
T-value from the gene-of-interest (atTOC75-III, atTIC110, SPP) C
T-value (ΔC
T). Then, ΔC
T values for the wild type were subtracted from those for the mutants to yield ΔΔC
T values, and these were used to estimate expression levels as 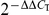 . Data shown are means (±SE) derived from four independent amplifications performed on three biological replicates. The ACTIN2 primers have been described previously [51]; the other primers used were: atTOC75-III sense 5′-TCG CAT CTC CAC TCA ATC-3′; atTOC75-III antisense, 5′-GTC TCT GTA TCT CGG TTA GG-3′; atTIC110 sense, 5′-CTC CTC AGG TGC CTT ATC AGA AG-3′; atTIC110 antisense, 5′-CGA GCA AGA GCA GCC GAG AAC-3′; SPP sense, 5′-AAG CTA GCC ATG ATT CTG CAA-3′; SPP antisense, 5′-CAT CAT GAG CAA CAG GAA GTT-3′. (B) Immunoblot analysis of protein levels. Total-protein was extracted from plant material samples equivalent to those employed in panel A, using previously-described procedures [51]. Three different protein amounts (80, 60 and 40 µg) of each genotype were analysed by immunoblotting, as described previously [51], using primary antibodies against atToc75-III, atTic110 and the plasma membrane H+-ATPase, PMA2 [51], [52], [53]; unfortunately, an SPP antibody was not available for this work. The secondary antibody was anti-rabbit IgG conjugated with horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and the detection reagent was ECL Plus (GE Healthcare, Chalfont St. Giles, UK). Chemiluminescence detection employed a Fujifilm LAS-4000 imager. Quantification of all images was performed using Aida software (Raytest, Straubenhardt, Germany). The atToc75-III and atTic110 data were normalized using equivalent PMA2 data. Values shown are means (±SE) derived from three independent measurements.
. Data shown are means (±SE) derived from four independent amplifications performed on three biological replicates. The ACTIN2 primers have been described previously [51]; the other primers used were: atTOC75-III sense 5′-TCG CAT CTC CAC TCA ATC-3′; atTOC75-III antisense, 5′-GTC TCT GTA TCT CGG TTA GG-3′; atTIC110 sense, 5′-CTC CTC AGG TGC CTT ATC AGA AG-3′; atTIC110 antisense, 5′-CGA GCA AGA GCA GCC GAG AAC-3′; SPP sense, 5′-AAG CTA GCC ATG ATT CTG CAA-3′; SPP antisense, 5′-CAT CAT GAG CAA CAG GAA GTT-3′. (B) Immunoblot analysis of protein levels. Total-protein was extracted from plant material samples equivalent to those employed in panel A, using previously-described procedures [51]. Three different protein amounts (80, 60 and 40 µg) of each genotype were analysed by immunoblotting, as described previously [51], using primary antibodies against atToc75-III, atTic110 and the plasma membrane H+-ATPase, PMA2 [51], [52], [53]; unfortunately, an SPP antibody was not available for this work. The secondary antibody was anti-rabbit IgG conjugated with horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and the detection reagent was ECL Plus (GE Healthcare, Chalfont St. Giles, UK). Chemiluminescence detection employed a Fujifilm LAS-4000 imager. Quantification of all images was performed using Aida software (Raytest, Straubenhardt, Germany). The atToc75-III and atTic110 data were normalized using equivalent PMA2 data. Values shown are means (±SE) derived from three independent measurements.
(TIF)