Abstract
Ornithine decarboxylase antizyme 3 (Oaz3) is expressed in spermatids, makes up the antizyme family of Oaz genes with Oaz1 and Oaz2, and was proposed to encode a 22 kDa antizyme protein involved in polyamine regulation similar to the 22 kDa OAZ1 and OAZ2 proteins. Here we demonstrate however that the major product encoded by Oaz3 is a 12 kDa protein, p12, which lacks the antizyme domain that interacts with ornithine decarboxylase. We show that p12 does not affect ornithine decarboxylase levels, providing an explanation for the surprising observation made in Oaz3 knock-out male mice, which do not display altered testis polyamine metabolism. This suggested a novel activity for Oaz3 p12. Using immuno-electron microscopy we localized p12 to two structures in the mammalian sperm tail, viz. the outer dense fibers and fibrous sheath, as well as to the connecting piece linking head and tail. We identified myosin phosphatase targeting subunit 3 (MYPT3), a regulator of protein phosphatase PP1β, as a major p12-interacting protein, and show that MYPT3 is present in sperm tails and that its ankyrin repeat binds p12. We show that MYPT3 can also bind protein phosphatase PP1γ2, the only protein phosphatase present in sperm tails, and that p12- MYPT3 interaction modulates the activity of both PP1β and PP1γ2. This is, to our knowledge, the first demonstration of a novel activity for an Oaz-encoded protein.
Keywords: Polyamines, Reproduction, Sperm, Spermatogenesis, Testis, Antizyme, Ornithine Decarboxylase
Introduction
Mammalian spermiogenesis is the final stage of male germ cell differentiation, during which the round haploid spermatids undergo complex morphological and biochemical changes and transform into spermatozoa. The process of spermiogenesis is extensively studied but only partially understood. One of the structures formed during spermiogenesis is the sperm flagellum, which contains beside the axoneme two unique structures, the outer dense fibers (ODF)2 throughout the tail and the fibrous sheath (FS) present in the principal piece, and is important in sperm motility. Our laboratory has characterized several proteins that are present in the sperm flagellum, which all play distinct roles in sperm structure and motility (1–3). Many sperm proteins are exclusively expressed after meiosis (such as ODF1), suggesting a specific function during spermiogenesis. Among them is the ornithine decarboxylase antizyme 3 gene (Oaz3) (4, 5).
Oaz3 belongs to the antizyme gene family and its mRNA is exclusively expressed in post-meiotic male germ cells (5). The other two family members Oaz1 and Oaz2 exhibit neuronal and ubiquitous expression, respectively (6), and regulate ornithine decarboxylase (ODC), which is involved in polyamine synthesis. Synthesis of OAZ1 and OAZ2 proteins requires a polyamine-induced translation frameshift which links open reading frame 1 (ORF1) with open reading frame 2 (ORF2). The translation of Oaz3 mRNA was proposed to start from the first AUG and also utilize the frameshift, which predicted that a 22 kDa protein could be made in spermatids (5). However, the frameshift efficiency of Oaz3 mRNA is very low compared with Oaz1 (7), suggesting that Oaz3 may encode a variant smaller than 22 kDa, resulting from a translation stop at the frameshift sequence, and may play a role in spermiogenesis different from a role as antizyme. Recently, Oaz3 knock-out mice were produced: homozygous male mice are infertile due to the separation of sperm heads and tails, whereas (as expected) female mice have no phenotype (8). The disengagement appears to take place during epididymal transition and occurs between the basal plate and the capitulum of the sperm. No gross structural abnormality was observed with the separated heads and tails. The mechanism of head-tail separation and the role for Oaz3 in maintaining the integrity of the sperm connecting piece are unknown. Surprisingly, the knock-out of Oaz3 did not result in alteration of polyamine levels in testis or sperm, supporting the possibility that Oaz3 has a function other than as an antizyme.
We previously reported that an abundant 12 kDa protein in the rat sperm flagellum is encoded by Oaz3 mRNA (9). We also showed that translation of Oaz3 p12 starts from a CUG codon upstream of the first AUG codon. In this study, we report that the p12 product of Oaz3 is encoded by ORF1. We did not detect either a 22 kDa OAZ3 protein or a putative 28 kDa precursor. p12 localizes to ODF, FS and capitulum of the rat sperm flagellum and does not regulate ODC. To uncover a role for p12 we identified interacting testis proteins using a yeast 2-hybrid assay. We discovered that p12 interacts with myosin phosphatase-targeting subunit 3 (MYPT3), which we show is also present in spermatids and sperm flagellum. p12 modulates protein phosphatase 1 activity through its binding to MYPT3.
EXPERIMENTAL PROCEDURES
Generation of p12 Antibodies
Rabbit polyclonal antibodies were raised against peptide RSITYKEQEDLTLRPH (anti-1017 antibody) and against purified MBP (maltose-binding protein)-ORF1 and MBP-ORF2 proteins.
Cell Transfections and Confocal Microscopy
Mouse NIH3T3 or human HEK293 cells were transfected with DNA constructs using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. 16 h after transfection, the cells on the coverslips were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, blocked with PBS and 2% BSA, and incubated with primary antibodies for 1 h (anti-HA dilution 1:500; anti-Myc dilution 1:500) at 37 °C. The cells were then gently washed and incubated with secondary antibody for 1 h (Cy3-anti-mouse IgG dilution 1:1000; Alex488-anti-rabbit IgG dilution 1:1000) at 37 °C. The cells were gently washed and mounted onto glass slides and observed and imaged using a Zeiss LSM510 confocal microscope.
Protein Analysis
Immunohistochemistry and immunoelectron microscopy were carried out as described previously (9, 10). Western blot analysis was carried out as follows: protein samples were boiled in loading buffer, separated on 10 or 15% polyacrylamide (SDS-PAGE) gels, transferred onto a nitrocellulose membrane (Amersham Biosciences), blocked overnight at 4 °C in blocking buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 0.1% Nonidet P-40, 0.1% Tween 20, 0.1% SDS, 5% dry nonfat dry milk), and analyzed using primary antibodies (anti-1017 dilution, 1:150; anti-ORF1 dilution 1:200; anti-ORF2 dilution 1:200) followed by goat anti-rabbit IgG antibody (Sigma) diluted 1:12,000. Prestained Protein Ladder SM0671 (Fermentas) was used as a size marker. The blot was developed with LumiGLO substrate (Kirkegaard & Perry Laboratories, Inc.) and exposed to Kodak films (Amersham Biosciences).
To perform immunoprecipitation, HEK293 cells were transfected with indicated plasmids. In indicated cases, co-transfected cultures were treated with 20 μg/ml cycloheximide for 3 h to block protein synthesis. Next, cells were lysed in IP buffer (50 mm Tris-HCl, pH 7.4; 300 mm NaCl; 1% Triton X-100; 0.5% IGEPAL, and protease inhibitor mixture (Roche)) for 30 min at 4 °C. The lysates were centrifuged at 14,000 rpm for 10 min to remove cell debris and insoluble particles. The supernatants were cleared with protein A (or protein G) beads (GE Healthcare) for 30 min at 4 °C with shaking. The cleared cell lysates were then incubated with anti-Myc, anti-HA, or anti-MBP-ORF1 antibodies for 2∼5 h at 4 °C with shaking. Immune complexes were incubated with protein A (or protein G) beads for 1 h at 4 °C with shaking, washed three times with IP buffer, and analyzed by SDS-PAGE and immunoblotting.
Yeast Two-hybrid Assays
A full-length rat testis two-hybrid cDNA library (cloned in the pEXP-AD502 vector) was custom made by Invitrogen. The bait strain is generated by transformation of MaV203 cells with pDBLeu vector with insertion of the first open reading frame (ORF1) of rat Oaz3 in frame with the GAL4 DNA-binding domain. The library transformation was performed using the LiAc/SS carrier DNA/PEG method adapted from Gietz and Schiestl (11). The transformed cells were plated on SC-Leu-Trp-His plates with 20 mm 3-amino-1,2,4-triazoleon (3-AT) for HIS3 growth assay. The plates were incubated at 30 °C for up to 8 days to allow growth of transformed cells with weak interactions. All colonies were also analyzed for LacZ reporter. Positive clones were further validated by purging the bait vector, retransforming with either empty vector or pDBLeu-ORF1, and testing the HIS3 and LacZ reporters.
PP1 Transfections and Activity Assays
NIH3T3 cells were maintained in DMEM supplemented with 10% fetal bovine serum and 10 mm sodium pyruvate (Invitrogen) and were plated on glass coverslips at 30–50% confluence for transfection. Cells were transfected with various constructs using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After fixation with 4% paraformaldehyde, cells were co-stained with anti-Myc or HA antibodies and Rhodamine-conjugated phalloidin (Cytoskeleton Inc.) for polymerized actin and Alex488-conjugated goat anti-rabbit (or mouse) IgG to visualize transfected cells. Stained cells were analyzed with an LSM510 laser scanning microscope and AxioCam MRm camera and LSM510 software (Zeiss) for stress fibers disassembly, indicative of enhanced PP1 activity. Three to four independent experiments with total counts of a minimum of 100 transfected cells were collected to calculate average percentage of cells possessing stress fibers and the standard deviation. Statistical analysis was achieved using the Student's t test.
RESULTS
The 12 kDa Protein in the Rat Sperm Flagellum Is Encoded by Oaz3
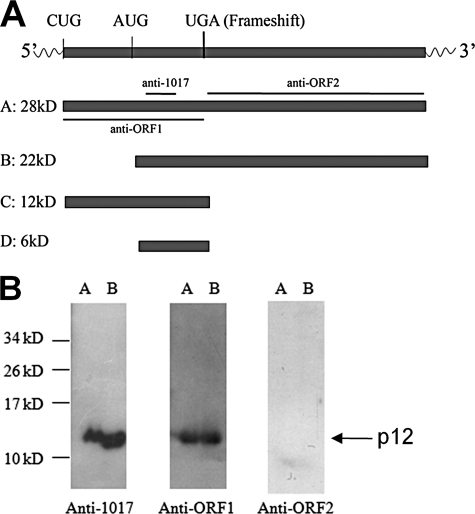
We showed that Oaz3 mRNA translation starts from CUG (9). One report suggested a start at AUG (5). Depending on the translation start site used and whether or not frameshift occurs, Oaz3 has the potential to encode four products (Fig. 1A). To distinguish these possible forms and to identify, which one(s) is (are) expressed in rat testis, we raised antibodies to different regions of the putative OAZ3 polypeptides (Fig. 1A). The specificities of the antibodies were confirmed with recombinant forms of ORF1 and ORF2 (not shown).
FIGURE 1.
Identification of p12 in testis and sperm. A, schematic representation of putative proteins that can be encoded by Oaz3 mRNA. CUG is the translation start site determined for p12. UGA is the translation stop codon present in the frameshift sequence. The locations of peptides used for generation of antisera are indicated. B, Western blot analysis of proteins from testis (lanes A) and sperm (lanes B) using the indicated antisera. Anti-1017 and anti-ORF1 sera detected only p12, not other OAZ3 proteins. No proteins were detected with anti-ORF2.
We used these antibodies to analyze total protein extracts from rat testis and sperm (Fig. 1B). Anti-1017 and anti-ORF1 antibodies recognized the 12 kDa protein in testis and sperm (Fig. 1B; lanes A and B). No proteins were detected at 6, 22, or 28 kDa with these antibodies in testis or sperm protein extracts. This demonstrates that these putative proteins are not present, or in quantities below the level of detection. p12 was not recognized by anti-ORF2 as expected (Fig. 1B). Anti-ORF2 also did not detect putative proteins of 22 and 28 kDa.
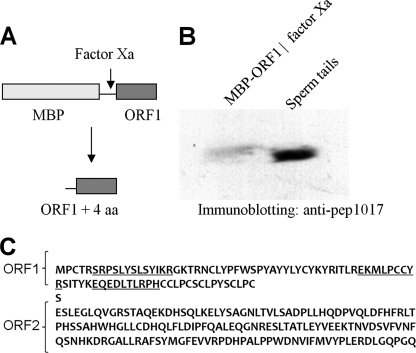
The size of p12 is similar to the calculated molecular mass of ORF1-encoded protein suggesting that the translation of p12 ends at the stop codon in the frameshift site. This was investigated in two ways: first, to analyze if ORF2 sequences contribute to p12, we compared the molecular mass of endogenous p12 to a recombinant ORF1 protein made in bacteria. A fusion protein of MBP-ORF1 was purified and digested with Factor Xa, which separated MBP from ORF1 and released a protein that was composed of ORF1 plus four amino acids at the N terminus (Fig. 2A). This protein was analyzed side-by-side with endogenous rat sperm p12 protein by SDS-PAGE. We observed that endogenous p12 protein in sperm tails migrates slightly faster than recombinant ORF1 (Fig. 2B). The small size difference (estimated to be less than 1 kDa) can be attributed to the four extra amino acids. Second, we analyzed p12 by mass spectrometry (Fig. 2C): we only observed peptides that are encoded by ORF1 (indicated in underline), not ORF2. Together, our data show that p12 does not contain amino acids encoded by ORF2 sequences and is the product of ORF1 of the Oaz3 gene (Fig. 1A).
FIGURE 2.
p12 is encoded by Oaz3 ORF1. A, a bacterial fusion protein was produced containing MBP linked to the ORF1 sequences of Oaz3. After purification, factor X was used to cleave the fusion protein generating MBP and the indicated peptide of ORF1 plus 4 amino acids. B, Western blotting analysis of recombinant, factor X-cleaved ORF1 and p12 in sperm tails using anti-peptide 1017 antibody. Sperm tail p12 migrates just ahead of the recombinant protein, which is larger by 4 amino acids. C, peptide sequence encoded by ORF1 and the putative sequence encoded by ORF2 are shown. Peptides indicated by underline were detected by mass spectrometry of trypsin-cleaved, purified sperm tail p12 protein. No peptides were detected stemming from ORF2.
Oaz3-encoded p12 Does Not Possess Antizyme Activity
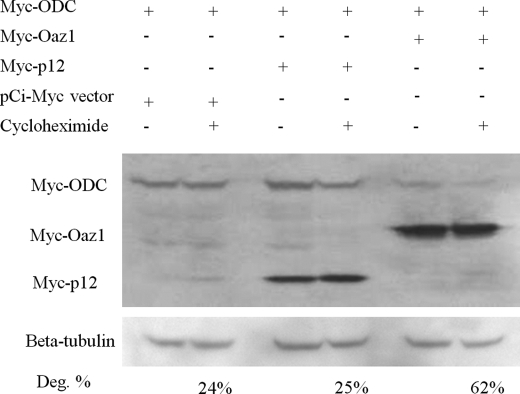
The ODC binding domain in OAZ1 and OAZ2 antizymes is localized to ORF2. ORF1 is dispensable for antizyme function (12). Therefore, p12 is not expected to bind to ODC or affect its expression levels. To investigate this, we examined whether p12 stimulates the degradation of ODC, using OAZ1 as positive control in this assay. Myc-tagged ODC, p12, and OAZ1 were co-transfected in various combinations and alone in HEK293 cells, which were next analyzed by Western blotting for protein levels. In indicated instances, co-transfected cultures were treated for 3 h with cycloheximide to block protein synthesis as done by Snapir et al. (13). The results (Fig. 3) show that, as expected, OAZ1 caused significant degradation of ODC (62%; lanes 5 and 6). In contrast, ODC levels in the presence of p12 (lanes 3 and 4) did not differ from the negative control (empty pCi-Myc vector; 24%; lanes 1 and 2). This demonstrates that p12 does not affect ODC, and indicates that p12 may have a function different from that of an antizyme.
FIGURE 3.
p12 does not regulate ODC. To analyze a possible effect of p12 on ODC levels, cells were co-transfected with the indicated DNA constructs. Myc-tagged OAZ1 was included as a positive control for ODC degradation. In indicated experiments, cultures were treated with cycloheximide for 3 h after co-transfection. Cell lysates were prepared and analyzed by Western blotting using anti-Myc antibodies. Protein loading in all lanes was analyzed using anti-β-tubulin antiserum. OAZ1 causes ODC degradation (lanes 5 and 6), while p12 OAZ3 protein has no effect (lanes 3 and 4) compared with the negative control (pCi-Myc DNA; lanes 1 and 2). Percentage degradation was determined by quantitation of ODC amounts: ODC levels were normalized for β-tubulin, setting the ODC level in cells transfected with empty vector and not treated with cycloheximide at 100%.
p12 Localizes to ODF, FS, and Capitulum of the Sperm Flagellum
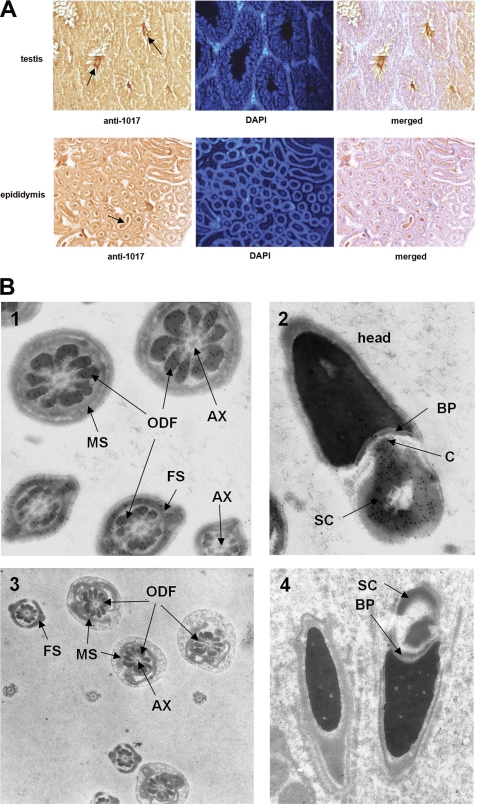
We next investigated the expression of p12 during spermatogenesis by immunohistochemistry and immuno-electron microscopy of mouse testis and epididymis, because while we had previously detected p12 in sperm tails, the recent Oaz3 knock-out results suggested, but did not show, that p12 may also be in the sperm connecting piece (8). As shown in Fig. 4A, p12 is abundantly expressed in flagella of spermatozoa and elongating spermatids in the lumen of the seminiferous tubules (arrows) and epididymis. Interstitial cells, Sertoli cells, spermatogonia, and spermatocytes only have background staining indicating that p12 protein is, as expected from the mRNA expression pattern, restricted to post-meiotic germ cells.
FIGURE 4.
p12 is present in ODF, FS, and the connecting piece of sperm. A, immunocytochemical analysis of p12 protein expression in testis sections and sections of epididymis, as indicated. To detect p12 we used anti-1017 serum. Cellular DNA was stained with DAPI. p12 is predominantly present in sperm tails (arrows). B, immuno-electron microscopic analysis of p12 in sperm tail and sperm connecting piece. Anti-peptide 1017 antiserum was used in immuno EM, followed by detection with gold-labeled secondary serum. Panel 1, in sperm tails, p12 is present in ODF and FS, but not in axoneme (AX) or mitochondrial sheath (MS). Panel 2, p12 is also detected in the capitulum (C) and striated collar (SC) of the connecting piece, but not in the basal plate (BP). Panels 3 and 4, immuno EM was done using pre-immune antiserum as negative control and to demonstrate specificity of the anti-1017 antisera used in panels 1 and 2. No stain was observed above background level.
To obtain ultrastructural details on the localization of p12 in sperm, we performed immuno-electron microscopy on rat spermatozoa. The p12 protein, detected with anti-peptide-1017 antibody and a gold-labeled secondary antibody is abundant in ODF as well as FS in elongated spermatids and epididymal sperm (Fig. 4B, panel 1). The signal is absent from the mitochondrial sheath, axoneme and sperm head (panel 1). We discovered that, in addition to its presence in the tail, p12 is also present at very high levels in the capitulum and striated collar of the connecting piece (Fig. 4B, panel 2). Control experiments using pre-immune antibodies in immuno-electron microscopy showed no gold label in the connecting piece or ODF and FS, indicating the specificity of the anti-peptide-1017 antibody (Fig. 4B, panels 3 and 4).
p12 Interacts with MYPT3
Our results suggested that p12 may play a role in spermiogenesis and/or the sperm tail different from that of a prototypic antizyme. To obtain initial clues as to this novel function, we investigated which testicular proteins can bind to p12 using a yeast 2-hybrid system. ORF1 was used as bait to identify interacting proteins. Table 1 lists the proteins we discovered in this assay that can bind p12, after eliminating false positives. One of the proteins represented by 7 independent clones was myosin phosphatase targeting subunit 3 (MYPT3). MYPT3 is expressed in many tissues including testis. Recombinant GST-MYPT3 was shown to inhibit the activity of protein phosphatase 1 (PP1) in vitro (14).
TABLE 1.
Results of yeast 2-hybrid screening for protein interactions with p12 protein as bait
p12 protein was used as bait in a yeast 2-hybrid screen of rat testis cDNA libraries.
| Analysis of interaction between the rat 12 kDa protein and: | Interaction |
|---|---|
| rat ODF1a | - |
| rat ODF2a | - |
| rat KLC3 | - |
| rat GGN1 | - |
| Interacting proteins highly expressed in testisb | # of coloniesc |
| Tubulin, beta 2c (Tubb2c) | 2 |
| rat family with sequence similarity 71, member D (Fam71d) | 1 |
| Kelch-like 10 (Drosophila) (Klhl10) | 1 |
| Izumo sperm-egg fusion 1 (Izumo1) | 1 |
| Similar to THUMP domain containing 3 | 1 |
| Rat arylsulfatase A | 1 |
| Rat 3-oxoacid CoA transferase 2A | 1 |
| Rat similar to gene model 711 (RGD1307355) | 1 |
| Interacting proteins expressed in testisb | # of coloniesc |
| Protein phosphatase 1, regulatory subunit 16A (MYPT3) | 7 |
| Similar to GE36 (RGD1305609) | 7 |
| Similar to SEC24, member C (LOC685144) | 1 |
| GDP dissociation inhibitor 2 (Gdi2) | 1 |
| Rat arsA arsenite transporter, ATP-binding, homolog 1 | 1 |
| Interacting proteins expression not determined in testisb | # of coloniesc |
| Rat similar to hypothetical protein FLJ23305 | 1 |
| Rat RNA-binding motif protein 8 (predicted) | 1 |
| Rat splicing factor 3b, subunit 2 (Sf3b2) | 1 |
a Interaction between ODF2 and ODF1 was used as positive control for the yeast 2-hybrid system.
b Proteins that were identified as potential 12 kDa protein-interacting partners were grouped according to expression level in testis as either “highly expressed,” “expressed,” or “expression not determined.”
c “# of colonies” indicates the number of times an independent clone was isolated for the same protein in the yeast 2-hybrid screening.
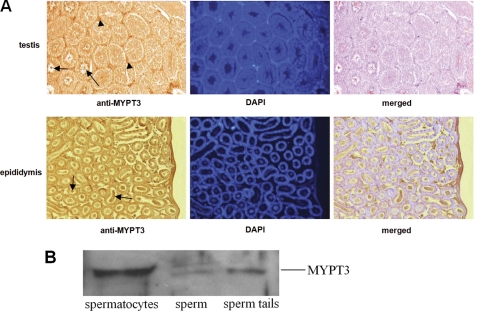
Because endogenous p12 in spermatids and sperm is Triton X-insoluble, it was not possible to isolate a complex of p12-MYPT3 from sperm. We therefore investigated the localization of MYPT3 in male germ cells and we analyzed the details of p12-MYPT3 complex formation in co-transfected somatic cells. Using a polyclonal MYPT3 antibody in immunohistochemistry we found that in testis, MYPT3 is present at high levels in interstitial cells (Fig. 5A, arrowheads), as well as in spermatids and sperm tail (Fig. 5A, arrows). In epididymis, MYPT3 signal is in sperm tails (Fig. 5A, arrows). The presence of MYPT3 in male germ cells was confirmed by immunoblotting (Fig. 5B). Thus, the expression patterns of p12 and MYPT3 overlap.
FIGURE 5.
MYPT3 expression in testis. A, immunocytochemical analysis of MYPT3 protein expression in testis sections and sections of epididymis, as indicated. Anti-MYPT3 antiserum detected the protein in interstitial cells (arrowheads) and sperm tails (arrows). Cellular DNA was stained with DAPI. B, Western blot analysis of spermatocytes, sperm and sperm tails using anti-MYPT3 antiserum indicated the presence of MYPT3.
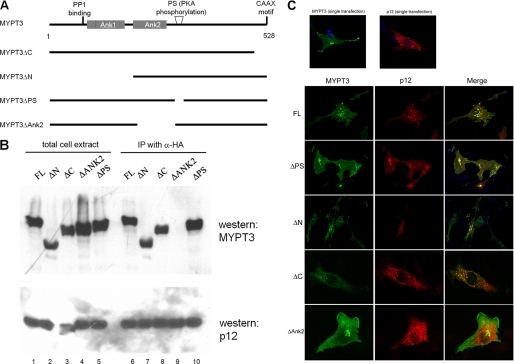
To confirm the specific interaction of p12 and MYPT3 and to uncover sequences crucial for the interaction of p12 and MYPT3, we used immunofluorescence and co-immunoprecipitation assays in co-transfected mammalian somatic cells. Somatic cells do not express the other abundant proteins present in the sperm tail outer dense fibers or fibrous sheath with which p12 and MYPT3 may interact and which may contribute to the insoluble characteristic. Thus, although they do not represent spermatids in several respects, they may provide for soluble complex formation to allow the protein interaction study of p12 and PYPT3, which are abundant in spermatids. MYPT3 has a PP1-binding motif, two ankyrin repeats (Ank1, Ank2), a PKA phosphorylation (PS) target site and a CAAX motif (Fig. 6A). The interaction of wt MYPT3 and several deletion mutants of Myc-tagged MYPT3 (Fig. 6A) with HA-tagged p12 was tested. Fig. 6B shows the co-immunoprecipitation-Western blot assays of p12-MYPT3 complexes from cells co-transfected with HA-tagged p12, wt Myc-tagged MYPT3 and the indicated mutant MYPT3 proteins. Lanes 1–5 show that wt MYPT3 and all deletion mutants are expressed, as was p12. The co-IP-western result (lanes 6–10) shows that the ΔAnk2 MYPT3 mutant completely lost the ability to bind p12 (Fig. 6B, lane 9), whereas all other mutants were able to bind p12.
FIGURE 6.
p12 interacts specifically with the ankyrin 2 repeat of MYPT3. A, schematic representation of MYPT3 motifs and MYPT3 deletion mutants made for the interaction study with p12. The binding site on MYPT3 for PP1 is indicated. The two ankyrin repeats are indicated, as well as the C-terminal CAAX motif and the PKA phosphorylation site (PS). B, protein interactions observed in yeast were independently confirmed in co-immunoprecipitation/Western blot analyses. Myc-tagged full-length MYPT3 (FL) and Myc-tagged mutants (as indicated) were co-transfected with HA-tagged p12, and cell lysates were analyzed for protein expression directly (lanes 1–5; total cell extract) or after immunoprecipitation with anti-HA antiserum and Western blot analysis of p12-bound MYPT3 using anti-Myc antiserum (lanes 6–10; IP with α-HA). Only the ΔAnk2 repeat MYPT3 deletion mutant failed to bind p12. C, protein localization of cells transfected with one or two constructs as indicated. Transfection of p12 alone results in appearance of cytoplasmic aggregates (top panel). MYPT3 alone displays a membrane-bound localization, mediated by the CAAX motif (top panel). Co-transfection of p12 with full-length (FL) MYPT3, the ΔPS mutant, the ΔN mutant and the ΔC mutant all result in colocalization of the two proteins in cytoplasmic aggregates. However, p12 does not colocalize with the ΔAnk2 repeat MYPT3 deletion mutant (which retains a membrane-bound localization), confirming the co-IP/Western blot results.
The interaction was next analyzed by immunofluorescence microscopy in co-transfected cells. The results are shown in Fig. 6C. The top two panels show the location of singly transfected p12 and MYPT3. The next panels show the localization of co-expressed p12 and wt MYPT3 (FL) or indicated MYPT3 deletion mutants. Wt MYPT3 and p12 colocalize. The immunofluorescence study demonstrated that binding to p12 changes the localization of MYPT3: MYPT3 accumulates with p12 in a punctuate pattern in the cytoplasm, away from the membrane, confirming the interaction observed in the co-IP Western blot assays. The ΔPS, ΔN, and ΔC mutants of MYPT3 also colocalized with p12 (Fig. 6C) indicating that, as expected from the co-IP study, the MYPT3 N terminus, C terminus or the PS region are not involved in p12 binding. However, deletion of the second group of ankyrin repeats (ΔAnk2) of MYPT3 disrupted its interaction with p12 (Fig. 6C). These data confirm the initial observations in yeast, and establish that p12 can bind MYPT3.
MYPT3 Can Bind to PP1γ2
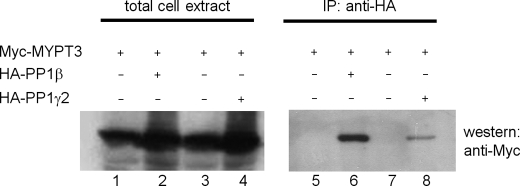
The binding of p12 to ankyrin repeat 2 (Ank2) of MYPT3 suggested a novel function for p12. MYPT3 had previously been shown to bind to and regulate activity of PP1β (15). It is thus possible that p12 may modulate the activity of PP1β through its interaction with MYPT3. However, the only PP1 isoform detected in sperm tails is PP1γ2. We therefore first needed to establish if MYPT3 can associate with PP1γ2. Myc-tagged MYPT3 and HA-tagged PP1β (as positive control) or HA-tagged PP1γ2 were co-expressed in HEK293 cells and their association was analyzed by co-immunoprecipitation-Western blotting. Our results (Fig. 7) show that Myc- MYPT3 was expressed in all experiments (lanes 1–4) and that MYPT3 can associate with both PP1β (lane 6) and PP1γ2 (lane 8). This result, together with the presence of p12, MYPT3, and PP1γ2 in sperm tails, suggested the possibility that p12 may modulate the activity of MYPT3 and PP1γ2.
FIGURE 7.
MYPT3 can bind to PP1γ2. It had been described that MYPT3 can bind PP1β. However, sperm tails only contain PP1γ2. To investigate if MYPT3 can bind PP1γ2 we co-transfected cells with the combinations indicated. Cell lysates were analyzed for MYPT3 expression directly (total cell extract) (lanes 1–4) or after immunoprecipitation with anti-HA antiserum (IP: anti-HA) followed by detection of PP1-bound MYPT3 by Western blotting. As expected, MYPT3 can bind PP1β (lane 6). Importantly, MYPT3 can also bind to PP1γ2 (lane 8).
p12 Modulates Activity of Both PP1β and PP1γ2
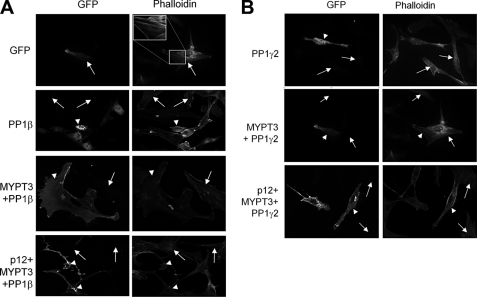
We next analyzed a possible effect of p12 on PP1 activity. As an indicator of PP1 activity, we measured the disassembly of actin stress fibers (visualized by phalloidin) resulting from dephosphorylation of myosin light chain, a convenient and commonly used test of myosin PP1 activity in transfected cells (15, 16). This assay allowed us to analyze effects of MYPT3 and of p12 or combinations of p12 and MYPT3 on PP1 activity (analyzing both PP1β and PP1γ2). In co-IP/Western and immunofluorescence experiments we had determined that p12 does not bind PP1 directly (not shown). Fig. 8A shows examples of results using p12, MYPT3, and PP1β and Fig. 8B shows examples of results for p12, MYPT3, and PP1γ2. Quantitation of all experiments is shown schematically in Fig. 9. First, as a control, we expressed PP1β alone in mouse fibroblasts: this resulted in a decrease in the % of transfected cells that exhibit stress fibers (Fig. 8A; PP1β; arrowhead shows examples of PP1-transfected cells that lost stress fibers), whereas the % of cells with stress fibers was not affected in untransfected cells or in cells expressing transfected GFP (Fig. 8A; GFP, PP1β; arrow shows examples of untransfected cells and of GFP-transfected cells that retain stress fibers). Co-expression of MYPT3 and PP1β resulted in a restoration of stress fibers in transfected cells to levels observed in untreated cells (Fig. 8A; MYPT3 + PP1β; arrowhead points to transfected cells that show stress fibers). This is consistent with the reported role of MYPT3 as an inhibitory subunit of PP1 (15). Importantly, introduction of p12 together with MYPT3 and PP1β resulted in a decrease of % of transfected cells that exhibit stress fibers to a level similar to that achieved by PP1 alone (Fig. 8A; p12+MYPT3+PP1β; arrowheads point to transfected cells that lost stress fibers) while untransfected cells retain stress fibers (arrows). The same results were observed for PP1γ2 (Fig. 8B) showing that PP1γ2 affects stress fiber formation and that this activity can be regulated by MYPT3 and p12. These results demonstrate that p12 counteracts the inhibitory effect of MYPT3 on PP1 activity.
FIGURE 8.
p12 modulates PP1 activity in the presence of MYPT3. A, possible effect of p12 and MYPT3 on PP1 activity was measured using the inhibition of stress fiber formation by active PP1. Stress fibers were visualized in transfected cells using phalloidin. Images are examples of areas in the cultures showing both transfected (GFP-positive) cells and untransfected cells. GFP has no effect on stress fiber formation (arrows): the top right hand panel shows an inset with a magnification of stress fibers in the GFP-positive cells as an example. PP1β inhibits stress fiber formation in cells as expected (arrowheads), whereas co-transfection of MYPT3 and PP1β restores stress fibers in cells (arrowheads). Co-transfection of p12, MYPT3 and PP1β causes loss of stress fibers (arrowheads) indicative of PP1β activation. Arrows indicate untransfected cells showing stress fibers. B, same analysis as in A was carried out for PP1γ2. The results are identical: PP1γ2 causes loss of stress fibers, and co-transfection of MYPT3 and PP1γ2 allows stress fiber formation in transfected cells (arrowheads) indicative of inactivation of PP1γ2 activity. Co-transfection of p12, MYPT3 and PP1γ2 results in loss of stress fibers (arrowheads) indicative of activation of PP1γ2 activation. Arrows indicate untransfected cells showing stress fibers.
FIGURE 9.
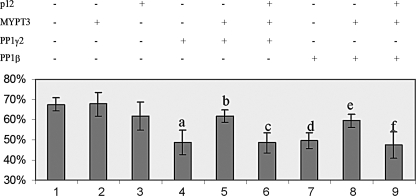
Quantitation of stress fiber formation in transfected cells. All data accumulated for the transfection experiments of which examples are shown in Fig. 8 were analyzed using the t test. Indicated are the combinations of the transfected DNA vectors, the average % of transfected cells with stress fibers, and the result of statistical analysis. The graph shows the mean and the standard deviation from four independent experiments. Statistical significance (t test): (a) p = 0.017; (b) p = 0.044; (c) p = 0.013; (d) p = 0.012; (e) p = 0.041; (f) p = 0.08.
The quantitation of experiments (Fig. 9) shows that the % of transfected cells that exhibit stress fibers decreases significantly upon transfection of PP1 (compare lanes 1, 4, and 7). Addition of MYPT3 to PP1 reverses this effect for both phosphatases (compare lanes 4 and 5 for PP1γ2, and lanes 7 and 8 for PP1β). Transfection of all three (p12, MYPT3 and PP1) results in a decrease in % transfected cells with stress fibers similar to that observed for transfection of only the PP1 (compare lanes 4 and 6 for PP1γ2, and lanes 7 and 9 for PP1β). These results represent the first demonstration of a novel activity for an Oaz gene, viz. the regulation of PP1 activity: p12 interferes with the inhibition of PP1 activity by MYPT3.
Mode of Action of p12
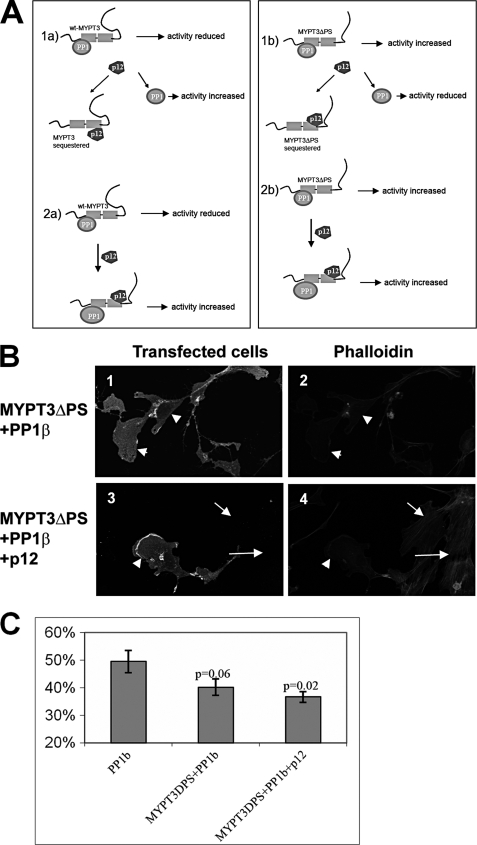
We developed two working models that each can explain the observed effect of p12 on MYPT3-PP1 activity (Fig. 10). These models take into consideration previous reports, which showed that MYPT3 and PP1 are in a complex in cells (15) (Fig. 10A), as well as the importance of the MYPT3 PKA phosphorylation site for the activity of MYPT3-PP1: the unphosphorylated PS binds the Ank domains resulting in inhibition of PP1 activity by MYPT3. Phosphorylation of the PS results in a MYPT3 conformational change resulting in enhanced PP1 activity. In model 1a, the interaction of p12 with MYPT3 disrupts the inactive MYPT3-PP1 complex and sequesters MYPT3. This results in increased PP1 activity. In model 2a, p12 does not disrupt the MYPT3-PP1 complex, but interferes with the inhibitory action of MYPT3 on PP1, resulting in increased PP1 activity. Models 1a and 2a cannot be distinguished using wt MYPT3, since both models predict activation of PP1 by p12. However, the MYPT3ΔPS deletion mutant had previously been shown to activate (not inactivate) PP1 (15) and as we demonstrate here binds p12: we used this mutant to distinguish the two models. In model 1b, p12 sequesters MYPT3ΔPS resulting in a decrease in PP1 activity as measured by an increase in % transfected cells with stress fibers. In model 2b) p12 does not disrupt the MYPT3ΔPS-PP1 complex, activity is maintained or enhanced, and one expects the presence of transfected cells without stress fibers. Cells were transfected with the constructs shown in Fig. 10, B and C. The results (quantitated in Fig. 10C) show that expression of PP1β results in a decrease to ∼50% of transfected cells displaying stress fibers similar to the results for PP1β shown in Fig. 9 (lane 7). Co-expression of MYPT3ΔPS and PP1β resulted in enhanced PP1 activity (only 40% of transfected cells displaying stress fibers), confirming the previously reported (15) positive effect of MYPT3ΔPS on PP1β activity. Co-expression of p12 with MYPT3ΔPS and PP1β resulted in a further, small increase of PP1 activity (as predicted in model 2b) with 37% of transfected cells displaying stress fibers. Thus, p12 does not appear to disrupt MYPT3-PP1 complexes, indicating that model 2 likely applies.
FIGURE 10.
p12 can form a tripartite complex with MYPT3 and PP1. A, schematic representation of two models that may explain the effect of p12 on MYPT3-PP1 activity. Models 1a and 2a show results for wild type MYPT3 (wt-MYPT3), whereas models 1b and 2b shows the predicted results for the MYPT3 PKA phosphorylation site deletion mutant MYPT3ΔPS. In contrast to wt-MYPT3, MYPT3ΔPS enhances PP1 activity. We show in Fig. 6 that the MYPT3ΔPS mutant binds p12. In model 1a p12 binds to MYPT3 and sequesters it, causing an increase PP1 activity. In model 2a p12 forms a complex with MYPT3-PP1 and prevents the inhibitory action of MYPT3 on PP1 activity, causing increased PP1 activity. In model 1b p12 binds to MYPT3ΔPS and sequesters it, resulting in a reduction in PP1 activity. In model 2b p12 forms a complex with MYPT3-PP1 and does not affect MYPT3-PP1 activity. B, to distinguish the two models, cells were transfected with PP1β, and with combinations of PP1β and MYPT3ΔPS (panels 1 and 2) or with p12, PP1β and MYPT3ΔPS (panels 3 and 4). Panels l and 2 show that transfected cells lost stress fibers (arrowheads), indicative of PP1 activity. This result confirmed earlier reports on the MYPT3ΔPS activity. Panels 3 and 4 show that addition of p12 to MYPT3ΔPS-PP1 did not change the result: transfected cells lost stress fibers (arrowheads) while untransfected cells have stress fibers. C, quantitation of independent experiments shows a significant (p = 0.02) decrease in stress fiber formation in cells co-transfected with all three constructs compared with cells transfected with only PP1β.
DISCUSSION
The function of ornithine decarboxylase antizymes is well established as an important regulator of the ODC/polyamine pathway (6, 17). The translational frameshift of antizyme mRNA is a unique feature of these genes. The first open reading frame ORF1 of the antizyme genes Oaz1 and Oaz2 are believed to lack a clear function, while the second open reading frame ORF2 contains the ODC binding sequences and mediates ODC regulation. The question remains how the ODC/polyamine pathway is regulated in testis where Oaz3 is expressed abundantly after meiosis. A previous report of testicular ODC activity during rat development indicated that while mRNA levels of ODC increase during sexual maturation, ODC activity decreased (18). It was also demonstrated that ODC transcripts were negatively regulated at the translational level in adult mouse testis, as most ODC mRNA was associated with non-polysomal fractions (19). These data suggest that the ODC/polyamine pathway is under tight regulation during spermatogenesis, but by a mechanism different from the canonical ODC-antizyme one. This conclusion recently received support from data from the Oaz3 knock-out mouse, where no disturbance of polyamine levels was found in homozygous male knockouts as well as by our current results, which show that the predominant Oaz3-encoded male germ cell product is p12, which does not regulate ODC. Instead, we discovered that p12 displays a novel activity, viz. the regulation of protein phosphatase 1.
Oaz3-encoded Protein p12
When mouse Oaz3 was discovered, a 22 kDa protein with antizyme activity was predicted based on studies of Oaz1 and Oaz2 genes (5). Tosaka et al. showed that a polyamine-induced frameshift of Oaz3 mRNA can be measured in a sensitive in vitro cell culture system and OAZ3 protein was detected in sperm flagella with an antibody raised to ORF2. Our results do not support the presence of a 22 kDa or 28 kDa OAZ3 protein in spermatids and sperm: we show that translation of p12 initiates from an upstream CUG and that p12 is encoded by ORF1. Our results support previous reports demonstrating that the translational frameshift of Oaz3 mRNA is inefficient and does not support production of detectable amounts of 22 or 28 kDa OAZ3 protein in male germ cells (7). Although it cannot be ruled out that a translational frameshift of Oaz3 gene occurs to generate very small amounts of the 22 kDa OAZ3 protein, we demonstrate that in spermatids p12 is the major gene product.
Oaz3-encoded p12 is not unique to rat: we have detected p12 in mouse and bull sperm using our antibodies (data not shown). The amino acid sequence of p12 is highly conserved in several mammalian species. However, using bioinformatics analysis we could not find a homolog of p12 in non-mammalian species including zebra fish, fruit fly, and nematode. This is in contrast with Oaz1 which is present in many organisms including yeast, and Oaz2 which is present in zebrafish. This result suggests that the acquisition of the Oaz3 gene may be a late event in evolution and that p12 may have evolved a specialized function.
p12, Protein Phosphatase Activity, and Sperm Function
The Oaz3 knock-out mice that were recently reported have an unexpected phenotype: rather than showing a disturbance of polyamine metabolism, homozygous males were infertile because of the presence of separated sperm heads and tails in cauda epididymis (8). Interestingly, the ultrastructure of separate heads and tails appeared normal. The breakage occurred at the basal plate and striated columns suggesting a fragile connecting piece. The tailless heads underwent acrosomal reactions normally and were able to fertilize eggs upon microinjection. Interestingly, the Oaz3− headless tails were observed to beat vigorously, even after 14 h of incubation, much more so than comparable normal Oaz3+ sperm.
It is unknown how loss of Oaz3 contributes to the separation of sperm heads and tails or to the vigorous beating of the separated sperm tails. Our observation of a role for p12 in the regulation of PP1 activity may provide clues. The caveat is that due to the insolubility of proteins in the outer dense fibers it has not been possible to demonstrate complexes of p12-MYPT3 and/or p12/MYPT3/PP1γ2 in sperm tails despite many different biochemical isolation strategies attempted. However we believe that our discovery of the novel activity of p12 is relevant to the study of spermatogenesis for the following reasons: (a) it is possible if not likely that p12-MYPT3 complexes may exist in vivo, because we show by immunofluorescence assays that these abundant proteins show similar localization in spermatids (supplemental Fig. S1) and both localize to the sperm tail, the site for PP1γ2. 2) The expression levels achieved in somatic cell co-transfection studies likely approach the abundant expression observed in vivo. 3) We suggest that p12 likely does not play a structural role in sperm: p12 is present in ODF and FS and connecting piece. Because no gross morphological abnormalities were observed in the connecting pieces or tails of Oaz3-null mice (8) (with the caveat that no ultrastructural data for ODF and FS were presented), it is not likely that p12 is an important structural component.
We propose that p12 plays a role in signaling by modulation of protein phosphatases. e.g. it is possible that p12 plays a role in the assembly of the head-tail coupling apparatus, without which the head and tail connection is fragile: e.g. p12 may modulate PP1 at these locations to ensure the correct phosphorylation status of important structural proteins, a process that may be disturbed in the knock-out spermatozoa. Our data show that p12 can regulate PP1γ2 activity, which is the only protein phosphatase present in the tail. PP1γ2 was shown to be crucial for sperm motility (20–22). It was demonstrated that phosphatase activity is important both during spermatogenesis and during epididymal transition. PP1 activity inhibits sperm tail motility during epididymal passage: inhibition of phosphatase activity in rat sperm by Calyculin A leads to the precocious activation of hyperactive-like motility (23). Our finding that p12 interacts with MYPT3 and positively modulates the activity of PP1γ2 indicates that p12 may have a role in maintaining phosphatase activity in sperm tails while sperm travels through the epididymis. In agreement with this model it has been documented that PP1γ2 phosphatase activity is high in caput epididymis, decreases during passage through the epididymal tract and is low in cauda epididymis (24). The gradient of protein phosphatase activity is possibly achieved by the association of PP1γ2 with the inhibitory subunit Sds22, which may replace MYPT3 (24, 25). In the Oaz3 knock-out male mice, p12 is not available during these critical stages, possibly resulting in inhibition of PP1 by MYPT3 and premature motility. The observed vigorous flagellar beating may then contribute to the separation of head and tail in the epididymis.
Supplementary Material
This work was supported by grants from the Canadian Institutes of Health Research (to F. A. v. d. H. and R. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- ODF
- outer dense fibers
- Oaz
- ornithine decarboxylase antizyme
- Mypt3
- myosin phosphatase targeting subunit
- PP1
- protein phosphatase 1
- ODC
- ornithine decarboxylase
- ORF
- open reading frame
- PKA
- protein kinase A
- Ank
- ankyrin repeat.
REFERENCES
- 1. Bhullar B., Zhang Y., Junco A., Oko R., van der Hoorn F. A. (2003) J. Biol. Chem. 278, 16159–16168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shao X., Tarnasky H. A., Schalles U., Oko R., van der Hoorn F. A. (1997) J. Biol. Chem. 272, 6105–6113 [DOI] [PubMed] [Google Scholar]
- 3. Zarsky H. A., Tarnasky H. A., Cheng M., van der Hoorn F. A. (2003) Biol. Reprod. 68, 543–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ivanov I. P., Rohrwasser A., Terreros D. A., Gesteland R. F., Atkins J. F. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4808–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tosaka Y., Tanaka H., Yano Y., Masai K., Nozaki M., Yomogida K., Otani S., Nojima H., Nishimune Y. (2000) Genes Cells 5, 265–276 [DOI] [PubMed] [Google Scholar]
- 6. Coffino P. (2001) Nat. Rev. Mol. Cell Biol. 2, 188–194 [DOI] [PubMed] [Google Scholar]
- 7. Howard M. T., Shirts B. H., Zhou J., Carlson C. L., Matsufuji S., Gesteland R. F., Weeks R. S., Atkins J. F. (2001) Genes Cells 6, 931–941 [DOI] [PubMed] [Google Scholar]
- 8. Tokuhiro K., Isotani A., Yokota S., Yano Y., Oshio S., Hirose M., Wada M., Fujita K., Ogawa Y., Okabe M., Nishimune Y., Tanaka H. (2009) PLoS. Genet. 5, e1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fitzgerald C., Sikora C., Lawson V., Dong K., Cheng M., Oko R., van der Hoorn F. A. (2006) J. Biol. Chem. 281, 38172–38180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ou Y., Ruan Y., Cheng M., Moser J. J., Rattner J. B., van der Hoorn F. A. (2009) Exp. Cell Res. 315, 2802–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gietz R. D., Schiestl R. H. (2007) Nat. Protoc. 2, 38–41 [DOI] [PubMed] [Google Scholar]
- 12. Ichiba T., Matsufuji S., Miyazaki Y., Murakami Y., Tanaka K., Ichihara A., Hayashi S. (1994) Biochem. Biophys. Res. Commun. 200, 1721–1727 [DOI] [PubMed] [Google Scholar]
- 13. Snapir Z., Keren-Paz A., Bercovich Z., Kahana C. (2009) Biochem. J. 419, 99–103 [DOI] [PubMed] [Google Scholar]
- 14. Skinner J. A., Saltiel A. R. (2001) Biochem. J. 356, 257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yong J., Tan I., Lim L., Leung T. (2006) J. Biol. Chem. 281, 31202–31211 [DOI] [PubMed] [Google Scholar]
- 16. Tan I., Ng C. H., Lim L., Leung T. (2001) J. Biol. Chem. 276, 21209–21216 [DOI] [PubMed] [Google Scholar]
- 17. Mangold U. (2005) IUBMB. Life 57, 671–676 [DOI] [PubMed] [Google Scholar]
- 18. Weiner K. X., Dias J. A. (1992) Biol. Reprod. 46, 617–622 [DOI] [PubMed] [Google Scholar]
- 19. Alcivar A. A., Hake L. E., Mali P., Kaipia A., Parvinen M., Hecht N. B. (1989) Biol. Reprod. 41, 1133–1142 [DOI] [PubMed] [Google Scholar]
- 20. Vijayaraghavan S., Chakrabarti R., Myers K. (2007) Soc. Reprod. Fertil. Suppl. 63, 111–121 [PubMed] [Google Scholar]
- 21. Chakrabarti R., Kline D., Lu J., Orth J., Pilder S., Vijayaraghavan S. (2007) Biol. Reprod. 76, 992–1001 [DOI] [PubMed] [Google Scholar]
- 22. Chakrabarti R., Cheng L., Puri P., Soler D., Vijayaraghavan S. (2007) Asian J. Androl 9, 445–452 [DOI] [PubMed] [Google Scholar]
- 23. Goto N., Harayama H. (2009) J. Reprod. Dev. 55, 327–334 [DOI] [PubMed] [Google Scholar]
- 24. Mishra S., Somanath P. R., Huang Z., Vijayaraghavan S. (2003) Biol. Reprod. 69, 1572–1579 [DOI] [PubMed] [Google Scholar]
- 25. Chun Y. S., Park J. W., Kim G. T., Shima H., Nagao M., Kim M. S., Chung M. H. (2000) Biochem. Biophys. Res. Commun. 273, 972–976 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.