Abstract
The N-linked oligomannose glycans of HIV gp120 are a target for both microbicide and vaccine design. The extent of cross-clade conservation of HIV oligomannose glycans is therefore a critical consideration for the development of HIV prophylaxes. We measured the oligomannose content of virion-associated gp120 from primary virus from PBMCs for a range of viral isolates and showed cross-clade elevation (62–79%) of these glycans relative to recombinant, monomeric gp120 (∼30%). We also confirmed that pseudoviral production systems can give rise to notably elevated gp120 oligomannose levels (∼98%), compared to gp120 derived from a single-plasmid viral system using the HIVLAI backbone (56%). This study highlights differences in glycosylation between virion-associated and recombinant gp120.
Introduction
The functional envelope spike of HIV is a trimer of non-covalently associated gp120/gp41 heterodimers [1], densely coated with N-linked carbohydrates that are essential for correct glycoprotein folding and shielding vulnerable protein surfaces from antibody recognition [2], [3], [4], [5], [6], [7], [8], [9]. These carbohydrates are attached to the envelope proteins via the host cell glycosylation pathway [9], [10]. However, the glycosylation processing of virion-associated gp120 is divergent from that of typical glycoproteins produced by the host cell: the extensive array of gp120 N-linked glycans contains an ‘intrinsic’ patch of densely packed oligomannose glycans which are inefficiently trimmed by host ER and Golgi α-mannosidases [5], [11]. Such clusters of oligomannose-type carbohydrates do not occur in mammalian glycosylation and they therefore provide a potential target for selective antibody recognition of the virus [12]. Indeed, one of the few known broadly neutralising anti-HIV-1 antibodies, 2G12, exploits this divergence in host and viral glycan processing and recognises Manα1→2Man-linked residues attached to oligomannose termini within the gp120 ‘intrinsic’ mannose patch [12], [13], [14], [15], [16]. Along with other broadly neutralising antibodies, 2G12 confers sterilizing immunity to primary viral challenge in non-human primates [3], [17], [18], [19]. The Manα1→2Man array, recognised by 2G12, has become the blueprint for a range of microbial [15], [20], [21], [22], synthetic [16], [23], [24], [25] and recombinant glycoconjugate [26], [27] vaccine candidates against HIV-1. Additionally a number of lectins, specific for Manα1→2Man structures, exhibit potent antiviral activity [28], [29]. The abundance and conservation of Manα1→2Man motifs on the functional envelope of primary viral isolates is therefore crucial for the applicability of a carbohydrate-based vaccine approach and is the focus of this study.
Two recent studies have shown that α1→2-mannosidase trimming is reduced by the steric constraints imposed by gp120 trimerisation [11], [30] leading to a ‘trimer-associated’ oligomannose population in addition to the ‘intrinsic’ mannose patch. Both studies observed that, compared to recombinant gp120, there is a greater abundance of Manα1→2Man terminating structures (Man6-9GlcNAc2) on trimeric envelope glycoprotein. We previously described that Env, derived mostly from pseudoviral systems, was almost entirely oligomannose with a predominant population of Man5GlcNAc2 [11]. Here, we examine a wider range of viral production systems and envelope expression levels, and report a greater range of abundances of oligomannose-type glycans, although in all cases there is an elevation of oligomannose on virion-associated Env compared to recombinant, monomeric gp120.
Results
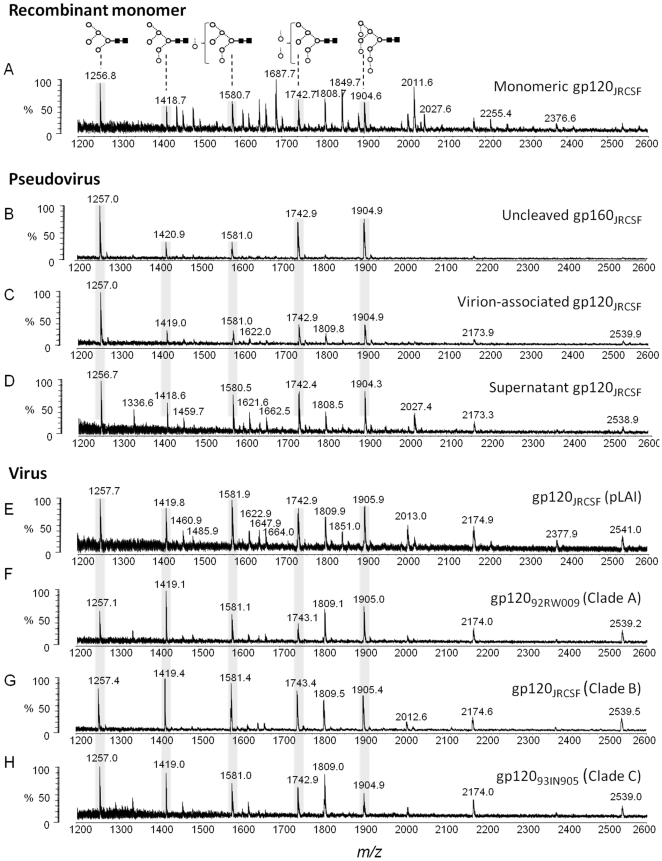
As previously reported, the matrix-assisted laser desorption/ionisation time of flight (MALDI-TOF) mass spectrometry (MS) spectrum for recombinant wild-type gp120JRCSF showed extensive complex-type glycosylation [11], with the intrinsic mannose patch forming around 29% of the total glycan population (Figure 1A). The abundances of oligomannose- and complex-type N-linked glycans released from gp120 in this and subsequent production systems are shown in Table 1.
Figure 1. Comparison of recombinant, pseudoviral and viral gp120.
MALDI-TOF MS analyses of released desialylated N-linked glycans ([M+Na]+ ions) from: (A) recombinant monomeric gp120JRCSF expressed in HEK 293T cells; (B, C and D) respectively gp160JRCSF, gp120JRCSF and soluble, non-virion associated envelope gp120JRCSF isolated from pseudoviral particle preparations generated by transfection of HEK 293T cells with the pSVIII-JRCSF and pSG3Δenv plasmids at a ratio of 1∶10; (E) gp120JRCSF isolated from replication competent viral particles generated by transfection of HEK 293T cells with pLAI-JRCSF env molecular clone; (F, G and H) respectively gp12092RW009, gp120JRCSF and gp12093IN905 isolated from virus obtained by infection of human PBMCs. Symbols used for the structural formulae in this and subsequent figures: ⋄ = Gal, ▪ = GlcNAc, ○ = Man,  = Fuc [46]. The linkage position is shown by the angle of the lines linking the sugar residues (vertical line = 2-link, forward slash = 3-link, horizontal line = 4-link, back slash = 6-link). Anomericity is indicated by full lines for β-bonds and broken lines for α-bonds [46]. The oligomannose series are highlighted.
= Fuc [46]. The linkage position is shown by the angle of the lines linking the sugar residues (vertical line = 2-link, forward slash = 3-link, horizontal line = 4-link, back slash = 6-link). Anomericity is indicated by full lines for β-bonds and broken lines for α-bonds [46]. The oligomannose series are highlighted.
Table 1. Abundances of released N-linked glycans obtained from recombinant (monomeric), pseudoviral, and viral gp120†.
| gp120 source | Cell-type | Man5-9GlcNAc2% | Man5GlcNAc2% | Complex% | Mannose content relative to rgp120‡ |
| Recombinant monomer (pHLsec JRCSF) | 293T | 29% | 7.7% | 71% | 1.0 |
| Pseudovirus (pSG3Δenv:pSVIII JRCSF, 2∶1) | 293T | 98% | 38% | 2% | 3.4 |
| Pseudovirus (pSG3Δenv:pSVIII JRCSF, 10∶1) | 293T | 85% | 39% | 15% | 2.9 |
| Supernatant (pSG3Δenv:pSVIII JRCSF, 10∶1) | 293T | 73% | 18% | 27% | 2.5 |
| Virus (pLAI-JRCSF env) | 293T | 56% | 10% | 44% | 1.9 |
| Virus JRCSF (clade B) | PBMC | 79% | 12% | 21% | 2.7 |
| Virus 92RW009 (clade A) | PBMC | 64% | 10% | 36% | 2.2 |
| Virus 93IN905 (clade C) | PBMC | 62% | 19% | 38% | 2.1 |
†Abundances obtained for desialylated N-linked glycans released from gp120 described in this study. Values were obtained from data presented in Figure 1 and Doores et al. [11].
‡Values represent the increase in oligomannose population (Man5-9GlNAc2) for pseudoviral and viral gp120 compared to monomeric, recombinant gp120.
Pseudoviral particles were prepared using human embryonic kidney (HEK) 293T cells with plasmids carrying JRCSF envelope gene (pSVIII-JRCSF) and the HIV-1 backbone (pSG3Δenv) at a ratio of 1∶10 respectively. A recent study by Crooks et al. has shown that pseudoviral production systems produce significant levels of non-functional uncleaved ‘gp160ER’ whose glycans are entirely sensitive to digestion by endoglycosidase H (endo H) [31]. In addition to ‘gp160ER’, a smaller population of partially endo H-resistant cleaved gp120/gp41 trimers (indicative of the presence of some complex-type glycans) was observed and it was proposed that only this more processed glycoform is shed from the functional envelope spike into the supernatant. We used mass spectrometry to determine the divergent glycosylation of these two species and showed that both gp160, and the less abundant virion-associated gp120, consisted predominantly of oligomannose glycans (94% and 85% respectively) (Figure 1B, C). Interestingly, increasing the Env:backbone plasmids ratio from 1∶10 to 1∶2 (constant DNA) resulted in an increased level of envelope expression [32] (data not shown) and an even higher oligomannose abundance (>98%, as previously reported [11]) suggesting envelope expression level might influence the glycosylation profile of gp120. We observed an unusual abundance of Man5GlcNAc2, indicating that most of the virion-associated material had not been exposed to the medial-Golgi-resident GlcNAc transferase I (GnT I). This lack of processing is also consistent with the abundance of uncleaved gp160, in this pseudoviral systems: the furin protease, responsible for gp160 cleavage into gp120/gp41, is proposed to be largely resident in the trans-Golgi apparatus [33], [34].
In contrast to virion-associated gp160/120, the gp120 shed into the supernatant, proposed to derive solely from cleaved functional trimers [31], contained more complex-type glycans (27%) (Figure 1D) but was nonetheless mostly oligomannose (73%). This elevated level of oligomannose glycans compared to recombinant monomeric gp120 (Figure 1A) is consistent with the reduced mannose trimming previously reported for recombinant, trimeric gp120 compared to recombinant, monomeric gp120 [11]. Moreover, the 27% complex-type glycans seen in this shed gp120 was matched by a corresponding reduction in the Man5GlcNAc2 peak compared to virion associated gp120 (Figure 1C) indicating this species does not evade processing by GnT I and subsequent Golgi-resident glycosidases and glycosyltranferases.
We next compared the glycosylation of pseudovirus-derived gp120 to replication competent virus-derived gp120. The glycans from gp120 derived from JRCSF virus prepared in HEK 293T cells using an infectious pLAI-JRCSF Env molecular clone [35] showed a more even division between oligomannose (56%) and complex-type glycans (44%), and a more equal distribution of abundances within the Man5–9GlcNAc2 structures (Figure 1E). The complex-type glycans were predominantly of the bi- or tri-antennary type with variable galactosylation and fucosylation typical for HEK 293T cells [36], [37]. We observed a reduced envelope expression level in these replication competent viral particles compared to the pseudoviral particles. This reduced envelope expression level and corresponding reduction in oligomannose abundance further suggests envelope expression levels may influence the glycosylation profile of virion-associated gp120. In addition to cleaved gp120, uncleaved, non-functional gp160 was also detected in the pLAI-JRCSF Env virus derived membrane-associated fraction. The analysis of gp160 glycosylation revealed, as for the pseudoviral derived gp160, less efficient processing by the Golgi α-mannosidases IA–C, with elevated populations of Manα1→2Man linked oligomannose glycans compared to gp120 (68% Man6–9GlcNAc2 for gp160 compared to 46% Man6–9GlcNAc2 for gp120; data not shown). This suggests that uncleaved gp160 adopts a quaternary arrangement with more occluded glycans compared to cleaved gp120/gp41.
Analyses of gp120 derived from virus prepared by infection of peripheral blood mononuclear cells (PBMCs) with viruses from clade A (92RW009), clade B (JRCSF), and clade C (93IN905) showed a predominantly oligomannose glycan composition (62–79% Man5–9GlcNAc2, Figure 1F, G, H) with a distribution similar to that previously reported for PBMC-derived gp120JRCSF [11]. In a previous study we noted the presence of some complex-type glycans but due to limitations of material we were unable to perform analysis of desialylated material required to distinguish these glycans from those of the capture antibodies [11]. Here, MALDI-TOF MS analysis of desialylated glycans revealed, in addition to the Man5–9GlcNAc2 glycans, a smaller series of branched, fucosylated complex-type glycans at m/z 1809 (11–24%), 2012 (1.5–3%), 2174 (3–5.5%) and 2539 (2.5–4.6%) in all three spectra corresponding to the neutral derivatives of sialylated bi, tri and tetra-antennary glycans.
Overall, the glycan distribution within the oligomannose series is similar to that observed for the single-plasmid infectious pLAI-JRCSF env clone (Figure 1E) and the shed material from the pseudoviral system (Figure 1D), with some complex-type glycans and without an elevated Man5GlcNAc2 peak. We note however that the distribution of the oligomannose series differs slightly between isolates: the ratio of oligomannose-type glycans that terminate with Manα1→2Man, compared to those that do not, is higher for 92RW009 (clade A, 5.2) and JRCSF (clade B, 5.6) than for 93IN905 (clade C, 2.7). A likely explanation for this difference in glycan processing, in clade C envelope, is the absence of key glycosylation site(s) which reduce the density of the intrinsic mannose patch and increase the processing of adjacent Manα1→2Man termini. Notably, the oligomannose glycan attached to Asn295 is absent in most clade C isolates, including HIV-1 93IN905, and is critical for efficient neutralisation by a number of mannose-specific ligands, including 2G12 [38].
Therefore, as for pseudoviral and viral particles obtained from HEK 293T cells (Figure 1C, E), the glycans on PBMC-derived virus from isolates from distinct antigenic and geographical backgrounds are predominantly oligomannose.
Discussion
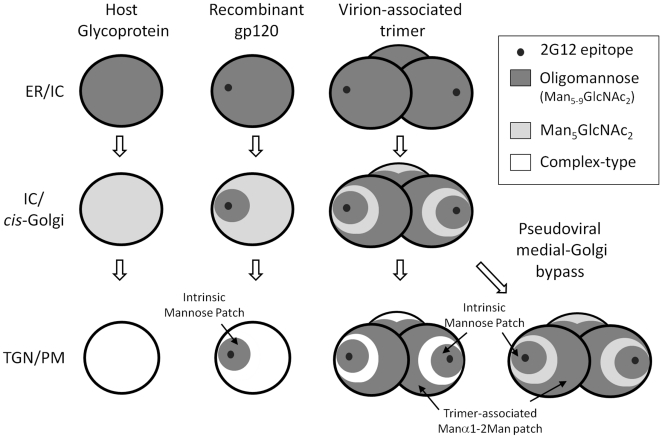
The HIV envelope is entirely processed by the glycosylation machinery of the host cell: the interaction of envelope with the spectrum of enzymatic activities present in the secretory pathway determines the types of glycans that will be presented on gp120 at the virion surface or as a recombinant protein. Although all N-linked glycosylation sites on gp120 are initially glycosylated with the same Glc3Man9GlcNAc2 precursor, these sites are not processed equivalently. We propose a model (Figure 2), based on the data reported here and integrating previous findings from our group and others, of how gp120 is processed as it traffics through the cell. First, the ER glycoform arises following the removal of the final glucose residue by α-glucosidase II to produce Man9GlcNAc2 (or depending on the cell type by the action of endomannosidase to yield D2,D3-Man8GlcNAc2). This natural gp120 glycoform, normally a transient biosynthetic intermediate, has been isolated in a number of studies using inhibitors of α-mannosidases such as kifunensine [14], [27], [39]. The Man8–9GlcNAc2 intermediates are then processed by the ER and Golgi α-mannosidases. This process is slower for glycans within the intrinsic mannose patch, and is further limited by the steric consequences of trimerisation [11], [30] (Figure 1 and 2). These two factors combine to yield an enhanced abundance of Manα1→2Man terminating glycans compared to recombinant monomeric gp120 which is largely insensitive to changes in expression system or envelope structure. Finally, the more exposed regions of gp120 are processed by the medial Golgi resident GnT I to form the hybrid-type glycan, GlcNAcβ1→2Man5GlcNAc2, and subsequent complex-type glycosylation found on cell surface and on virions. These complex-type N-glycans are processed in a tissue-specific manner, consistent with observations that they are not essential for viral function but may modulate infectivity and accessibility of some antibody epitopes [40]. The predominance of the biosynthetic intermediate, Man5GlcNAc2 (and the absence of complex-type glycans), and reduced gp160 processing are both markers for a lack of processing in the medial-Golgi apparatus. Both these phenomena are observed in envelope glycoproteins isolated from pseudoviral particles, and the mechanism for this Golgi by-pass, which is consistent with a recent study showing an abundance of ‘gp160ER’ on pseudoviral particles [31] is unknown, but might reasonably be attributed to either an alteration of compartmentalisation or to a substrate saturation of Golgi-resident envelope processing enzymes.
Figure 2. Multiple divergences of gp120 glycosylation from host cell glycosylation.
Following removal of terminal α-linked glucose residues in the ER, folded glycoproteins contain exclusively oligomannose glycans. During transit through the ER, intermediate compartment (IC) and cis-Golgi apparatus, Manα1→2Man termini are removed by ER Mannosidase I and Golgi Mannosidases A–C to yield Man5GlcNAc2. However, the oligomannose cluster intrinsic to monomeric gp120 [5], [14] limits glycan processing on both monomeric and oligomeric gp120 [11], [30]. The steric consequences of trimerisation further limit Manα1→2Man trimming [30] leading to an additional ‘trimer-associated’ population of Man5–9GlcNAc2. The exposed Man5GlcNAc2 glycans on gp120 that passage through the full extent of the Golgi apparatus and trans Golgi network (TGN) to the plasma membrane (PM) are processed by GnT I and subsequent enzymes to form complex-type glycans. However, envelope glycoprotein that does not follow this route to the PM is characterized by an elevated abundance of Man5GlcNAc2 (and closely resembles gp120 expressed in GnT I-deficient cells [11], [30]), and reduced furin cleavage. Thus the intrinsic mannose patch, which includes the 2G12 epitope, persists from the earliest stages of glycan processing whilst other elements of the glycan shield exhibit variably processed glycans depending on oligomerization state and, at least in the case of pseudoviral gp160/gp120, cellular trafficking.
Overall, the data presented here and in our previous study [11] indicate that the glycosylation of HIV envelope glycoproteins diverges from typical host-cell glycosylation on at least three levels. First, the clustering of N-glycans gives rise to an ‘intrinsic’ mannose patch (Figure 1A). Second, the steric constraints of trimerisation result in an additional population of oligomannose glycans. Third, in pseudoviral systems, a majority of envelope glycoproteins bypass the Golgi-resident enzymes responsible for complex glycan biosynthesis and protein cleavage, leading to an unusual elevation of Man5GlcNAc2 on non-functional envelope gp160. The ‘intrinsic’ and ‘trimer-associated’ mannose patches give rise to a predominance of oligomannose-type glycans on virion-associated gp120 that is conserved regardless of virus production system, envelope expression level, 2G12 sensitivity or envelope sequence.
Materials and Methods
Ethics statement
Human blood samples from healthy donors were obtained from The Normal Blood Donor service at The Scripps Research Institute. The collection of human blood samples for isolation of PBMCs and subsequent propagation of HIV-1 virus was approved by the Institutional Review Board at The Scripps Research Institute (protocol number HSC-06-4604).
Recombinant protein expression
HEK 293T (ATCC number CRL-1573) were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum, penicillin and streptomycin. Transient transfection using the pHLsec vector followed that of Aricescu [41]. Briefly, for each T175 flask, 90 µg of polyethyleneimine (PEI) and 50 µg of DNA were incubated for 10 min in 5 mL of serum free media; then added to 80–90% confluent cells cultured in 25 mL of serum free media. Culture supernatant was collected at 4 days post-transfection, and subsequently centrifuged, sterile filtered and then concentrated by centrifugal filtration using Vivaspin 20 devices.
Pseudovirus and virus preparation in 293T cells
Pseudovirus was generated in HEK 293T cells as described [42]. Briefly, HEK 293T cells were transfected with plasmids carrying the reporter gene expressing the virus backbone (pSG3Δenv) and the functional envelope clone (pSVIII-JRCSF) at a ratio of 2∶1 or 10∶1 (total DNA, 60 µg per 7×106 cells) using Fugene (Roche) according to the manufacturer's instructions. Virus supernatants were harvested after 3 days. Fully replicative JRCSF virus capable of multiple round infection was made in 293T cells by transfection with a single plasmid construct (pLAI-JRCSF env) using Fugene [35].
Virus preparation in PBMCs
Human PBMCs were obtained from healthy individuals and isolated and stimulated as previously described [43]. HIV-1JRCSF, HIV-192RW009 and HIV-193IN905 virus stocks were grown and titered on CD8+-depleted PBMCs [44]. Virus production was monitored by p24 ELISA (Aalto Bioreagents, Dublin, Eire).
Envelope Isolation
Virus preparations were pre-cleared by low speed centrifugation. Virus particles were pelleted by ultracentrifugation (22,000 rpm, 1 hour). Virus pellets were lysed with NP-40 (1% in PBS with protease inhibitors, 20 mins at 4°C). The debris was removed by centrifugation and the envelope protein was immunoprecipitated with HIV envelope specific monoclonal antibodies (D7324, b12, b6, F425-b4e8, VRC01, VRC03, PGV04) depending on virus isolate). Protein A and G beads were added and incubated overnight at 4°C. The beads were washed 5 times with PBS and then the protein was eluted by heating in loading buffer (containing dithiothreitol) for 10 mins at 100°C and resolved by SDS-PAGE. The envelope band was confirmed by western blot (primary antibodies; 2G12, F425-b4e8, PGV04, HIVIG (depending on strain), secondary antibody, goat-anti-human-Fcγ-HRP) and cut to use directly in glycan analysis. The ‘soluble non-virion associated fraction’ is the envelope protein isolated by immunoprecipation of the supernatant after the virus has been removed by ultracentrifugation.
MALDI-TOF mass spectrometry
Oligosaccharides were released from target glycoproteins with Peptide-N-Glycosidase (PNGase) F (New Englands Biolabs) from Coomassie blue-stained NuPAGE [45]. Excised bands were washed five times alternatively with acetonitrile and deionised water, and rehydrated with a 3000 Units/ml of PNGase F water solution. After incubation for 12 hours at 37°C, the enzymatically released N-linked glycans were eluted with water. Samples were analysed by positive ion matrix-assisted laser desorption/ionization (MALDI) time-of-flight (TOF) mass spectra with a Shimazu AXIMA TOF2 MALDI TOF/TOF mass spectrometer (Kratos Analytical, Manchester, UK) fitted with delayed extraction and a nitrogen laser (337 nm). Samples were cleaned on a Nafion 117 membrane (Aldrich), and then prepared for mass spectrometry by adding 0.5 µL of an aqueous solution of the glycans to the matrix solution (0.3 µL of a solution of 2,5-dihydroxybenzoic acid in acetonitrile:water (1∶1, v:v) on the stainless steel target plate and allowing it to dry at room temperature. The sample/matrix mixture was then recrystallized from ethanol. Samples were examined after removal of any potential sialic acids by heating at 80°C for 1 hr with 1% acetic acid.
Acknowledgments
We thank Pascal Poignard for helpful discussion.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by a project grant from the International AIDS Vaccine Initiative (#UOXFORCOA1101). CB was supported by a Glycobiology Institute graduate scholarship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJ, et al. Molecular Architectures of Trimeric SIV and HIV-1 Envelope Glycoproteins on Intact Viruses: Strain-Dependent Variation in Quaternary Structure. PLoS Pathog. 2010;6:e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pantophlet R, Burton DR. GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 3.Scanlan CN, Offer J, Zitzmann N, Dwek RA. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature. 2007;446:1038–1045. doi: 10.1038/nature05818. [DOI] [PubMed] [Google Scholar]
- 4.Wei X, Decker JM, Wang S, Hui H, Kappes JC, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 5.Zhu X, Borchers C, Bienstock RJ, Tomer KB. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry. 2000;39:11194–11204. doi: 10.1021/bi000432m. [DOI] [PubMed] [Google Scholar]
- 6.Go EP, Chang Q, Liao HX, Sutherland LL, Alam SM, et al. Glycosylation site-specific analysis of clade C HIV-1 envelope proteins. J Proteome Res. 2009;8:4231–4242. doi: 10.1021/pr9002728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Go EP, Irungu J, Zhang Y, Dalpathado DS, Liao HX, et al. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes' accessibility. J Proteome Res. 2008;7:1660–1674. doi: 10.1021/pr7006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizuochi T, Spellman MW, Larkin M, Solomon J, Basa LJ, et al. Carbohydrate structures of the human-immunodeficiency-virus (HIV) recombinant envelope glycoprotein gp120 produced in Chinese-hamster ovary cells. Biochem J. 1988;254:599–603. doi: 10.1042/bj2540599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, et al. Essentials of Glycobiology: Cold Spring Harbour Laboratory Press; 2009. [PubMed] [Google Scholar]
- 10.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 11.Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, et al. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci U S A. 2010;107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, et al. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol. 2002;76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 14.Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, et al. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1,2 mannose residues on the outer face of gp120. J Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunlop DC, Bonomelli C, Mansab F, Vasiljevic S, Doores KJ, et al. Polysaccharide mimicry of the epitope of the broadly neutralising anti-HIV antibody, 2G12, induces enhanced antibody responses to self oligomannose glycans. Glycobiology. 2010;20:812–823. doi: 10.1093/glycob/cwq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee HK, Scanlan CN, Huang CY, Chang AY, Calarese DA, et al. Reactivity-based one-pot synthesis of oligomannoses: defining antigens recognized by 2G12, a broadly neutralizing anti-HIV-1 antibody. Angew Chem Int Ed Engl. 2004;43:1000–1003. doi: 10.1002/anie.200353105. [DOI] [PubMed] [Google Scholar]
- 17.Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, et al. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 18.Montefiori DC, Mascola JR. Neutralizing antibodies against HIV-1: can we elicit them with vaccines and how much do we need? Curr Opin HIV AIDS. 2009;4:347–351. doi: 10.1097/COH.0b013e32832f4a4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009;15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 20.Dunlop DC, Ulrich A, Appelmelk BJ, Burton DR, Dwek RA, et al. Antigenic mimicry of the HIV envelope by AIDS-associated pathogens. AIDS. 2008;22:2214–2217. doi: 10.1097/QAD.0b013e328314b5df. [DOI] [PubMed] [Google Scholar]
- 21.Luallen RJ, Lin J, Fu H, Cai KK, Agrawal C, et al. An engineered Saccharomyces cerevisiae strain binds the broadly neutralizing human immunodeficiency virus type 1 antibody 2G12 and elicits mannose-specific gp120-binding antibodies. J Virol. 2008;82:6447–6457. doi: 10.1128/JVI.00412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luallen RJ, Agrawal-Gamse C, Fu H, Smith DF, Doms RW, et al. Antibodies against Manα1,2-Manα1,2-Man oligosaccharide structures recognize envelope glycoproteins from HIV-1 and SIV strains. Glycobiology. 2010;20:280–286. doi: 10.1093/glycob/cwp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Astronomo RD, Lee HK, Scanlan CN, Pantophlet R, Huang CY, et al. A glycoconjugate antigen based on the recognition motif of a broadly neutralizing human immunodeficiency virus antibody, 2G12, is immunogenic but elicits antibodies unable to bind to the self glycans of gp120. J Virol. 2008;82:6359–6368. doi: 10.1128/JVI.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabanova A, Adamo R, Proietti D, Berti F, Tontini M, et al. Preparation, characterization and immunogenicity of HIV-1 related high-mannose oligosaccharides-CRM197 glycoconjugates. Glycoconj J. 2010;27:501–513. doi: 10.1007/s10719-010-9295-0. [DOI] [PubMed] [Google Scholar]
- 25.Doores KJ, Fulton Z, Hong V, Patel MK, Scanlan CN, et al. A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity. Proc Natl Acad Sci U S A. 2010;107:17107–17112. doi: 10.1073/pnas.1002717107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luallen RJ, Fu H, Agrawal-Gamse C, Mboudjeka I, Huang W, et al. A yeast glycoprotein shows high-affinity binding to the broadly neutralizing human immunodeficiency virus antibody 2G12 and inhibits gp120 interactions with 2G12 and DC-SIGN. J Virol. 2009;83:4861–4870. doi: 10.1128/JVI.02537-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scanlan CN, Ritchie GE, Baruah K, Crispin M, Harvey DJ, et al. Inhibition of mammalian glycan biosynthesis produces non-self antigens for a broadly neutralising, HIV-1 specific antibody. J Mol Biol. 2007;372:16–22. doi: 10.1016/j.jmb.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 28.Balzarini J. Targeting the glycans of glycoproteins: a novel paradigm for antiviral therapy. Nat Rev Microbiol. 2007;5:583–597. doi: 10.1038/nrmicro1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bewley CA, Otero-Quintero S. The potent anti-HIV protein cyanovirin-N contains two novel carbohydrate binding sites that selectively bind to Man(8) D1D3 and Man(9) with nanomolar affinity: implications for binding to the HIV envelope protein gp120. J Am Chem Soc. 2001;123:3892–3902. doi: 10.1021/ja004040e. [DOI] [PubMed] [Google Scholar]
- 30.Eggink D, Melchers M, Wuhrer M, van Montfort T, Dey AK, et al. Lack of complex N-glycans on HIV-1 envelope glycoproteins preserves protein conformation and entry function. Virology. 2010;401:236–247. doi: 10.1016/j.virol.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crooks ET, Tong T, Osawa K, Binley JM. Enzyme Digests Eliminate Non-Functional Env from HIV-1 Particle Surfaces Leaving Native Env Trimers Intact and Viral Infectivity Unaffected. J Virol. 2011 doi: 10.1128/JVI.00154-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Provine NM, Puryear WB, Wu X, Overbaugh J, Haigwood NL. The infectious molecular clone and pseudotyped virus models of human immunodeficiency virus type 1 exhibit significant differences in virion composition with only moderate differences in infectivity and inhibition sensitivity. J Virol. 2009;83:9002–9007. doi: 10.1128/JVI.00423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bosshart H, Humphrey J, Deignan E, Davidson J, Drazba J, et al. The cytoplasmic domain mediates localization of furin to the trans-Golgi network en route to the endosomal/lysosomal system. J Cell Biol. 1994;126:1157–1172. doi: 10.1083/jcb.126.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shapiro J, Sciaky N, Lee J, Bosshart H, Angeletti RH, et al. Localization of endogenous furin in cultured cell lines. J Histochem Cytochem. 1997;45:3–12. doi: 10.1177/002215549704500102. [DOI] [PubMed] [Google Scholar]
- 35.Leaman DP, Kinkead H, Zwick MB. In-solution virus capture assay helps deconstruct heterogeneous antibody recognition of human immunodeficiency virus type 1. J Virol. 2010;84:3382–3395. doi: 10.1128/JVI.02363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowden TA, Crispin M, Harvey DJ, Aricescu AR, Grimes JM, et al. Crystal structure and carbohydrate analysis of Nipah virus attachment glycoprotein: a template for antiviral and vaccine design. J Virol. 2008;82:11628–11636. doi: 10.1128/JVI.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crispin M, Chang VT, Harvey DJ, Dwek RA, Evans EJ, et al. A human embryonic kidney 293T cell line mutated at the Golgi α-mannosidase II locus. J Biol Chem. 2009;284:21684–21695. doi: 10.1074/jbc.M109.006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexandre KB, Gray ES, Lambson BE, Moore PL, Choge IA, et al. Mannose-rich glycosylation patterns on HIV-1 subtype C gp120 and sensitivity to the lectins, Griffithsin, Cyanovirin-N and Scytovirin. Virology. 2010;402:187–196. doi: 10.1016/j.virol.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agrawal-Gamse C, Luallen RJ, Liu B, Fu H, Lee FH, et al. Yeast-elicited cross-reactive antibodies to HIV Env glycans efficiently neutralize virions expressing exclusively high-mannose N-linked glycans. J Virol. 2011;85:470–480. doi: 10.1128/JVI.01349-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Binley JM, Ban YE, Crooks ET, Eggink D, Osawa K, et al. Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. J Virol. 2010;84:5637–5655. doi: 10.1128/JVI.00105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 42.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mann AM, Rusert P, Berlinger L, Kuster H, Gunthard HF, et al. HIV sensitivity to neutralization is determined by target and virus producer cell properties. AIDS. 2009;23:1659–1667. doi: 10.1097/QAD.0b013e32832e9408. [DOI] [PubMed] [Google Scholar]
- 44.Rusert P, Fischer M, Joos B, Leemann C, Kuster H, et al. Quantification of infectious HIV-1 plasma viral load using a boosted in vitro infection protocol. Virology. 2004;326:113–129. doi: 10.1016/j.virol.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 45.Küster B, Wheeler SF, Hunter AP, Dwek RA, Harvey DJ. Sequencing of N-linked oligosaccharides directly from protein gels: in-gel deglycosylation followed by matrix-assisted laser desorption/ionization mass spectrometry and normal-phase high-performance liquid chromatography. Anal Biochem. 1997;250:82–101. doi: 10.1006/abio.1997.2199. [DOI] [PubMed] [Google Scholar]
- 46.Harvey DJ, Merry AH, Royle L, Campbell MP, Dwek RA, et al. Proposal for a standard system for drawing structural diagrams of N- and O-linked carbohydrates and related compounds. Proteomics. 2009;9:3796–3801. doi: 10.1002/pmic.200900096. [DOI] [PubMed] [Google Scholar]