Abstract
Purpose
To identify mutations in the paired box 6 (PAX6) gene of 33 probands with aniridia and to reveal the mutational spectrum in the Chinese population.
Methods
Unrelated probands with aniridia from 27 newly selected families and six previously analyzed families participated in this study. The coding regions of PAX6 in the 27 new families were analyzed using cycle sequencing. Families that lacked detectable variations based on sequencing (14 new and six previously analyzed) were further analyzed using multiplex ligation-dependent probe amplification (MLPA).
Results
Fifteen mutations were identified in 16 of the 33 families: c.[65_94del30; 99_105dup7], c.101_102insA, c.177delG, c.238_239insGCGA, c.1033–42_1033–26del17insG, c.1A>G, c.120C>A, c.718C>T, c.949C>T, c.1062C>A, c.1183G>A, c.1268A>T, and three gross deletions involving exons 1–14, exons 8–14, and exons 9–14. The first five mutations were novel and the c.1268A>T mutation was present in two families. Phenotypic variations were observed between families and between different affected patients within the families.
Conclusions
The PAX6 mutation spectrum in Chinese aniridia patients is comparable to that reported in other ethnic groups. Further studies of the 17 families with no detected mutations may provide additional information to improve the understanding of the molecular genetics of aniridia.
Introduction
Aniridia (OMIM 106210) is a bilateral, panocular disorder characterized by the absence of the iris. It is usually accompanied by developmental defects of the cornea, lens, retina, optic nerve, and/or the anterior chamber angle [1]. Approximately two thirds of all cases are transmitted as an autosomal dominant trait among families. The remaining one third are sporadic, with no family history [1-3]. Mutations in PAX6 (OMIM 607108) have been shown to be responsible for aniridia in most patients [4-10]. In rare cases, PAX6 mutations may cause other ocular abnormalities, such as cataracts [11], Peters anomaly [12,13], foveal dysplasia, microphthalmia, and optic nerve malformations [14,15]. To date, at least 334 mutations in PAX6 have been identified [16]. However, the absence of PAX6 mutations in some cases of aniridia implies that mutations in additional genes cannot be excluded [6-10]. This possibility is supported by recent reports: 1) mutations in the forkhead box C1 gene (FOXC1, OMIM 601090) are associated with aniridia in two families [17,18] and 2) aniridia in patients with preserved visual function are not related to PAX6 mutations [19].
Understanding the mutation spectrum and frequency will not only provide biomarkers for clinical practice but will also represent a fundamental basis for identifying any additional causative genes. The spectrum and frequency of PAX6 mutations in Chinese aniridia patients has not been well identified for the following reasons [20-27]: 1) most reports only involved a single or a limited number of families and 2) mutations were usually detected by cycle sequencing (detection of small variations), in which a larger deletion involving part or all of the gene could not be detected. Cycle sequencing combined with multiplex ligation-dependent probe amplification (MLPA) has been shown to effectively detect small mutations and large deletions in PAX6 [10]. In this study, both cycle sequencing and MLPA were used to detect mutations in the PAX6 gene of 33 Chinese probands with aniridia.
Methods
Patients
Thirty-three unrelated probands with aniridia participated in this study, including six families previously analyzed by sequencing [22] and 27 newly selected families. Patients suspected of having Peters anomaly or Rieger syndrome were excluded. All aniridia patients in this study were recruited from our Pediatric and Genetic Eye Clinic at the Eye Hospital of the Zhongshan Ophthalmic Center, Guangzhou, China. Written informed consent that complied with the tenets of the Declaration of Helsinki and following the Guidance of Sample Collection of Human Genetic Diseases (863-Plan) by China’s Ministry of Public Health was obtained from each participant before the study. Genomic DNA was prepared from venous blood.
Detection of PAX6 mutations
Thirteen pairs of primers (Table 1) were used to amplify the 14 exons (3 noncoding exons and 11 coding exons) and their adjacent intronic sequences of PAX6 (NCBI human genome build 37.2, NC_000011.9 for gDNA, NM_001604.4 for cDNA, and NP_001595.2 for protein). The PCR products of individual exons for each patient were sequenced using the ABI BigDye Terminator v3.1 Cycle Sequencing Kit (ABI Applied Biosystems, Foster City, CA) and the ABI 3100 Genetic Analyzer (ABI Applied Biosystems) according to the manufacturer’s recommendations. Sequencing results from patients’ sequences and PAX6 consensus sequences from the National Center for Biotechnology Information (NCBI) human genome database (NC_000011.9) were imported into the SeqManII program of the Lasergene package (DNAStar Inc., Madison, WI) and aligned to identify variations. Each mutation was confirmed by bidirectional sequencing. Mutation descriptions followed the nomenclature recommended by the Human Genomic Variation Society (HGVS) [28].
Table 1. Primers used for amplification and sequencing of PAX6.
| Primer ID | Sequence (5′-3′) | Product length (bp) | Annealing Temperature (°C) |
|---|---|---|---|
| Extra-E1-F* |
GAGCTGTGCCCAACTCTAGC |
|
|
| Extra E1-R |
TCCATCTTTGTATGCCTCCTTT |
399 |
56 |
| Exon1F |
GGAGAGGGAGCATCCAATC |
|
|
| Exon1R |
TCCTGGGAAGGAGACAGAGA |
396 |
56 |
| Exon2F |
ACACACTTGAGCCATCACCA |
|
|
| Exon2R |
CTCCTGCGTGGAAACTTCTC |
467 |
56 |
| Exon3F |
AGAGAGCCCATGGACGTATG |
|
|
| exon3R |
CCCAATCTGTTTCCCCTACA |
318 |
56 |
| Exon4F |
TGCAGCTGCCCGAGGATTA |
|
|
| Exon4R |
GCACCCCGAGCCCGAAGTC |
144 |
66 |
| Exon5F |
TCCCTCTTCTTCCTCTTCACT |
|
|
| Exon5R |
GGGGTCCATAATTAGCATC |
301 |
61 |
| Exon6–7F# |
GCTCTCTACAGTAAGTTCTC |
|
|
| Exon6–7R |
AGGAGAGAGCATTGGGCTTA |
457 |
61 |
| Exon8F |
GATTTGCAGGTGTCATCAAT |
|
|
| Exon8R |
ATATGGAGAGCTGCGTGGAT |
212 |
65 |
| Exon9F |
TTTGGTGAGGCTGTCGGGA |
|
|
| Exon9R |
TCTTTGTACTGAAGATGTGGC |
339 |
58 |
| Exon10F |
GTAGTTCTGGCACAATATGG |
|
|
| Exon10R |
GTACTCTGTACAAGCACCTC |
206 |
62 |
| Exon11–12F |
GGCTCGACGTAGACACAGT |
|
|
| Exon11–12R |
TGCAGACACAGCCAATGAGG |
500 |
62 |
| Exon13F |
GCTGTGTGATGTGTTCCTCA |
|
|
| Exon13R |
AAGAGAGATCGCCTCTGTG |
245 |
62 |
| Exon14F |
CATGTCTGTTTCTCAAAGGG |
|
|
| Exon14R | CCATAGTCACTGACTGAATTAACAC | 202 | 61 |
#Exon numbers are based on the current version of gene structure and NM_001604.4, where the original exon 5a is numbered as exon 6. *This extra exon is based on another transcript variant NM_001127612.1, which is not present in the trancript variant of NM_001604.4.
MLPA analysis
For patients who were determined not to have a PAX6 mutation based on sequencing analysis, MLPA was used to detect deletions of part or all of PAX6, according to the manufacturer’s instructions (SALSA MLPA Kits P219-B1 PAX6; MRC-Holland bv, Amsterdam, the Netherlands) [10]. Briefly, 100 ng DNA samples were denatured for 5 min at 98 °C and then cooled to 25 °C. Probes were mixed and hybridized with DNA samples at 60 °C overnight and were then reacted with ligase 65 at 54 °C for 15 min, followed by 5 min at 98 °C and then held at 4 °C. Finally, all probes and sample ligations were amplified by PCR using specific carboxyfluorescein (FAM) labeled PCR primers. PCR products were separated by electrophoresis using the ABI PRISM 3100 Analyzer. Data analysis was performed using GeneMarker V1.5 software. A peak area between 0.7 and 1.3 times was considered normal; however, peak areas below 0.7 represent deletions and those above 1.3 represent duplications.
Results
Sequencing of the 14 exons of PAX6 of the 27 probands revealed 12 mutations in 13 patients, including five novel deletion/insertion mutations and seven known point mutations, as follows: c.[65_94del30; 99_105dup7] (p.P22LfsX25), c.101_102insA (p.Q34fsx21), c.177delG (R59SfsX19), c.238_239insGCGA (p.S80fsx12), c.1033–42_1033–26del17insG (splice change), c.1A>G (Initiation codon abolished), c.120C>A (p.C40X), c.718C>T (p.R240X), c.949C>T (p.R317X), c.1062C>A (p.Y354X), c.1183G>A (p.G395R), and c.1268A>T (p.X423LeuextX*15; Table 2, Figure 1 and Figure 2). Of the 12 observed mutations, eight resulted in a premature stop codon. The other four were involved in the splicing acceptor, initial codon, or the stop codon. No putative mutations were detected in the other 14 probands. No variants were detected in the three noncoding exons (exons 1~3).
Table 2. Clinical data and PAX6 mutations in the 16 probands.
| ID |
Age (years) |
Gender |
Inheritance |
Visual acuity |
Clinical manifestation# |
Mutations detected in PAX6 |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cornea | Iris | Lens | Exon | Sequence variations | Effect | |||||
| QT183 |
8 |
M |
sporadic |
0.1; 0.1 |
normal |
complete aniridia |
inferior opacity; upward dislocation |
E5 |
c.120C>A |
C40X |
| QT314 |
3/12 |
F |
sporadic |
NA |
normal |
complete aniridia |
transparent |
E9 |
c.718C>T |
R240X |
| QT322 |
12 |
M |
AD |
0.1; 0.1 |
normal |
subtotal aniridia |
punctate opacity |
E5 |
c.65_94del30.c.99_105dup7 |
P22LfsX25 |
| QT346 |
3 |
M |
sporadic |
NA |
inferior leucoma |
complete aniridia |
transparent |
E12 |
c.1183G>A |
G395R |
| QT350 |
4 |
M |
sporadic |
0.1; 0.1 |
normal |
complete aniridia |
transparent |
E9–14 |
E9–14 del. |
One copy deletion |
| QT374 |
7 |
M |
sporadic |
0.1; 0.1 |
normal |
complete aniridia |
transparent |
E10 |
c.949C>T |
R317X |
| QT462 |
4 |
F |
sporadic |
NA |
normal |
complete aniridia |
transparent |
E8–14 |
E8–14 del. |
One copy deletion |
| QT464 |
19 |
M |
AD |
0.2; 0.2 |
normal |
complete aniridia |
punctate opacity |
E5 |
c.101_102insA |
p.H34QfsX21 |
| QT467 |
3 |
F |
AD |
NA |
normal |
subtotal aniridia |
punctate opacity |
E1–14 |
E1–14 del. |
One copy deletion |
| QT468 |
8 |
M |
AD |
0.2; 0.2 |
normal |
circumpupillary aplasia |
punctate opacity |
E6 |
c.238_239dupGCGA |
T80Sfsx12 |
| QT517 |
2/12 |
M |
sporadic |
NA |
normal |
complete aniridia |
transparent |
E6 |
c.177delG |
R59SfsX19 |
| QT522 |
18 |
F |
AD |
0.1; 0.1 |
microcornea |
complete aniridia |
punctate opacity |
E4 |
c.1A>G |
Initiation codon abolished |
| QT572 |
17 |
F |
AD |
0.2; 0.3 |
normal |
complete aniridia |
punctate opacity |
E12 |
c.1033–42_1033–26del17insG |
Splice change |
| QT602 |
14 |
M |
AD |
0.1; 0.1 |
normal |
circumpupillary aplasia |
transparent |
E12 |
c.1062C>A |
Y354X |
| QT609 |
5 |
F |
AD |
0.1; 0.2 |
normal |
complete aniridia |
transparent |
E13 |
c.1268A>T |
X423LeuextX*15 |
| QT634 | 7 | F | AD | 0.1; 0.1 | normal | full iris* | punctate opacity | E13 | c.1268A>T | X423LeuextX*15 |
Note: # Nystagmus and foveal hypoplasia were present in all patients with PAX6 mutations. AD: Autosomal dominant. NA: Not available. *Her father had the same mutation but had complete aniridia.
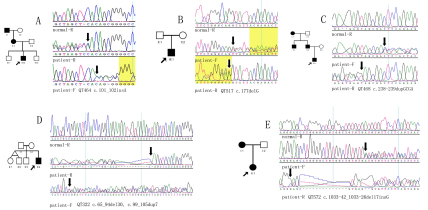
Figure 1.
Frameshift mutations detected in PAX6. Five novel deletion/insertion mutations were identified in five probands with aniridia from unrelated families. Pedigrees (left) are accompanied with sequence chromatography (right). Arrows indicate the probands. R represents reverse sequence.
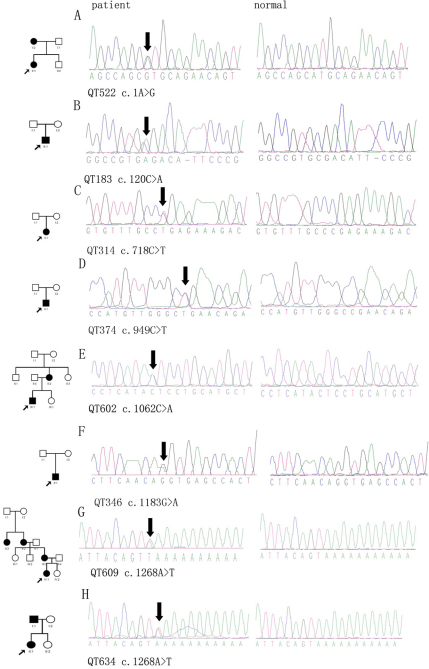
Figure 2.
Point mutations detected in PAX6. One missense and six nonsense mutations in PAX6 were found in eight probands with aniridia from unrelated families. From left to the right, the columns represent pedigrees, sequencing results from probands with aniridia, and corresponding sequences from normal controls.
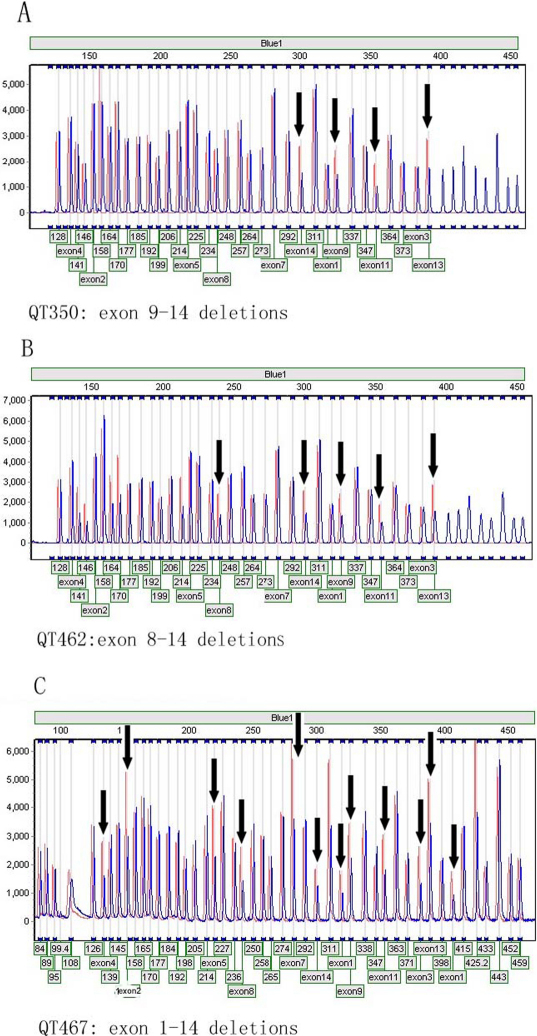
For the 20 probands in whom PAX6 mutations were not detected by sequencing, including the remaining 14 probands from this study and six probands from our previous study [22], we detected PAX6 deletions involving multiple exons (exons 1–14, 8–14, or 9–14, respectively) in three probands detected by MLPA (Table 2, Figure 3).
Figure 3.
PAX6 mutations detected by MLPA. Three gross deletions were involved in exons 9–14, 8–14, and 1–14, respectively. Black arrows indicate the exons with deletions, in which each peak area is below 0.7 compared to internal controls.
Of the 16 probands with PAX6 mutations, all had congenital nystagmus and foveal dysplasia. Complete aniridia, subtotal aniridia, circumpupillary aplasia, or a full iris in both eyes was recorded in probands 11, 2, 2, and 1, respectively (Table 2). One proband (QT634) had a full iris, but his father harbored the same heterozygous PAX6 mutation and had complete aniridia. Additional ocular abnormalities included lens opacities (eight probands), inferior corneal leukoma (one proband with the missense mutation), and microcornea (one proband). The visual acuity of all probands ranged from 0.1 to 0.3, except for five children who were too young to be evaluated (Table 2). Phenotypic variations in the family harboring the c.1A>G mutation were observed: one proband (QT522) had complete absence of the iris in both eyes, although her mother with the same PAX6 mutation only had iris defects similar to iris coloboma (Figure 4). The patient with the missense mutation (c.1183G>A, p.G395R) had typical aniridia and inferior corneal leukoma.
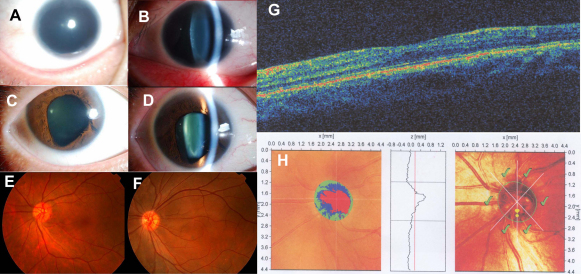
Figure 4.
Photos showing the clinical phenotype of patients with a heterozygous c.1A>G mutation in PAX6. A and B: Complete absence of iris and microcornea (9.5 mm in diameter) was observed in both eyes of the proband, an 18-year-old girl. C and D: A partial defect of the bilateral iris mimicking iris coloboma was present in the proband’s mother, who also had the mutation. E and F: Foveal hypoplasia was observed in the mother (E) and the proband (F). G and H: A flat fovea (G) and optic disc hypoplasia (H) in the mother were demonstrated by optical coherence tomography and Heidelberg retinal tomography, respectively.
Discussion
The mutation frequency of PAX6 in Chinese aniridia patients is similar to that in Caucasian aniridia patients. In this study, PAX6 mutations were identified in 16 of the 33 families tested. When the results of this study are combined with those of our previous study [22], the PAX6 mutations have been identified in 21 of 38 unrelated patients using cycle sequencing and MLPA. Of the 21 patients, mutations in 18 patients were identified by analyzing PAX6 coding regions using direct sequencing, and mutations in 3 patients were detected using MLPA. For PAX6 mutations in Chinese aniridia patients, the overall mutation detection rates detected with cycle sequencing, MLPA, or both were 47% (18/38), 8% (3/38), and 55% (21/38), respectively. In a similar study of Caucasian aniridia patients [10], PAX6 mutations were detected in 49% (34/70), 11% (8/70), and 60% (42/70) patients with cycle sequencing, MLPA, or both, respectively. Several other studies have detected aniridia-associated PAX6 mutations in 30% (9/30) of Mexican patients [29], 56% of Indian patients [30], 67% (4/6) of Thai patients [31], 38%–58% (3/8–14/24) of German patients [32,33], 50% (2/4) of Japanese patients [34], 79% (30/38) of Danish patients [35], and 83%–94% (10/12–67/71) of British patients [9,36]. The detection of these mutations was based solely on sequence analysis in most studies, but chromosomal analysis was additionally performed in a few studies. These reports demonstrate that while PAX6 mutations were prevalent, they were not detected in all patients with aniridia. One reason that may account for this is the possibility that small variations outside the exons, such as intronic regions [10], may not be detectable by the techniques used to analyze PAX6. Furthermore, frequent chromosomal rearrangements have been described in aniridia patients previously [37], which may not be detectable by cycle sequencing and MLPA. It is also possible that there are mutations in other genes which contribute to aniridia given that mutations in FOXC1 are associated with aniridia [17,18] and that no PAX6 mutations were detected in aniridia patients with preserved visual function [19].
The spectrum of PAX6 mutations in aniridia is similar within both Chinese and Caucasian patient cohorts. The majority of PAX6 mutations reported so far would lead to truncation of encoded proteins (such as nonsense, splicing, insertion, or deletion mutation) and only about 2%–11.7% are missense mutations [38,39]. In one review [38], 257 aniridia-associated mutations were classified as nonsense mutations (38.9%), splice mutations (13.2%), frame-shifting insertions or deletions (25.3%), in-frame insertions or deletions (6.2%), missense mutations (11.7%), and run-on mutations (4.7%). For the 21 mutations in the Chinese patients analyzed in the present study, these percentages are 33.3%, 14.3%, 19.0%, zero, 9.5%, and 9.5%, with an additional 14.3% being gross deletions of the PAX6 gene. In addition, seven point mutations detected in this study are known mutations, suggesting common mutations. Of the seven, the p.R240X and p.R317X mutations involving CpG dinucleotides are the most common nonsense mutations in PAX6. The p.C40X mutation was detected in one patient in this study and another patient in our previous study [22]. The run-on mutation, X423LeuextX*15, was detected in two unrelated Chinese patients.
In this study, we detected five novel small insertion/deletion mutations, seven known point mutations, and three known gross deletions in 33 unrelated aniridia patients. In this and one of our previous studies [22], the PAX6 gene was analyzed by cycle sequencing and MLPA in 38 unrelated aniridia patients. However, PAX6 mutations were only detected in 55% (21/38) patients. Further studies of the 17 families without PAX6 mutations may elucidate the molecular basis of aniridia in these families.
Acknowledgments
The authors thank all patients and family members for their participation. We thank Liying Huang and Lijuan Deng for their excellent technical assistance with MLPA. This study was supported by the National Natural Science Foundation of China (30725044, 30800615) and the Fundamental Research Funds of State Key Lab of Ophthalmology, Sun Yat-sen University.
References
- 1.Nelson LB, Spaeth GL, Nowinski TS, Margo CE, Jackson L. Aniridia. A review. Surv Ophthalmol. 1984;28:621–42. doi: 10.1016/0039-6257(84)90184-x. [DOI] [PubMed] [Google Scholar]
- 2.Brauner SC, Walton DS, Chen TC. Aniridia. Int Ophthalmol Clin. 2008;48:79–85. doi: 10.1097/IIO.0b013e318169314b. [DOI] [PubMed] [Google Scholar]
- 3.Ton CC, Hirvonen H, Miwa H, Weil MM, Monaghan P, Jordan T, van Heyningen V, Hastie ND, Meijers-Heijboer H, Drechsler M, Royer-Pokora B, Collins F, Swaroop A, Strong LC, Saunders GF. Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell. 1991;67:1059–74. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 4.Jordan T, Hanson I, Zaletayev D, Hodgson S, Prosser J, Seawright A, Hastie N, van Heyningen V. The human PAX6 gene is mutated in two patients with aniridia. Nat Genet. 1992;1:328–32. doi: 10.1038/ng0892-328. [DOI] [PubMed] [Google Scholar]
- 5.Glaser T, Walton DS, Maas RL. Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nat Genet. 1992;2:232–9. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- 6.Grønskov K, Rosenberg T, Sand A, Brondum-Nielsen K. Mutational analysis of PAX6: 16 novel mutations including 5 missense mutations with a mild aniridia phenotype. Eur J Hum Genet. 1999;7:274–86. doi: 10.1038/sj.ejhg.5200308. [DOI] [PubMed] [Google Scholar]
- 7.Vincent MC, Pujo AL, Olivier D, Calvas P. Screening for PAX6 gene mutations is consistent with haploinsufficiency as the main mechanism leading to various ocular defects. Eur J Hum Genet. 2003;11:163–9. doi: 10.1038/sj.ejhg.5200940. [DOI] [PubMed] [Google Scholar]
- 8.Neethirajan G, Krishnadas SR, Vijayalakshmi P, Shashikant S, Sundaresan P. PAX6 gene variations associated with aniridia in south India. BMC Med Genet. 2004;5:9. doi: 10.1186/1471-2350-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson DO, Howarth RJ, Williamson KA, van Heyningen V, Beal SJ, Crolla JA. Genetic analysis of chromosome 11p13 and the PAX6 gene in a series of 125 cases referred with aniridia. Am J Med Genet A. 2008;146A:558–69. doi: 10.1002/ajmg.a.32209. [DOI] [PubMed] [Google Scholar]
- 10.Redeker EJ, de Visser AS, Bergen AA, Mannens MM. Multiplex ligation-dependent probe amplification (MLPA) enhances the molecular diagnosis of aniridia and related disorders. Mol Vis. 2008;14:836–40. [PMC free article] [PubMed] [Google Scholar]
- 11.Mihelec M, St Heaps L, Flaherty M, Billson F, Rudduck C, Tam PP, Grigg JR, Peters GB, Jamieson RV. Chromosomal rearrangements and novel genes in disorders of eye development, cataract and glaucoma. Twin Res Hum Genet. 2008;11:412–21. doi: 10.1375/twin.11.4.412. [DOI] [PubMed] [Google Scholar]
- 12.Churchill AJ, Yeung A. A compound heterozygous change found in Peters' anomaly. Mol Vis. 2005;11:66–70. [PubMed] [Google Scholar]
- 13.Hanson IM, Fletcher JM, Jordan T, Brown A, Taylor D, Adams RJ, Punnett HH, van Heyningen V. Mutations at the PAX6 locus are found in heterogeneous anterior segment malformations including Peters' anomaly. Nat Genet. 1994;6:168–73. doi: 10.1038/ng0294-168. [DOI] [PubMed] [Google Scholar]
- 14.Dansault A, David G, Schwartz C, Jaliffa C, Vieira V, de la Houssaye G, Bigot K, Catin F, Tattu L, Chopin C, Halimi P, Roche O, Van Regemorter N, Munier F, Schorderet D, Dufier JL, Marsac C, Ricquier D, Menasche M, Penfornis A, Abitbol M. Three new PAX6 mutations including one causing an unusual ophthalmic phenotype associated with neurodevelopmental abnormalities. Mol Vis. 2007;13:511–23. [PMC free article] [PubMed] [Google Scholar]
- 15.Azuma N, Tadokoro K, Asaka A, Yamada M, Yamaguchi Y, Handa H, Matsushima S, Watanabe T, Kohsaka S, Kida Y, Shiraishi T, Ogura T, Shimamura K, Nakafuku M. The Pax6 isoform bearing an alternative spliced exon promotes the development of the neural retinal structure. Hum Mol Genet. 2005;14:735–45. doi: 10.1093/hmg/ddi069. [DOI] [PubMed] [Google Scholar]
- 16.Brown A, McKie M, van Heyningen V, Prosser J. The Human PAX6 Mutation Database. Nucleic Acids Res. 1998;26:259–64. doi: 10.1093/nar/26.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan AO, Aldahmesh MA, Al-Amri A. Heterozygous FOXC1 mutation (M161K) associated with congenital glaucoma and aniridia in an infant and a milder phenotype in her mother. Ophthalmic Genet. 2008;29:67–71. doi: 10.1080/13816810801908152. [DOI] [PubMed] [Google Scholar]
- 18.Ito YA, Footz TK, Berry FB, Mirzayans F, Yu M, Khan AO, Walter MA. Severe molecular defects of a novel FOXC1 W152G mutation result in aniridia. Invest Ophthalmol Vis Sci. 2009;50:3573–9. doi: 10.1167/iovs.08-3032. [DOI] [PubMed] [Google Scholar]
- 19.Traboulsi EI, Ellison J, Sears J, Maumenee IH, Avallone J, Mohney BG. Aniridia with preserved visual function: a report of four cases with no mutations in PAX6. Am J Ophthalmol. 2008;145:760–4. doi: 10.1016/j.ajo.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Song SJ, Liu YZ, Cong RC, Jin Y, Hou ZQ, Ma ZZ, Ren GC, Li LS. Mutation analysis of PAX6 gene in a large Chinese family with aniridia. Chin Med J (Engl) 2005;118:302–6. [PubMed] [Google Scholar]
- 21.Sun DG, Yang JH, Tong Y, Zhao GJ, Ma X. A novel PAX6 mutation (c.1286delC) in the patients with hereditary congenital aniridia. Yi Chuan. 2008;30:1301–6. doi: 10.3724/sp.j.1005.2008.01301. [DOI] [PubMed] [Google Scholar]
- 22.Wang P, Guo X, Jia X, Li S, Xiao X, Zhang Q. Novel mutations of the PAX6 gene identified in Chinese patients with aniridia. Mol Vis. 2006;12:644–8. [PubMed] [Google Scholar]
- 23.Zhu HY, Wu LQ, Pan Q, Liang DS, Long ZG, Dai HP, Xia K, Xia JH. Analysis of PAX6 gene in a Chinese aniridia family. Chin Med J (Engl) 2006;119:1400–2. [PubMed] [Google Scholar]
- 24.Yuan H, Kang Y, Shao Z, Li Y, Yang G, Xu N. Two novel PAX6 mutations identified in northeastern Chinese patients with aniridia. Mol Vis. 2007;13:1555–61. [PubMed] [Google Scholar]
- 25.Baum L, Pang CP, Fan DS, Poon PM, Leung YF, Chua JK, Lam DS. Run-on mutation and three novel nonsense mutations identified in the PAX6 gene in patients with aniridia. Hum Mutat. 1999;14:272–3. doi: 10.1002/(SICI)1098-1004(1999)14:3<272::AID-HUMU21>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 26.Li PC, Yao Q, Ren X, Zhang MC, Li H, Liu JY, Sheng SY, Wang Q, Liu MG. Analysis of PAX6 gene in a Chinese family with congenital aniridia. Zhonghua Yan Ke Za Zhi. 2009;45:931–4. [PubMed] [Google Scholar]
- 27.Cai F, Zhu J, Chen W, Ke T, Wang F, Tu X, Zhang Y, Jin R, Wu X. A novel PAX6 mutation in a large Chinese family with aniridia and congenital cataract. Mol Vis. 2010;16:1141–5. [PMC free article] [PubMed] [Google Scholar]
- 28.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 29.Villarroel CE, Villanueva-Mendoza C, Orozco L, Alcantara-Ortigoza MA, Jimenez DF, Ordaz JC, Gonzalez-del Angel A. Molecular analysis of the PAX6 gene in Mexican patients with congenital aniridia: report of four novel mutations. Mol Vis. 2008;14:1650–8. [PMC free article] [PubMed] [Google Scholar]
- 30.Neethirajan G, Nallathambi J, Krishnadas SR, Vijayalakshmi P, Shashikanth S, Collinson JM, Sundaresan P. Identification of novel mutant PAX6 alleles in Indian cases of familial aniridia. BMC Ophthalmol. 2006;6:28. doi: 10.1186/1471-2415-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atchaneeyasakul LO, Trinavarat A, Dulayajinda D, Kumpornsin K, Thongnoppakhun W, Yenchitsomanus PT, Limwongse C. Novel and de-novo truncating PAX6 mutations and ocular phenotypes in Thai aniridia patients. Ophthalmic Genet. 2006;27:21–7. doi: 10.1080/13816810500481667. [DOI] [PubMed] [Google Scholar]
- 32.Zumkeller W, Orth U, Gal A. Three novel PAX6 mutations in patients with aniridia. Mol Pathol. 2003;56:180–3. doi: 10.1136/mp.56.3.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf MT, Lorenz B, Winterpacht A, Drechsler M, Schumacher V, Royer-Pokora B, Blankenagel A, Zabel B, Wildhardt G. Ten novel mutations found in Aniridia. Hum Mutat. 1998;12:304–13. doi: 10.1002/(SICI)1098-1004(1998)12:5<304::AID-HUMU3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 34.Kondo-Saitoh A, Matsumoto N, Sasaki T, Egashira M, Saitoh A, Yamada K, Niikawa N, Amemiya T. Two nonsense mutations of PAX6 in two Japanese aniridia families: case report and review of the literature. Eur J Ophthalmol. 2000;10:167–72. doi: 10.1177/112067210001000213. [DOI] [PubMed] [Google Scholar]
- 35.Grønskov K, Olsen JH, Sand A, Pedersen W, Carlsen N, Bak Jylling AM, Lyngbye T, Brondum-Nielsen K, Rosenberg T. Population-based risk estimates of Wilms tumor in sporadic aniridia. A comprehensive mutation screening procedure of PAX6 identifies 80% of mutations in aniridia. Hum Genet. 2001;109:11–8. doi: 10.1007/s004390100529. [DOI] [PubMed] [Google Scholar]
- 36.Axton R, Hanson I, Danes S, Sellar G, van Heyningen V, Prosser J. The incidence of PAX6 mutation in patients with simple aniridia: an evaluation of mutation detection in 12 cases. J Med Genet. 1997;34:279–86. doi: 10.1136/jmg.34.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crolla JA, van Heyningen V. Frequent chromosome aberrations revealed by molecular cytogenetic studies in patients with aniridia. Am J Hum Genet. 2002;71:1138–49. doi: 10.1086/344396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tzoulaki I, White IM, Hanson IM. PAX6 mutations: genotype-phenotype correlations. BMC Genet. 2005;6:27. doi: 10.1186/1471-2156-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanson I, Churchill A, Love J, Axton R, Moore T, Clarke M, Meire F, van Heyningen V. Missense mutations in the most ancient residues of the PAX6 paired domain underlie a spectrum of human congenital eye malformations. Hum Mol Genet. 1999;8:165–72. doi: 10.1093/hmg/8.2.165. [DOI] [PubMed] [Google Scholar]