Abstract
Despite the sequence and structural conservation between cryptochromes and photolyases, members of the cryptochrome/photolyase (flavo)protein family, their functions are divergent. Whereas photolyases are DNA repair enzymes that use visible light to lesion-specifically remove UV-induced DNA damage, cryptochromes act as photoreceptors and circadian clock proteins. To address the functional diversity of cryptochromes and photolyases, we investigated the effect of ectopically expressed Arabidopsis thaliana (6-4)PP photolyase and Potorous tridactylus CPD-photolyase (close and distant relatives of mammalian cryptochromes, respectively), on the performance of the mammalian cryptochromes in the mammalian circadian clock. Using photolyase transgenic mice, we show that Potorous CPD-photolyase affects the clock by shortening the period of behavioral rhythms. Furthermore, constitutively expressed CPD-photolyase is shown to reduce the amplitude of circadian oscillations in cultured cells and to inhibit CLOCK/BMAL1 driven transcription by interacting with CLOCK. Importantly, we show that Potorous CPD-photolyase can restore the molecular oscillator in the liver of (clock-deficient) Cry1/Cry2 double knockout mice. These data demonstrate that a photolyase can act as a true cryptochrome. These findings shed new light on the importance of the core structure of mammalian cryptochromes in relation to its function in the circadian clock and contribute to our further understanding of the evolution of the cryptochrome/photolyase protein family.
Introduction
Life is subject to the 24-hour rotation cycle of the earth, which imposes rhythmic changes in light and temperature conditions. In order to anticipate these environmental changes, most organisms have developed a circadian clock with a period of approximately 24 hours that allows them to adjust behavior, physiology and metabolism to the momentum of the day. To keep pace with the day/night cycle, this internal clock needs to be reset every day, using light (the most predictable environmental cue) as the strongest Zeitgeber (German for “time giver” or synchronizer). The mammalian master clock is located in the suprachiasmatic nuclei (SCN) of the hypothalamus, and receives light-induced signals from the retina via the retino-hypothalamic tract [1]. In turn, this master clock sends humoral and neuronal signals that synchronize peripheral oscillators, located in virtually every cell or tissue [2]–[5].
The mammalian cryptochrome proteins (CRY1 and CRY2) belong to the cryptochrome/photolyase family (CPF) of flavoproteins and were initially identified as homologues of photolyase [6], [7]. In view of their strong resemblance to plant cryptochrome proteins, which act as blue light photoreceptors, the mammalian CRY proteins were hypothesized to act as photoreceptors for resetting of the circadian clock [6], [8]. Unexpectedly however, inactivation of the Cry1 and Cry2 genes in the mouse was shown to shorten or lengthen the period length of the circadian clock respectively, whereas in the absence of both genes circadian rhythmicity was completely lost [9]–[11]. This observation, together with the finding that the Cry genes encode the most potent inhibitors of the circadian transcription activator CLOCK/BMAL1 [12], positioned the mammalian CRY proteins at the heart of the circadian core oscillator.
The mammalian circadian clock consists of a molecular oscillator, composed of a set of clock genes that act in transcription-translation-based feedback loops. The CLOCK/BMAL1 heterodimer activates transcription of the Period (Per1, Per2) and Cryptochrome (Cry1, Cry2) clock genes through E-box elements in their promoter. Following synthesis, the PER and CRY proteins will gradually accumulate in the nucleus and ultimately repress CLOCK/BMAL1, and thereby transcription of their own gene [13]–[15]. A second loop is formed by REV-ERBα, which cyclically inhibits RORα-driven transcription of the Bmal1 gene [16]–[19]. Adding to this transcription/translation feedback loop mechanism is a network of post-translational modifications of clock proteins (phosphorylation, (de)acetylation, sumoylation and ubiquitylation) that fine-tune the period length of the circadian oscillator and confer robustness and persistence to the molecular clock [20]–[27].
Photolyases, the other members of the CPF, are DNA repair enzymes that use visible light to lesion-specifically remove ultraviolet light-induced cyclobutane pyrimidine dimers (CPDs) or (6-4) pyrimidine-pyrimidone photoproducts ((6-4)PPs) from the DNA in a reaction called photoreactivation [28], [29]. Placental mammals have lost photolyase genes during evolution and solely rely on nucleotide excision repair for removal of CPDs and (6-4)PPs [30]. Nevertheless, when expressed in the mouse, CPD and (6-4)PP photolyases rapidly remove these UV-induced lesions in a light-dependent manner and protect the animal from sunburn, mutation induction, and skin cancer development [31]–[33].
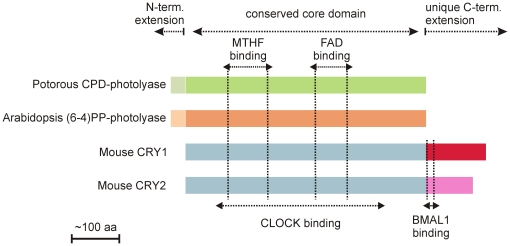
Phylogenetic analysis has shown that the CPF is divided in two major subgroups. The first subgroup encompasses (i) class I CPD photolyases, (ii) (6-4)PP photolyases and animal cryptochromes, (iii) plant cryptochromes, and (iv) DASH cryptochromes, whereas the second subgroup is solely composed of class II CPD photolyases [29]. It is accepted that all members of the photolyase/cryptochrome protein family evolved from a common ancestor CPD photolyase by multiple gene duplications [34]. Cryptochromes and photolyases on the one hand share a common backbone, the core domain, which binds two chromophoric cofactors (i.e. FAD and either 5,10-methenyl-tetrahydrofolate or 8-hydroxy-5-deazaflavin), but on the other hand differ in the presence of N- and C-terminal extensions (see Figure 1). Whereas eukaryotic photolyases have an N-terminal extension, containing nuclear and mitochondrial localization signals, cryptochromes contain a unique C-terminal extension of variable length and amino acid composition. It is currently accepted that the functional diversity among cryptochromes (i.e. photoreceptor, circadian photoreceptor, or core clock protein) is achieved by the diversity of their C-terminal extensions.
Figure 1. Cryptochromes and photolyases.
Schematic representation of PtCPD-PL, At(6-4)PP-PL and mouse CRY1 and 2. Conserved and unique domain are indicated above, chromophore binding site are indicated by vertical doted lines. CLOCK and BMAL1 binding regions in mouse CRY and indicated below.
Detailed structure/function analysis of the C-terminal region of mammalian CRY1 allowed us to identify a putative coiled-coil domain at the beginning of the C-terminal extension as a potential PER and BMAL1 binding site [35]. Deletion of the complete C-terminal extension (aa 471-606 of mouse CRY1) abolished the CLOCK/BMAL1 transcription inhibitory potential of CRY1, Similarly, Green and co-workers have demonstrated that the C-terminal extension of Xenopus laevis CRY proteins is crucial for transcription repression [36]. Interestingly, specific deletion of either the coiled-coil domain (aa 471-493) or the downstream tail region (aa 494-606) of mammalian CRY1 failed to eliminate its ability to inhibit CLOCK/BMAL1-mediated transcription [35], likely because these mutant proteins can still bind to CLOCK via a yet unidentified region of the core domain. This finding lead us to suggest that an interaction between the C-terminal extension and the core domain is mandatory for the clock function of mammalian cryptochromes, possibly by providing structure to the latter [35].
In the present study, we have explored the importance of the core domain of mammalian CRY proteins for core oscillator function by addressing the question to what extent photolyase enzymes affect circadian core oscillator function. Using in vivo (photolyase transgenic mice) and in vitro (cellular clock reporter and CLOCK/BMAL1 transcription assays) approaches, we show that Potorous tridactylus CPD photolyase (hereafter referred to as PtCPD-PL) not only displays cryptochrome-associated functions, but also can replace the CRY proteins in the mammalian circadian core oscillator.
Materials and Methods
Ethics statement
Mice were kept at the Animal Resource Center (Erasmus University Medical Center), which operates in compliance with the European guidelines (European Community 1986) and The Netherlands legislation for the protection of animals used for research, including ethical review. Animal studies at Erasmus University Medical Center were approved by DEC Consult, an independent Animal Ethical Committee (Dutch equivalent of the IACUC) under permit numbers 139-09-02 (EUR1702) 139-09-11 (EUR1760) and 139-09-12 (EUR1761).
Mouse lines and monitoring of circadian behavior
β-actin::At(6-4)PP-PL [33], β-actin::PtCPD-PL [31], and Per2::Luc transgenic mice (generation described below), as well as Cry1 -/-/Cry2 -/- knockout mice [9], all in a C57BL/6J background, were housed under standard conditions and fed ad libitum. All mouse lines were backcrossed at least 11 times to a C57BL/6J background. For the monitoring of locomotor activity rhythms, male mice (12–16 weeks) were individually housed in a light-proof chamber in cages (30×45 cm) equipped with a running wheel (14 cm in diameter) and a sensor system to detect wheel revolutions. Animals were maintained in a cycle of 12 h light (150 lux) and 12 h darkness (LD) or in continuous darkness (DD) in constant ambient temperature with water and food available ad libitum. Voluntary wheel running (wheel revolutions per unit of time) was continuously recorded by an online computer using the ERS program. Activity records were plotted as actograms and the period of locomotor activity was determined by the chi-square method. Unpaired Student's t-tests were used to make statistical comparisons between the different genotypes.
Generation of Per2::Luc transgenic mice
The construct used to generate the Per2::Luc transgenic mice consists of the luciferase gene under control of the mPer2 promoter, cloned in pBS (Figure S1). The primers used to amplify the 4.2 kb mPer2 promoter fragment are indicated in Figure S1. Intronic sequences from the rabbit β-globin locus were included in the expression construct for messenger stability. The expression construct fragment was excised from the plasmid using appropriate restriction enzymes, separated from the vector DNA by agarose gel electrophoresis, isolated from the gel with the GeneClean II kit (Bio101), and further purified using Elutip-D-mini columns (Schleicher and Schuell, Dassel, Germany). The fragment was dissolved in injection buffer (10 mM Tris–HCl pH 7.5, 0.08 mM EDTA) and injected in the pronucleus of fertilized eggs derived from FVB/N intercrosses as described [37]. Animals were backcrossed in a C57BL/6J background. Genotyping was performed by PCR using primers located in the luciferase gene (Figure S1). Annealing was performed at 55°C. DNA derived from transgenic mice rendered a PCR product of 475 bp, whereas no product was detected using DNA from wild type litter mates.
RNA isolation and quantitative PCR
Coronal cryrosections (25 µm) mounted on 1 mm PALM pen-membraneTM slides were rapidly thawed, fixed for 30 seconds in 70% EtOH and immediately stained with haematoxylin for 3 minutes. Following staining sections were rinsed in DEPC treated dH2O and dehydrated by several rinses in 100% EtOH. Laser catapult microdissection (LCM) of the SCN was accomplished using the PALM Microlaser system on freshly prepared sections. Isolated SCN was dissolved immediately in Lysis Buffer (Qiagen) and stored at −80°C for subsequent RNA purification. RNA was purified with the inclusion of ‘on-column’ DNase treatment using the Qiagen RNeasy ‘Micro’ kit according the manufacturers protocol, except that an additional elution was performed in the final step to maximize RNA yield. RNA eluted with RNase-free dH2O was vacuum evaporated for immediate amplification and cDNA generation. Quality of the freshly purified RNA was assayed using the Agilent BioAnalyzer in combination with the RNA ‘Pico’ chip. When intact 18S/28S ribosomal RNA peaks were evident, the sample was considered worthy of assay by Q-PCR.
Amplification was accomplished using the Ovation™ RNA Amplification System V2 according to the manufactures protocols (Nugen Technologies Inc). Efficiency of the amplification was assayed quantitatively by 260/280 nm estimation of cDNA concentration, where a yield of at least 4.8 µg cDNA was deemed sufficient for specific amplification of the RNA template; and qualitatively using the BioAnalyzer with the RNA ‘Nano’ chip to confirm that the majority of unfragmented, amplified cDNA is approximately of 900 bp in length. Quantitative PCR for the determination of At(6-4)PP-PL and PtCPD-PL mRNA levels was performed in triplicate using an iCycler iQ™ Real-Time PCR Detection System (BioRad), SYBR-green and primers [31], [33] generating intron-spanning products of 150-300 bp. Expression levels were normalized to Hprt (hypoxanthine guanine phosphoribosyl transferase) mRNA levels. The generation of specific products was confirmed by melting curve analysis, and primer pairs were tested with a logarithmic dilution of a cDNA mix to generate a linear standard curve, which was used to calculate primer pair efficiencies.
Cell culture and transfection
COS7 [35], NIH3T3 (American Type Culture Collection), and HEK293T (American Type Culture Collection) cells, as well as primary wild type and PtCPD-PL mouse dermal fibroblasts (MDFs) and immortalized Cry1-/-/Cry2-/- MDFs, were cultured in Dulbecco's modified Eagle's medium-F10-Pen/Strep-10% fetal calf serum. To generate MDFs, mice were sacrificed by cervical dislocation, and a small piece of back skin of the mouse was removed and cut into pieces with a razor blade). Skin pieces were washed in ethanol, rinsed in phosphate-buffered saline, and incubated overnight in medium supplemented with 1.6 mg/ml collagenase type II. Single cells were obtained by passing through a cell strainer, and collected by centrifugation for 5 min at 500 g, resuspended in culture medium, and seeded onto a 10 cm dish. MDFs were cultured in a low-oxygen incubator (5% CO2, 3% O2). Transient expression studies were performed by transfecting cells with plasmids using Fugene reagent (Boehringer) according to the manufacturer's instructions. The following pCDNA3-based plasmids (Invitrogen) were used: pcDNA-HA-mCry1, pcDNA-PtCPD-PL, pcDNA-Bmal1 and pcDNA-Clock. pcDNA-PtCPD-PL is based on the construct used to generate the transgenic mice [31]. For luminescence measurements pGl4.11-Bmal1::luciferase (kindly provided by Dr. U. Schibler, Geneva) was used as a reporter.
Real time bioluminescence monitoring
To monitor circadian oscillations in cultured cells in real time, cells were cultured in medium buffered with 25 mM HEPES and containing 0.1 mM luciferin (Sigma). After synchronization of intracellular clocks by treatment of confluent cultures with forskolin (dissolved in 100% ethanol, added to the culture medium at a final concentration of 30 µM), bioluminescence was recorded for 7 days (75 sec measurements at 10 min intervals) with a LumiCycle 32-channel automated luminometer (Actimetrics) placed in a dry, temperature-controlled incubator at 37°C. Data was analyzed with the Actimetrics software and two sample comparisons were done using a Students T-test. Amplitudes were calculated both with Actimetrics software, on base-line subtracted data, and by comparing peak versus trough values for RAW data. Control amplitude was set at 100%. Both methods were comparable.
CLOCK/BMAL1 transcription reporter assay
To determine the capacity of PtCPD-PL to inhibit CLOCK/BMAL1 driven transcription, we used a luciferase reporter assay as previously described [12], [35]. COS7 cells were transfected with 200 ng of the mPer1::luciferase reporter construct and 15 ng of null-Renilla luc, which was used as an internal control. Clock, Bmal1, Cry1 and PtCPD-PL plasmids were added as indicated in the figure legend. The total amount of DNA transfected was kept constant at 2 µg by supplementing with empty pcDNA3.1 vector (Invitrogen). Transcriptional activity was assessed with the Dual-Luciferase 10 Reporter Assay System (Promega) by measuring a ratio of firefly luciferase activity to Renilla luciferase activity in each cellular lysate.
Co-immunoprecipitation experiments
Co-immunoprecipitation studies were performed as described previously [21]. In short, we transiently expressed a PtCPD-PL and either Flag-Bmal1 and/or Flag-Clock in HEK 293T cells and used anti-FLAG antibodies (Sigma) and anti-PtCPD-PL [31] antibodies for the immunoprecipitation and immunoblot analysis step (1∶1000 dilution). As secondary antibody, we used horseradish peroxidase conjugated anti-mouse IgG (DAKO) and anti-rabbit IgG (BioSource) at a 1∶1000 dilution. Chemoluminescence was detected using the ECL system (Pharmacia Biotech).
Hydroporation experiments
Hydrodynamic tail vein injection experiments were performed as described [38]. In brief, a sterile Ringers Solution (0.9% NaCl, 0.3% KCl, 0.13% CaCl2) containing a total of 10 µg plasmid DNA was rapidly injected (8–10 sec) in the tail vein of the mouse under isoflurane anesthesia. This induces uptake of DNA by the liver, which is initially transient and a small proportion will integrate upon regeneration of the liver. Expression of the hydroporated constructs was non-invasively analyzed using an IVIS® Spectrum imaging device (Caliper/Xenogen) (Figure S2), and positive mice were selected for liver isolation and slicing. Animals were sacrificed 24 h after injection (transient expression) and the livers were rapidly removed and placed in ice cold Hank's balanced salt solution supplemented with 50 mM glucose, 4 mM sodium bicarbonate, 10 mM HEPES, 10.000 unit/ml penicillin/10.000 µg/ml streptomycin [5]. Liver slices (200 µm) were prepared using an automated Krumdieck tissue slicer (Alabama R&D). Individual slices were placed on a membrane insert (Millipore) in a 35-mm dish in imaging medium (DMEM supplemented with 0.1 mM luciferin, 2% B27 supplement, 4 mM sodium bicarbonate, 10 mM HEPES, 2.5 ml 10.000 units/ml penicillin/10.000 µg/ml streptomycin). Real time imaging and synchronization were performed as described above. The plasmids used were pGl4.11-Bmal1::luciferase and pcCry1::PtCPD-PL. Cloning and characterization of the Cry1 promoter will be described elsewhere (Saito and van der Horst, unpublished data).
Results
Potorous tridactylus CPD photolyase transgenic mice have a short period circadian clock
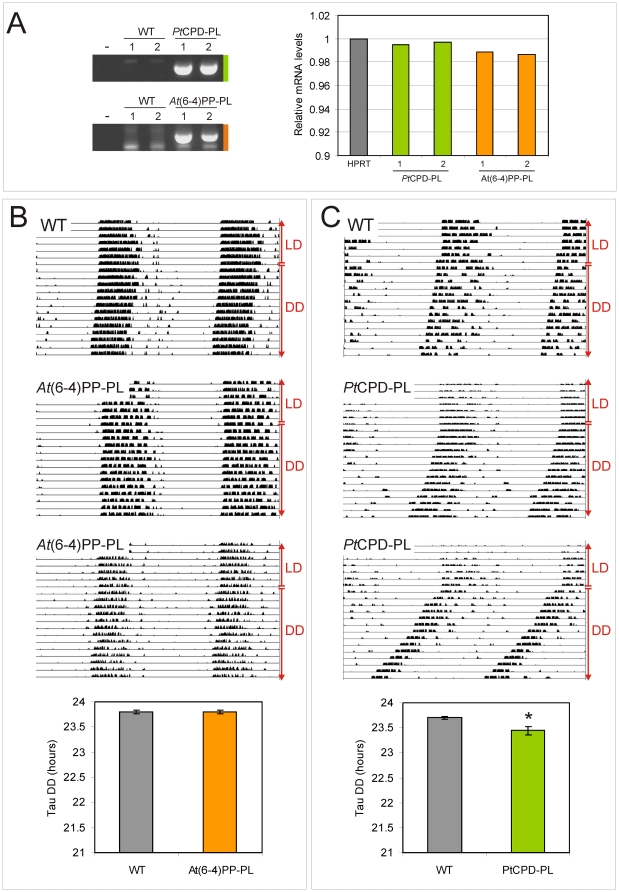
To investigate whether their strong structural resemblance to cryptochromes (see Figure 1) allows photolyases to interfere with mammalian circadian core oscillator function, we took advantage of the availability of β-actin promoter-driven Potorous tridactylus CPD photolyase and Arabidopsis thaliana (6-4)PP photolyase transgenic mice (hereafter referred to as PtCPD-PL and At(6-4)PP-PL mice), previously generated in our laboratory. These animals carry 3 copies of the PtCPD-PL and At(6-4)PP-PL transgene, respectively, and express an active photolyase, capable of removing DNA lesions from the DNA in a light-dependent manner [31], [33]. As shown in Figure 2A, and as could be expected on the basis of the ubiquitous expression of the β-actin promoter, quantitative RT-PCR analysis of mRNA derived from laser microdissected SCN revealed that PtCPD-PL and At(6-4)PP-PL mice express the photolyase transgene in the SCN at comparable levels to the Hprt gene.
Figure 2. Circadian behavior of photolyase transgenic mice.
(A) Quantitative RT-PCR analysis of photolyase mRNA levels in the laser-microdissected SCN of PtCPD-PL and At(6-4)PP-PL transgenic mice. Left panel: Ethidium bromide stained gel of PCR amplified cDNA, obtained from two independent transgenic mice and corresponding wild type littermates (sacrificed at ZT3). Right panel: graphic representation of quantitative RT-PCR amplification data from two independent animals per genotype (see Experimental procedures for details). The Y-axis represents the PtCPD-PL and At(6-4)PP-PL mRNA levels relative to that of Hprt. (B, C) Circadian behavior of At(6-4)PP-PL (B) and PtCPD-PL (C) transgenic mice and corresponding littermates (n = 10 per genotype). Animals were kept under normal light conditions (LD 12∶12 h) and subsequently exposed to constant darkness (DD) (indicated on the right side of the panels). Shown are representative examples of double-plotted actograms and graphic representations of the free-running period (τ) in constant darkness (bottom panels). Error bars represent the standard error of the mean (SEM); the asterisk indicates significance (p = 0.03).
We next addressed the question whether expression of photolyase in the SCN would affect the circadian behavior of the mouse. To this end, we measured circadian wheel-running behavior of photolyase transgenic mice and sex and age-matched control littermates under normal light/dark (LD) cycles and in constant darkness (dark/dark; DD). As shown in Figure 2B, the period length (tau or τ) of circadian behavior of At(6-4)PP-PL transgenic mice is indistinguishable from that of the corresponding wild type littermates. In marked contrast, PtCPD-PL transgenic mice revealed a small but significant shortening of the period length of (15 to 20 min, p<0.05), as compared to wild type littermates (Figure 2C). In addition, the tau of the PtCPD-PL mice has a larger (2-fold) variation than that of control littermates, possibly derived from small individual differences in expression levels.
These findings strongly suggest that expression of Potorous tridactylus CPD photolyase in the SCN interferes with circadian clock performance. In contrast, Arabidopsis thaliana (6-4)PP photolyase does not appear to influence the circadian clock. This observation is in agreement with our finding that At(6-4)PP-PL is not able to inhibit CLOCK/BMAL1 [35]. We therefore further will focus on the effect of PtCPD-PL on circadian rhythms.
Potorous tridactylus CPD photolyase dampens the circadian core oscillator
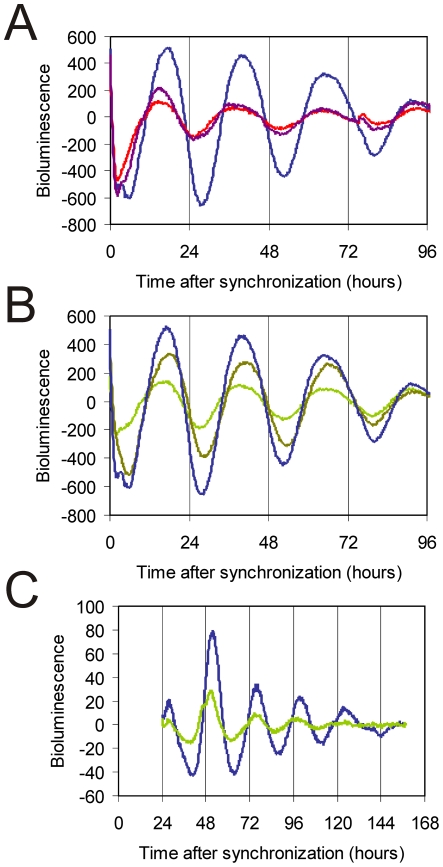
As photolyases structurally resemble cryptochromes [29], [34], and given the observation that transient constitutive overexpression of the CRY1 protein suppresses the rhythmic expression of a cotransfected Bmal1::Luc reporter gene (Figure 3A), we next investigated the effect of transient overexpression of PtCPD-PL on the circadian clock of cultured fibroblasts. After synchronization of the individual intracellular circadian clocks with forskolin [39], cells cotransfected with the Bmal1::Luc reporter construct and empty pcDNA3 vector (used as a negative control) were shown to oscillate with a period of 25.6±0.2 hr (n = 11). Interestingly, overexpression of PtCPD-PL reduces the amplitude of the oscillations in a dose-dependent manner (Figure 3B; Table 1).
Figure 3. PtCPD photolyase dampens circadian oscillations.
(A, B) Representative examples of bioluminescence rhythms in NIH3T3 cells co-transfected with a mBmal1::luciferase reporter construct and either empty pcDNA3 (blue line), pcDNA-Cry1, (100 ng, purple line; 200 ng, red line) or pcDNA-CPD photolyase (200 ng, olive line; 400 ng, green line). Empty pcDNA3 vector was added to correct for the amount of DNA transfected. (C) Representative example of bioluminescence rhythms in primary MDFs, derived from PtCPD photolyase transgenic mice (green line) and wild type littermates (blue line), transiently expressing the Bmal1::luciferase reporter gene. Bioluminescence recordings were started immediately after forskolin synchronization of the individual cellular clocks. The Y-axis represents base line subtracted bioluminescence values.
Table 1.
| NIH 3T3 cells | ||
| Tau (hours) | Amplitude (%) | |
| control | 25.60±0.16 | 100 |
| PtCPD-PL | 25.57±0.16 | 65±7 |
To study the influence of CPD photolyase on core oscillator performance under physiological conditions, PtCPD-PL mice were interbred with Per2::Luc mice to obtain primary CPD-photolyase mouse dermal fibroblast (MDF) lines containing a clock reporter. Bioluminescence rhythms in PtCPD-PL/Per2::Luc MDFs show a reduction in amplitude (38±5%) when compared to those in Per2::luc (control) fibroblasts (Figure 3C; Table 1), thus confirming the data obtained in the transient expression studies. Interestingly, and in line with the animal studies, the period of oscillations in PtCPD-PL/Per2::Luc MDFs (22.7±0.4 hr; n = 4) is approximately 50 min shorter (p<0.05) than that of Per2:Lluc fibroblasts that do not carry the CPD photolyase transgene (23.8±0.2 hr; n = 4).
Taken together, these data demonstrate that the PtCPD-PL exerts a dominant negative effect on the circadian clock by dampening the oscillations and shortening the period length, likely by interfering with CRY mediated functions.
Potorous tridactylus CPD photolyase inhibits CLOCK/BMAL1-driven transcription
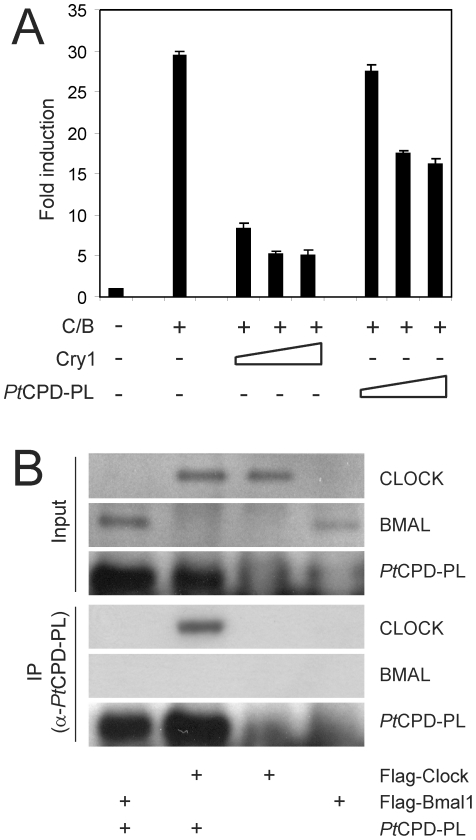
The dominant negative effect of CPD photolyase on cellular clock performance and circadian behavior, as evident from the in vivo and in vitro studies, prompted us to investigate the underlying mode of action. Since CRY proteins are strong inhibitors of the CLOCK/BMAL1 transcription activator [12], we used a COS7 cell based reporter assay to analyze the ability of PtCPD-PL to inhibit CLOCK/BMAL1-driven transcription of the mPer1 promoter-driven luciferase reporter gene. Consistent with previous studies [12], [35] simultaneous expression of CLOCK and BMAL1 causes a 30-fold induction in transcription of the luciferase gene, which is strongly repressed in the presence of CRY1 (Figure 4A). Interestingly, notwithstanding the fact that the protein should be expressed at high level, the PtCPD-PL is also capable of significantly suppressing CLOCK/BMAL1 activity.
Figure 4. PtCPD photolyase represses CLOCK/BMAL1-driven transcription and interacts with CLOCK.
(A) COS7 cell-based CLOCK/BMAL1 transcription assay using a mPer1 E-box promoter-luciferase reporter construct. Luminescence, shown as x-fold induction from the basal expression level (set to 1), is indicated on the Y axis. pcDNA3, pRL-CMV, and the mPer2::luc were added in all reactions. The presence or absence of Cry1 (10-100 ng) and PtCPD-PL (100-300 ng) expression plasmids is indicated below the graph. Empty pcDNA3 vector was added to correct for the amount of DNA transfected. Mean and standard deviation of triplicate samples are shown. (B) Identification of photolyase-binding proteins. PtCPD-PL was precipitated from HEK293T cells, transfected with PtCPD-PL, Flag-Clock or Flag-Bmal1 or double transfected with PtCPD-PL and either Flag-Clock or Flag-Bmal1. Upper panels: Immunoblot analysis of total cell lysates, confirming the presence of the various transiently expressed proteins. Lower panels (IP): Immunoblot analysis of precipitated PtCPD-PL (anti-PtCPD-PL antibodies) and CLOCK and BMAL1 (anti-FLAG antibodies).
From these data we conclude that PtCPD-PL is able to interfere with mammalian core oscillator performance by exerting a cryptochrome-like function, i.e. inhibition of CLOCK/ BMAL1-mediated transcription.
Potorous tridactylus CPD photolyase interacts with CLOCK
We have previously shown that the inhibition of CLOCK/BMAL1 mediated transcription by CRY1 requires a complex network of interactions with CLOCK and BMAL1, involving the CRY1 core domain and C-terminal extension [35] (see also Figure 1). We therefore next asked the question whether PtCPD-PL, like CRY proteins, can physically interact with CLOCK and BMAL1. To this end, we performed a co-immunoprecipitation experiment using HEK293T cells overexpressing the FLAG-CLOCK or FLAG-BMAL1 proteins alone, or in combination with PtCPD-PL (Figure 4B). In the absence of PtCPD-PL neither CLOCK nor BMAL1 were pulled down with anti-PtCPD-PL antibodies, which excludes non-specific binding of the antibodies to these proteins. However, in the presence of PtCPD-PL, the CLOCK protein is shown to co-precipitate with PtCPD-PL, whereas the BMAL1 protein is not. This result indicates that PtCPD photolyase inhibits CLOCK/BMAL1-mediated transcription through direct interaction with CLOCK.
Potorous tridactylus CPD photolyase can replace cryptochromes in the mammalian circadian oscillator
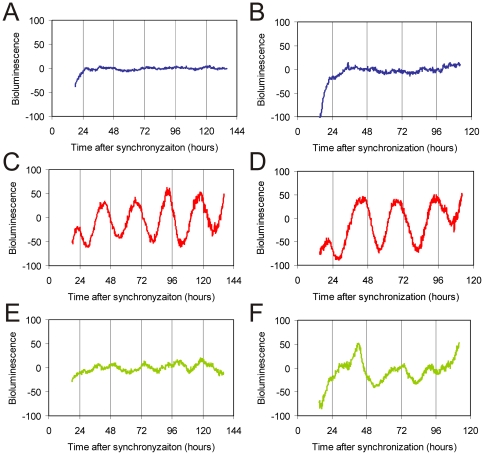
Having shown that PtCPD-PL can interact with CLOCK, leading to inhibition of CLOCK/BMAL1-driven transcription, the intriguing question arises whether this photolyase can actually replace the CRY proteins in the mammalian circadian oscillator and rescue the arrhythmicity of Cry1-/-/Cry2-/- mice. Generation of a new Cry1 promoter-driven PtCPD-PL transgenic mouse line in a Cry-deficient background and subsequent behavioral analysis would be extremely time-consuming. As peripheral circadian clocks form a good model for the master clock in the SCN [40], [41], we chose to generate stable fibroblast lines, derived from Cry1-/-/Cry2-/- mice, transfected with pCry1::PtCPD-PL, pCry1::Cry1 or the empty vector. These cells were then transiently transfected with Bmal1::Luc reporter construct, synchronized with forskolin, and subjected to real time luminescence monitoring. As expected, expression of pCry1::Cry1 (C, D) restores circadian rhythms in arrhythmic Cry1-/-/Cry2-/- fibroblasts (Figure 5C and D) , whereas the empty vector has no effect (Figure 5A and B). Interestingly, pCry1::PtCPD-PLinduces oscillations (Figure 5E and F). Although the period appears in the circadian range, the irregular character of the oscillations precluded calculation of tau.
Figure 5. Correction of the circadian clock in fibroblast lines derived from CRY-deficient mice.
Representative examples of bioluminescence rhythms in immortalized MDF lines, derived from Cry1-/-/Cry2-/- mice, and stably expressing either empty pcDNA3 (A, B), pcCry1::Cry1 (C, D) or pcCry1::PtCPD-PL (E, F), and transfected with the reporter construct. Y-axis represents base line subtracted bioluminescence values.
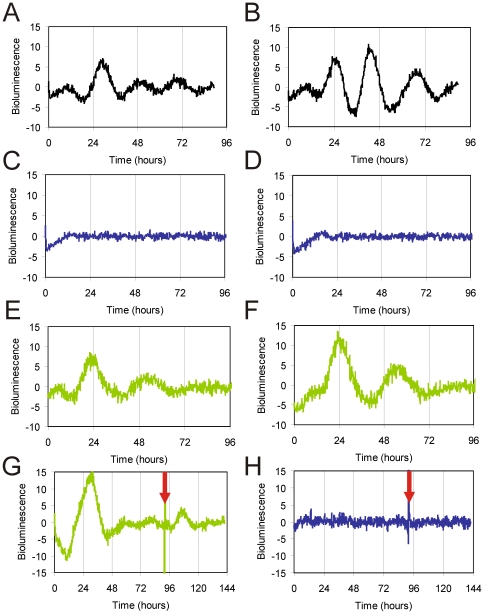
Taking into account the irregular character of the above mentioned oscillations, we took an alternative approach to further to study the capacity of PtCPD-PL to rescue circadian rhythms in tissues. To this end, we used the hydroporation technique [38] to introduce clock reporter and CPD photolyase expression constructs in the mouse liver. Twenty-four hours after co-injection of pGL4.11-Bmal1::Luc and either pCry1::PtCPD-PL or the empty vector in the tail vein of the mouse, liver slices were prepared and clock performance was monitored by real-time imaging of bioluminescence. Co-injection of the Bmal1::Luc reporter plasmid and empty vector in Cry1-/-/Cry2-/- mice and Cry1+/-/Cry2+/- littermate controls resulted in rhythmic expression of the luciferase reporter gene in Cry1+/-/Cry2+/- liver slices (Figure 6A and B), whereas, in line with the absence of a circadian clock, bioluminescence levels remained flat in liver slices from Cry1-/-/Cry2-/- mice (Figure 6C and D), thus validating the hydroporation approach. Interestingly, upon co-injection of the Bmal1::Luc reporter plasmid and pCry1::PtCPD-PL and PtCPD-PL expression construct in Cry1-/-/Cry2-/- animals, we observed a reinitiation of circadian rhythmicity in Cry-deficient liver slices for at least two cycles (Figure 6E and F). This oscillation dampened rapidly, but could be revived for at least one cycle by forskolin treatment of the liver slices (Figure 6G). As in the absence of PtCPD-PL forskolin did not exert any effect on bioluminescence levels in Cry1-/-/Cry2-/- slices (Figure 6H), the forskolin-induced bioluminescence rhythm in PtCPD-PL expressing Cry1-/-/Cry2-/- slices can only be explained by resynchronization of (running) intracellular clocks.
Figure 6. PtCPD photolyase corrects the circadian clock in the liver of CRY-deficient mice.
Representative examples of bioluminescence rhythms in liver slices obtained from mice hydroporated with pGL4.11-Bmal1::Luc (black line), together with either empty pcDNA3 (blue line) or pcDNA-CPD photolyase constructs (green line). (A, B )Liver slices from control mice, injected with the reporter construct only. (C-H) Liver slices from Cry1-/-/Cry2-/- mice injected with either empty pcDNA3 (C, D, H) or pcCry1::PtCPD-PL (E-G), in addition to the reporter construct. In some experiments (G and H) the slices were treated with Forskolin at 96 h to resynchronize the circadian clock. The Y-axis represents base line subtracted bioluminescence values.
From these data we conclude that rhythmically expressed PtCPD photolyase can functionally substitute for CRY proteins in the mammalian circadian oscillator and that such a PtCPD-PL-driven molecular oscillator can still respond to non-photic clock-synchronizing stimuli (i.e. forskolin).
Discussion
In the present study, we analyzed the capacity of two different photolyases to interfere with circadian clock performance: the Class II CPD photolyase from Potorous tridactylus (PtCPD-PL), which is only distantly related to CRY1, and the (6-4)PP photolyase from Arabidopsis thaliana (At(6-4)PP-PL), which is closely related to CRY1. In line with our observation that At(6-4)PP-PL does not inhibit CLOCK/BMAL1 transcriptional activity [35], analysis of the circadian behavior of β-actin promoter-driven At(6-4)PP-PL transgenic mice (ubiquitously expressing the transgene, including the SCN) did not reveal any dominant negative effect of the photolyase on circadian period length. In marked contrast, β-actin promoter-driven PtCPD-PL transgenic mice showed a small but significant reduction of the period length of circadian behavior. In support of this in vivo observation, we also obtained a shortening of the period length of the molecular oscillator in cultured PtCPD-PL dermal fibroblasts. This accelerated clock in PtCPD-PL transgenic mice is unlikely an artifact resulting from unintended inactivation of known (core) clock genes, as analysis of the sequences flanking the integration site of the transgene excluded the presence of such genes (data not shown). Moreover, transiently overexpressed PtCPD-PL was able to dampen the circadian oscillator in NIH3T3 cells. In this respect PtCPD-PL resembles CRY1, which when overexpressed also blunts circadian rhythms. The effect of PtCPD-PL on circadian rhythms is mediated by repression of CLOCK/BMAL1-mediated transcription via direct physical interaction with CLOCK, but not with BMAL1.
Previously, we have shown that removal of the complete C-terminal extension of CRY1 abolishes repressor activity towards CLOCK/BMAL1 driven transcription of E-box containing clock genes and clock-controlled genes [35]. In the same study, we demonstrated that whereas At(6-4)PP-PL by itself has no effect on CLOCK/BMAL1, fusion of the last 100 aa of the CRY1 core domain in conjunction with its C-terminal extension (aa 371-606) to At(6-4)PP-PL) resulted in a chimeric protein which is still able to inhibit CLOCK/BMAL1-mediated transcription. Based on these findings, we hypothesized that acquirement of C-terminal extensions (to the core domain) during evolution functionally separated cryptochromes from photolyase and conferred a clock function to the CRY proteins [35]. We now provide evidence that PtCPD-PL harbors core clock features that allow it to repress CLOCK/BMAL1 transcriptional activity and function as a true cryptochrome. Considering that At(6-4)PP-PL is more homologous to CRY1 than PtCPD-PL, our findings suggest that it is not the primary amino acid sequence per sé, but rather the overall structure of the core domain, that makes a photolyase repressing CLOCK/BMAL1-mediated transcription. Our results indicate that the PtCPD-PL by itself has the proper structure, whereas At(6-4)PP-PL gains such a structure after fusion with a C-terminal extension of mammalian CRY1 [35]. In addition, as PtCPD-PL fails to bind BMAL1, interaction with CLOCK is sufficient to inhibit CLOCK-BMAL1-mediated transcription, which is in complete agreement with our previous observation that the BMAL1-binding coiled-coil domain in the C-terminal extension of CRY1 can be deleted without major consequences [35].
Given the dominant negative effect of constitutive PtCPD-PL expression on circadian behavior of the mouse (in vivo data), it is tempting to speculate on the underlying molecular mechanism by which PtCPD-PL can inhibit CLOCK/BMAL1-driven transcription (in vitro data) and its impact on circadian core oscillator performance. By interacting with CLOCK, PtCPD-PL may prevent the formation of CLOCK/BMAL1 heterodimers and/or binding of the CLOCK/BMAL1 heterodimer to E-box promoters in the DNA. In this scenario, the photolyase reduces the efficiency at which E-box containing clock (controlled) genes are transcribed by reducing the number of available transcription activators. However, as we have shown that a Chrysodeixis chalcites nucleopolyhedroviral photolyase can bind to mammalian CLOCK without affecting CLOCK/BMAL1 transcription potential, binding of a CPF protein per se does not prevent CLOCK/BMAL1 heterodimerization and DNA binding (Biernat and Chaves, submitted for publication). Therefore, a more plausible explanation would be that binding of PtCPD-PL to CLOCK inhibits transcription activation of E-box promoter-bound CLOCK/BMAL1 heterodimers in a cryptochrome like manner. Strikingly, using an in vivo hydroporation approach, we show that PtCPD-PL can rescue the lost circadian oscillator in CRY-deficient cells. When expressed from the Cry1 promoter, PtCPD-PL revived rhythmic expression of the Bmal1::luciferase reporter gene in the Cry1-/-/Cry2-/- mouse liver explants. Moreover, the period of oscillations was in the same range as that of a CRY-driven oscillator and responds to non-photic phase synchronizing stimuli (i.e. forskolin). We therefore conclude that the PtCPD-PL protein has the potential to act as a true mammalian CRY protein.
While this work was in progress, two other members of the CPF have been shown to maintain dual functions: the PtCPF1 protein from the marine diatom Phaeodactylum tricornutum and the OtCPF1 protein from the green algae Ostreococcus tauri. These proteins hold (6-4)PP photolyase activity and (like Potorous CPD-PL) can inhibit CLOCK/BMAL1 driven transcription in a heterologous mammalian system [42], [43] and are therefore considered a missing link in evolution. Interestingly, we found that Arabidopsis thaliana (6-4)PP photolyase does not inhibit CLOCK/BMAL1 [35] or affect circadian behavior, whereas the distantly related Potorous CPD photolyase does. Moreover, we show that this marsupial class II CPD-photolyase can actually substitute for CRY proteins in the mammalian circadian oscillator. In a parallel study, we have shown that the Chrysodeixis chalcites nucleopolyhedrovirus PHR2 protein, another class II CPD photolyase, is able to interact with CLOCK and affect circadian rhythms in vitro (Biernat and Chaves, unpublished data).
These findings contribute to understanding the functional evolution of cryptochromes and photolyases. So far, the identified CPF members with a dual function are either (6-4)PP photolyases from lower eukaryotes [42], [43] or class II CPD photolyases (this manuscript; Biernat and Chaves, submitted for publication). Ancestral CPF members were likely proteins with both DNA repair and circadian clock function. We propose that after the divergence of classes I and II, class II CPD photolyases have kept this dual function throughout evolution. Class I CPD photolyases and (6-4)PP photolyases, however, have lost the circadian function in time, which was taken by cryptochromes. In view of this hypothesis, it will be challenging to study molecular clocks in organisms that have both a photolyase with a dual function and cryptochromes, as is the case of marsupials, such as Potorous and Monodelphis. The genome of Monodelphis domesticus has been sequenced and reveals the presence of cryptochrome genes, as well as photolyase. It will be of interest to determine how the clock of non-placental mammals will respond to the loss of photolyase. On the basis of our data, one would predict a change in tau, suggesting that the circadian clock of placental mammals has adapted to the loss of photolyase by adjusting period. Studying the marsupial circadian system at the cellular and molecular level will answer these questions and shed light on the functional evolution of the CPF.
The present study has identified the Potorous tridactylus CPD photolyase as an attractive candidate for further structure-function studies, aiming at understanding the functional diversity between cryptochromes and photolyases. Analogous to our previous study with mammalian CRY1 - Arabidopsis (6-4)PP photolyase chimeric proteins, it will be informative to swap domains between Potorous CPD photolyase and CRY proteins for functional mapping. Such studies will ultimately reveal how nature uses the same core sequence for completely different functions (e.g photoreactivation by photolyases vs clock function of cryptochromes).
Supporting Information
Schematic representation of the mPer2::Luc construct used to generate transgenic clock reporter mice. (A) The luciferase gene is cloned in front of the mPer2 promoter, using pBS as backbone. Intronic sequences from the rabbit β-globin locus were included in the expression construct for messenger stability. Restriction enzyme sites are indicated. (B) Sequence of the primers used to amplify the 4.2 kb mPer2 promoter fragment. (C) Sequence of the luciferase primers used to genotype mPer2::Luc mice.
(TIF)
Detection of luminescence in the liver of hydroporated mice. Representative examples of dorsal luminescence images, obtained 24 hour after hydroporation of mice with either the Bmal1::Luc reporter construct (left) or the empty vector (right). Expression of the hydroporated constructs was non-invasively monitored in isoflurane anesthetized animals using an IVIS® Spectrum imaging device (Caliper/Xenogen). Colors indicate signal intensity. Note that the reporter is prominently expressed in the liver.
(TIF)
Footnotes
Competing Interests: This work was in part funded by a SenterNovem research grant. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: This work was supported in part by grants from the Netherlands Organization for Scientific Research (ZonMW Vici 918.36.619, NWO-CW 700.51.304, NWO-CW ECHO 700.57.012), SenterNovem (Neuro-Bsik BSIK03053) and the European Community (EUCLOCK LSHG-CT-2006-018741) to GTJvdH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rusak B, Meijer JH, Harrington ME. Hamster circadian rhythms are phase-shifted by electrical stimulation of the geniculo-hypothalamic tract. Brain Res. 1989;493:283–91. doi: 10.1016/0006-8993(89)91163-3. [DOI] [PubMed] [Google Scholar]
- 2.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–37. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 3.McNamara P, Seo SB, Rudic RD, Sehgal A, Chakravarti D, et al. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell. 2001;105:877–89. doi: 10.1016/s0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- 4.Pando MP, Morse D, Cermakian N, Sassone-Corsi P. Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell. 2002;110:107–17. doi: 10.1016/s0092-8674(02)00803-6. [DOI] [PubMed] [Google Scholar]
- 5.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–46. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Todo T, Ryo H, Yamamoto K, Toh H, Inui T, et al. Similarity among the Drosophila (6-4)photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science. 1996;272:109–12. doi: 10.1126/science.272.5258.109. [DOI] [PubMed] [Google Scholar]
- 7.van der Spek PJ, Kobayashi K, Bootsma D, Takao M, Eker AP, et al. Cloning, tissue expression, and mapping of a human photolyase homolog with similarity to plant blue-light receptors. Genomics. 1996;37:177–82. doi: 10.1006/geno.1996.0539. [DOI] [PubMed] [Google Scholar]
- 8.Hsu DS, Zhao X, Zhao S, Kazantsev A, Wang RP, et al. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry. 1996;35:13871–7. doi: 10.1021/bi962209o. [DOI] [PubMed] [Google Scholar]
- 9.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–30. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 10.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96:12114–9. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, et al. Photic induction of mPer1 and mPer2 in Cry-deficient mice lacking a biological clock. Science. 1999;286:2531–4. doi: 10.1126/science.286.5449.2531. [DOI] [PubMed] [Google Scholar]
- 12.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 13.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–76. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 14.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–15. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 15.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–7. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 16.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–60. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 17.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–82. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 18.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–37. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 19.Tsai TY, Choi YS, Ma W, Pomerening JR, Tang C, et al. Robust, tunable biological oscillations from interlinked positive and negative feedback loops. Science. 2008;321:126–9. doi: 10.1126/science.1156951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–3. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 21.Yagita K, Tamanini F, Yasuda M, Hoeijmakers JH, van der Horst GT, et al. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 2002;21:1301–14. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardone L, Hirayama J, Giordano F, Tamaru T, Palvimo JJ, et al. Circadian clock control by SUMOylation of BMAL1. Science. 2005;309:1390–4. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Lee Y, Lee MJ, Park E, Kang SH, et al. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol Cell Biol. 2008;28:6056–65. doi: 10.1128/MCB.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 25.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–48. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 27.Vanselow K, Kramer A. Role of phosphorylation in the mammalian circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:167–76. doi: 10.1101/sqb.2007.72.036. [DOI] [PubMed] [Google Scholar]
- 28.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103:2203–37. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 29.Eker APM, Quayle C, Chaves I, van der Horst GTJ. Direct DNA damage reversal: elegant solutions for nasty problems. Cell Mol Life Sci. 2009;66:968–980. doi: 10.1007/s00018-009-8735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yasui A, Eker AP, Yasuhira S, Yajima H, Kobayashi T, et al. A new class of DNA photolyases present in various organisms including aplacental mammals. EMBO J. 1994;13:6143–51. doi: 10.1002/j.1460-2075.1994.tb06961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schul W, Jans J, Rijksen YM, Klemann KH, Eker AP, et al. Enhanced repair of cyclobutane pyrimidine dimers and improved UV resistance in photolyase transgenic mice. EMBO J. 2002;21:4719–29. doi: 10.1093/emboj/cdf456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jans J, Schul W, Sert YG, Rijksen Y, Rebel H, et al. Powerful skin cancer protection by a CPD-photolyase transgene. Curr Biol. 2005;15:105–15. doi: 10.1016/j.cub.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Jans J, Garinis GA, Schul W, van Oudenaren A, Moorhouse M, et al. Differential role of basal keratinocytes in UV-induced immunosuppression and skin cancer. Mol Cell Biol. 2006;26:8515–26. doi: 10.1128/MCB.00807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanai S, Kikuno R, Toh H, Ryo H, Todo T. Molecular evolution of the photolyase-blue-light photoreceptor family. J Mol Evol. 1997;45:535–48. doi: 10.1007/pl00006258. [DOI] [PubMed] [Google Scholar]
- 35.Chaves I, Yagita K, Barnhoorn S, Okamura H, van der Horst GT, et al. Functional evolution of the photolyase/cryptochrome protein family: importance of the C terminus of mammalian CRY1 for circadian core oscillator performance. Mol Cell Biol. 2006;26:1743–53. doi: 10.1128/MCB.26.5.1743-1753.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Schalie EA, Conte FE, Marz KE, Green CB. Structure/function analysis of Xenopus cryptochromes 1 and 2 reveals differential nuclear localization mechanisms and functional domains important for interaction with and repression of CLOCK-BMAL1. Mol Cell Biol. 2007;27:2120–9. doi: 10.1128/MCB.01638-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hogan B, Beddington R, Costantini F, Lacy E. 217–52. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. Manipulating the Mouse Embryo Section E: Production of Transgenic Mice. [Google Scholar]
- 38.Liu F, Song YK, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Therapy. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 39.Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr. Biol. 2000;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- 40.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 41.Yagita K, Tamanini F, van der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 42.Coesel S, Mangogna M, Ishikawa T, Heijde M, Rogato A, et al. Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Rep. 2009;10:655–61. doi: 10.1038/embor.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heijde M, Zabulon G, Corellou F, Ishikawa T, Brazard J, et al. Characterization of two members of the cryptochrome/photolyase family from Ostreococcus tauri provides insights into the origin and evolution of cryptochromes. Plant Cell Environ. 2010;33:1614–26. doi: 10.1111/j.1365-3040.2010.02168.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of the mPer2::Luc construct used to generate transgenic clock reporter mice. (A) The luciferase gene is cloned in front of the mPer2 promoter, using pBS as backbone. Intronic sequences from the rabbit β-globin locus were included in the expression construct for messenger stability. Restriction enzyme sites are indicated. (B) Sequence of the primers used to amplify the 4.2 kb mPer2 promoter fragment. (C) Sequence of the luciferase primers used to genotype mPer2::Luc mice.
(TIF)
Detection of luminescence in the liver of hydroporated mice. Representative examples of dorsal luminescence images, obtained 24 hour after hydroporation of mice with either the Bmal1::Luc reporter construct (left) or the empty vector (right). Expression of the hydroporated constructs was non-invasively monitored in isoflurane anesthetized animals using an IVIS® Spectrum imaging device (Caliper/Xenogen). Colors indicate signal intensity. Note that the reporter is prominently expressed in the liver.
(TIF)