Abstract
Emerging evidence has implicated G protein-coupled receptors, such as CXCR4 and PAR2, in breast cancer progression and the development of metastatic breast cancer. However, the role of proteins that regulate the function of these receptors, such as arrestins, in breast cancer has yet to be determined. Examination of the expression of the two nonvisual arrestins, arrestin2 and 3, in various breast cancer cell lines revealed comparable expression of arrestin3 in basal and luminal lines while arrestin2 expression was much higher in the luminal lines compared to the more aggressive basal lines. Analysis of normal human breast tissue revealed that arrestin2 and 3 were expressed in both luminal and myoepithelial cells of mammary epithelia with arrestin2 highest in myoepithelial cells and arrestin3 comparable in both cell types. Quantitative immunofluorescence-based examination of primary breast tumors revealed that arrestin2 expression significantly decreased with cancer progression from ductal carcinoma in situ to invasive carcinoma and further to lymph node metastasis (P < 0.001). Moreover, decreased arrestin2 expression was associated with decreased survival (P = 0.0007) as well as positive lymph node status and increased tumor size and nuclear grade. In contrast, arrestin3 expression significantly increased during breast cancer progression (P < 0.001) and increased expression was associated with decreased survival (P = 0.014). Arrestin3 was also an independent prognostic marker of breast cancer with a hazard ratio of 1.65. Overall, these studies demonstrate that arrestin2 levels decrease while arrestin3 levels increase during breast cancer progression and these changes correlate with a poor clinical outcome.
Keywords: Arrestin, Breast carcinoma, Immunohistochemistry, Metastasis, Survival
Introduction
Breast cancer is the second most common and second deadliest cancer diagnosed in women each year [1]. Environmental and genetic factors influence the development of breast cancer causing it to be a disease of heterogeneity in which progression and prognosis differ significantly among individuals [2–4]. Hundreds of genes are thought to be involved in the etiology of breast cancer and it is now well recognized that the gene signature expressed by each tumor can greatly influence its invasive and metastatic potential [4, 5]. Through microarray and immunohistochemical analysis, breast cancer has been categorized into several major groups: luminal, which includes luminal A (ER+, PR−/+, and HER2−) and luminal B (ER+, PR−/+, and HER2+); basal-like or triple negative (ER−, PR−, and HER2−); and HER2+ (ER−, PR−, and HER2+) [2–4, 6]. The genes expressed within these subgroups directly impact clinical outcome, and individuals with HER2+ and basal-like subtypes have the shortest survival time and are more prone to disease recurrence than individuals with luminal subtypes [2, 4, 6]. However, with many genes potentially involved in breast cancer development and progression, the discovery of new prognostic markers to further categorize breast cancer into distinct subgroups will help our understanding and treatment of this complicated disease.
G protein-coupled receptors (GPCRs) comprise the largest and most diverse family of cell surface proteins [7]. GPCRs respond to a variety of stimuli and regulate a range of physiological functions such as sensory and cardiac function, immune response, metabolism, hormone release, and cell growth, survival, and migration. GPCRs also represent the most heavily targeted signaling molecules by the pharmaceutical industry, because alterations in normal function often result in disease [8, 9]. It is now becoming well accepted that GPCRs play a major role in the development and progression of cancer. Cancer cells often utilize normal GPCR function to foster growth, angiogenesis, transformation, and metastatic potential [8]. The three main mechanisms whereby cancers exploit normal GPCR physiology include activating mutations, such as in endocrine tumors, expression of constitutively active GPCRs by viruses, and overexpression of GPCRs, which is most common to breast cancer [8].
Although GPCRs have a clear involvement in cancer, the proteins that regulate GPCR function, such as GPCR kinases and arrestins, have not been well studied in human cancer. The arrestin family is composed of four members that include two visual arrestins (arrestin1 and 4), which are expressed in the retina, and two nonvisual arrestins (arrestin2 and 3 also known as β-arrestin1 and 2, respectively), which are ubiquitously expressed. Arrestins were first discovered for their ability to shut off or ‘arrest’ G protein signaling by binding to agonist-activated, phosphorylated GPCRs, resulting in the uncoupling of the receptor from G protein [10]. However, it is now recognized that the nonvisual arrestins are also multifunctional adaptor proteins and facilitate receptor trafficking via interaction with the endocytic machinery [11] and regulate signaling via interaction with a variety of signaling proteins [12]. Recently, it has been shown that the nonvisual arrestins can also translocate to the nucleus and regulate transcriptional events [13].
A few studies have suggested a potential role for arrestins in the development of cancer. For example, nonvisual arrestins can regulate cell migration through their interaction with proteins such as ERK2 [14], p38 [15], cofilin [16, 17], and PI-3 kinase [18], suggesting that they could have a potential role in metastasis. Indeed, Buchanan et al. [19] used a mouse colorectal cancer model to show that arrestin2 mediates metastasis by the EP4 receptor through Src transactivation of the epidermal growth factor receptor. The nonvisual arrestins may also influence tumor aggressiveness via their ability to act as co-repressors. For example, arrestin3 was shown to function as a co-repressor of the androgen receptor in androgen-dependent prostate cancer, through its ability to scaffold MDM2 and facilitate receptor degradation [20]. Arrestin2 and 3 are also components of the centrosome and loss of arrestin promotes centrosomal abnormalities such as an increase in centrosome number, multinucleation, and microtubule nucleation capacity [21]. Interestingly, it was shown that arrestin overexpression reduced, while arrestin knock-down enhanced, centrosomal defects in MDA-MB-231 breast cancer cells, suggesting the possibility that altered arrestin expression might contribute to centrosomal abnormalities and chromosomal instability often seen in breast cancer [21]. Nonvisual arrestins also appear to foster tumor growth and aggressiveness through the creation of a favorable tumor microenvironment. In a murine lung cancer model, arrestin3 knock-out mice experience enhanced angiogenesis and metastasis because of increased CXCR2 and NFκB activity [22], whereas transgenic mice overexpressing arrestin2 displayed enhanced tumor growth through an increase in angiogenesis because of increased MMP9 activity and VEGF secretion in a murine liver cancer model [23].
Arrestin expression in primary tumors has not been thoroughly investigated. To date, only one study analyzed arrestin expression in primary tumors, specifically in a few prostate cancers [20]. However, these studies did not determine whether changes in arrestin expression correlated with disease progression or were associated with clinical outcome data. In this study, we performed a comprehensive examination of arrestin expression in breast cancer utilizing cell lines and primary human breast tumors. We found that expression of arrestin2 decreased with increasing breast cancer aggressiveness and was associated with positive node status, increased tumor size and nuclear grade, and decreased survival. In contrast, increased arrestin3 expression occurred with increasing breast cancer aggressiveness and was associated with decreased survival. Arrestin3 was also found to be an independent marker for prognosis through multivariate analysis. This is the first study to demonstrate that alterations in arrestin2 and 3 expression are associated with disease progression and clinical outcome and that arrestin3 expression is an independent prognostic marker of breast cancer.
Materials and methods
Cell culture
MDA-MB-231 (a gift from Dr. Tracy Handel, University of California, San Diego), T47D, BT474, and BT549 (ATCC) cells were grown in RPMI supplemented with 10% fetal bovine serum (FBS). MCF7 and MDA-MB-361 cells (ATCC) were grown in Modified Eagle’s Medium supplemented with sodium bicarbonate, nonessential amino acids, sodium pyruvate, and 10% FBS. SKBR3, MDA-MB-436, and HS578t (ATCC) were grown in Dulbecco’s Modified Eagle’s Medium supplemented with 10% FBS.
Western blotting
Samples were run on a 10% SDS polyacrylamide gel, transferred to nitrocellulose, and blocked in 5% nonfat milk in TBS (20 mM Tris–HCl, pH 7.5, 150 mM NaCl) with 0.1% Tween-20. Blots were incubated with arrestin2, arrestin3, e-cadherin, or vimentin antibodies (1:500–1:1,000 dilution) overnight at 4°C, washed with TBS with 0.1% Tween, and then incubated with the appropriate secondary antibody for 1 h at room temperature. Blots were washed and developed with either a West PICO or DURA chemiluminescence assay (Thermo Scientific, Pierce Protein Research Products, Rockford, IL). The loading control used was tubulin, which was detected with the secondary antibody IRDye 800 goat anti-mouse (Rockland Immunochemicals) and the ODYSSEY® infrared imaging system (Li-Cor® Biosciences).
Breast tumor tissues
Progression array
A human breast cancer progression array was constructed using cutting edge matrix assembly (CEMA) [24]. In brief, this array was constructed with human breast tissues from the Thomas Jefferson University Hospital archive from 1970 to 1972 as described [25]. In total, the array comprised 180 deidentified patient tissues of which 40 were unmatched normal breast tissues, 20 ductal carcinoma in situ (DCIS), 100 invasive ductal carcinomas (IDC), and 20 lymph node breast cancer metastases.
Clinical array
A tissue microarray developed at Yale University has been described in previous studies [26]. The array contains 651 human breast cancer tissues from the Yale University Department of Pathology archive from 1961 to 1983. The patient cohort used to generate the array contains approximately half node-positive specimens and half node-negative specimens. Although complete clinical treatment information was not available for this cohort, the node-negative patients were largely treated only by surgical resection while most node-positive patients were treated with local radiation and none received Herceptin. Approximately, 15% of the node-positive patients were treated with chemotherapy (primarily Adriamycin, cytoxan, and 5-fluorouracil) and subsequently ~27% of the cases from 1979 to 1982 received tamoxifen (<5% overall).
Immunohistochemistry
Antibody specificity
Tissue arrays were deparaffinized in xylene and rehydrated in an ethanol gradient. Antigens were retrieved by heating slides in a pressure cooker in sodium citrate buffer (pH 6.0) (Biogenix, San Ramon, CA). Following antigen retrieval, slides were washed in washing buffer (TBS with 0.05% Tween-20; DAKO, Carpinteria, CA) and incubated with a peroxidase-blocking buffer (DAKO) for 15 min at room temperature. Slides were washed in washing buffer and blocked with protein-blocking buffer (normal goat serum, Biogenix) for 20 min at room temperature. Tissues were incubated overnight with the following primary antibodies diluted in antibody dilutent buffer (DAKO): arrestin2 (rabbit polyclonal generated and purified in the lab, 1:1,000) alone or with saturating amounts of glutathione S-transferase (GST) or GST-arrestin2 blocking peptide (BP), and arrestin3 (rabbit polyclonal generated and purified in the lab, 1:500) alone or with saturating amounts of GST or GST-arrestin3 BP. Following overnight incubation, slides were washed in washing buffer and incubated with horseradish peroxidase (HRP) conjugated antirabbit secondary antibodies (DAKO) for 20 min. Slides were washed with washing buffer, incubated with substrate-chromagen solution, 3,3′-diaminobenzidine (DAB) (DAKO) for 5 min, washed in distilled water, and counterstained with hematoxylin for 10 min. Slides were washed in washing buffer followed by distilled water and dehydrated in ethanol gradient and xylene baths. Cover slips were mounted and evaluated for positive staining.
Progression array
Antigen retrieval and blocking of peroxidase activity and nonspecific binding were performed as described above. Slides were incubated overnight with the following primary antibodies diluted in antibody dilutent buffer (DAKO): arrestin2 (1:1,000) and pan Cytokeratin (DAKO, AE1/AE3, mouse, 1:100) for detection of tumor mask. Following overnight incubation, slides were washed in washing buffer and incubated with HRP-conjugated antirabbit antibody (DAKO) for 30 min at room temperature, washed with washing buffer, and incubated with Cy5-tyramide (Perkin Elmer, Waltham, MA). Slides were then incubated with Alexa-Fluor 488-goat anti-mouse (Molecular Probes) for 30 min at room temperature. Slides were washed with washing buffer and mounted with 4′,6-diamidino-2-phenylindole (DAPI)-containing mounting media.
Clinical outcome array and arrestin3 progression array
Slides were deparaffinized and antigens retrieved using a low-pH and a high-pH retrieval for arrestin2 and arrestin3, respectively, with the DAKO PT-module. The DAKO Autolink Plus autostainer was used for staining. Slides were first incubated with a peroxidase-blocking buffer (DAKO FLEX Peroxidase Block) for 10 min and subsequently with protein block for 30 min. To stain for arrestin2, slides were incubated for 20 min with arrestin2 (1:1,000) and pan Cytokeratin (1:100) primary antibodies. To stain for arrestin3, slides were incubated overnight with arrestin3 (1:400) and pan Cytokeratin (1:100) primary antibodies. Following incubation with primary antibodies, slides were washed in DAKO washing buffer to remove unbound primary antibodies and incubated with HRP-conjugated antirabbit antibody (DAKO) and Alexa-Fluor 488 or 555 goat anti-mouse (molecular probes). Slides were washed with washing buffer, incubated with Cy5-tyramide, washed with washing buffer, and mounted with DAPI-containing mounting media.
Automated quantitative analysis
To quantify immunohistochemical immunofluorescence of arrestin2 and 3, the AQUA/PM2000 platform (HistoRx, New Haven, CT) was utilized because it objectively determines intensities [27]. For the progression array, three to six spots were manually selected per tissue while the clinical array slides were automatically scanned for spots. Images were captured for AQUA analysis in the following channels: FITC/Alexa-488/555 (pan cytokeratin), Cy5 (arrestin2 or 3), or DAPI (nuclei). Tumor tissue was identified and tumor mask created using pan cytokeratin positive cells only. Arrestin2 and arrestin3 intensities were measured only in tumor mask (cytokeratin-positive cells) and mean logarithmic signal intensities were reported. It should be noted that AQUA scores are relative values defined by average pixel intensity of target signal within the cytokeratin-positive tumor mask, and are not directly translatable into absolute protein equivalents. All images were validated manually.
Statistical analysis
For the progression array, a one-way ANOVA test was used to determine overall differences in the various breast cancer stages, whereas Tukey’s post hoc test was utilized to determine difference between stages (GraphPad Prism Version 4.0, GraphPad Software, Inc, San Diego, CA). For the clinical array, a one-way ANOVA test was utilized to determine overall differences in tumor size and nuclear grade (SPSS version 16.0, SPSS, Inc, Chicago, IL). Tukey HSD post hoc test was utilized to determine differences between groups. Student’s t test was used for clinical marker comparisons, such as estrogen receptor (ER), progesterone receptor (PR), HER2+, and triple negative status (SPSS version 16.0). Kaplan–Meier curves and adjusted Cox proportional hazards analysis were utilized for survival analysis with an endpoint of breast cancer–specific death (SPSS version 16.0). Cut points for the survival analysis were determined using X-tile software which uses cross-validation to produce P-values for multiple cutpoints [28]. Univariate and multivariate Cox proportional hazards regression models were used to determine the hazard ratio (HR; SPSS version 16.0), and P values <0.05 (*), <0.01 (**), and <0.001 (***) were considered statically significant. Results are reported following the guidelines for the reporting of tumor marker studies (REMARK) [29].
Results
Arrestin2 is differentially expressed in breast cancer cell lines
Previous studies have demonstrated that breast cancer cell lines are representative of the different breast tumor subtypes and that studies carried out with these lines are directly applicable to primary breast tumors [5]. In addition, the profile of genes expressed in breast cancer cell lines directly correlates to invasive potential. For example, basal-like lines migrate efficiently, display a poorly differentiated phenotype, lack substantial cell–cell adhesion and nuclear organization when grown in 3D culture, and exhibit a mesenchymal morphology with invasive projections [5, 30]. In contrast, luminal breast cancer cell lines migrate poorly, are well differentiated, and the majority grow in 3D culture with acceptable adhesion characteristics and nuclear organization [5, 30].
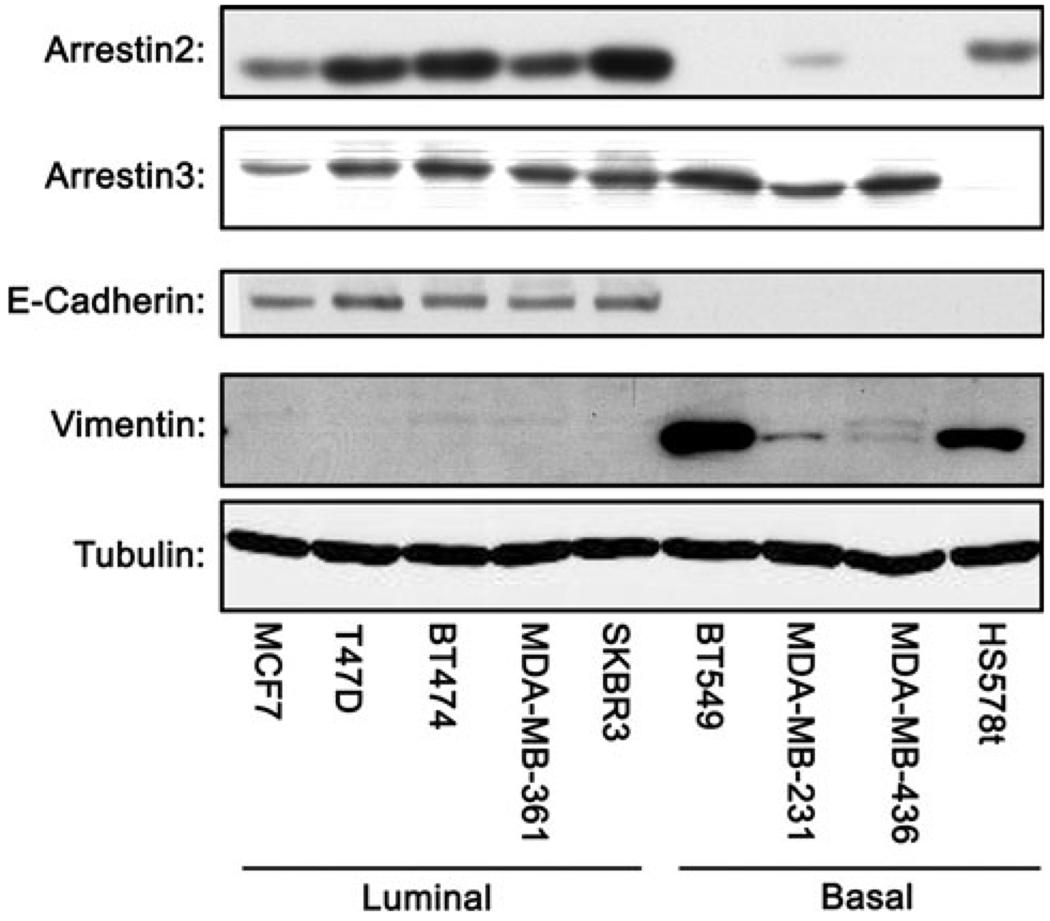
To determine whether arrestin expression varied with cell lines of varying aggressiveness, we analyzed the expression of arrestin2 and 3 using subtype-specific antibodies in five luminal and four basal-like breast cancer cell lines (Fig. 1). Interestingly, we found that although the expression of arrestin3 did not vary substantially between the luminal and basal-like breast cancer lines, the expression pattern of arrestin2 was dramatically different. In general, the luminal lines expressed more arrestin2 than the more aggressive basal-like lines suggesting that a loss of arrestin2 expression may affect parameters associated with invasive cancers, such as de-differentiation and increased metastatic potential. One exception to this expression pattern was the basal-like line HS578t, which had arrestin2 levels similar to the luminal lines as well as very low levels of arrestin3.
Fig. 1.
Arrestin expression in breast cancer cell lines. Using Western blot analysis, arrestin2 and 3 expression was evaluated in five luminal and four basal-like breast cancer cell lines. E-cadherin and vimentin were used as markers of luminal and basal-like breast cancer, respectively. Expression of arrestin3 was similar between the subtypes. However, arrestin2 expression was subtype dependent, with expression greatest in luminal breast cancer cell lines
Expression of arrestin in vivo
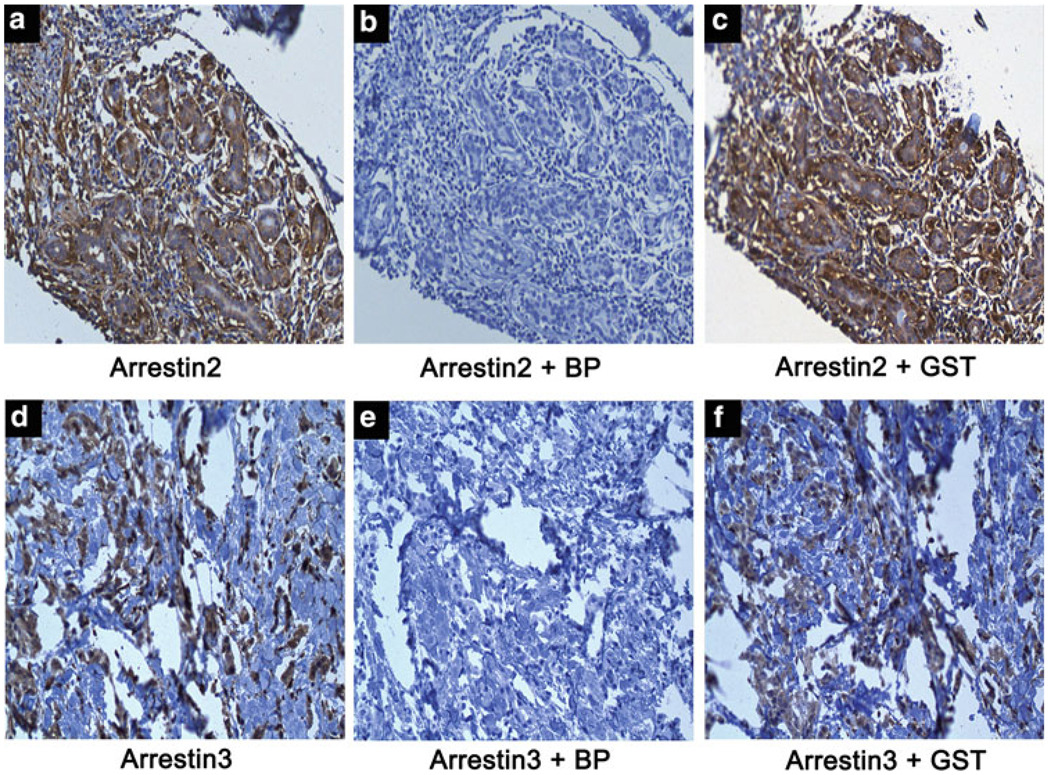
To determine if arrestin expression can be detected in vivo, we stained for arrestin2 and 3 in primary human breast ductal type carcinomas by immunohistochemistry (IHC). Utilization of either the arrestin2 (Fig. 2, panel a) or arrestin3 (Fig. 2, panel d) antibody alone yielded positive staining as denoted by DAB-chromagen reactivity. Incubation of the antibodies with the GST-immunizing blocking peptides effectively blocked arrestin2 (Fig. 2, panel b) and arrestin3 (Fig. 2, panel e) staining, whereas incubation of the antibodies with GST did not affect the staining pattern (Fig. 2, panels c and f). Previous characterization of these antibodies using arrestin2 and arrestin3 depleted cells (using either RNA interference or cells from knock-out animals) validated the specificity of these antibodies [21].
Fig. 2.
Arrestin antibody specificity. Sample breast tumor tissues were used to test arrestin2 and arrestin3 antibody specificity. Incubation with the arrestin2 antibody alone (panel a) or the arrestin3 antibody alone (panel d) yielded specific staining in tumor tissue. Co-incubation of the arrestin2 antibody with the GST-arrestin2 blocking peptide (panel b) and the arrestin3 antibody with the GST-arrestin3 blocking peptide (panel e) completely blocked staining. Co-incubation of either arrestin2 (panel c) or arrestin3 (panel f) antibody with GST yielded the same staining pattern as antibody alone
Arrestin2 and 3 are expressed in the luminal and myoepithelial cells of the normal mammary duct
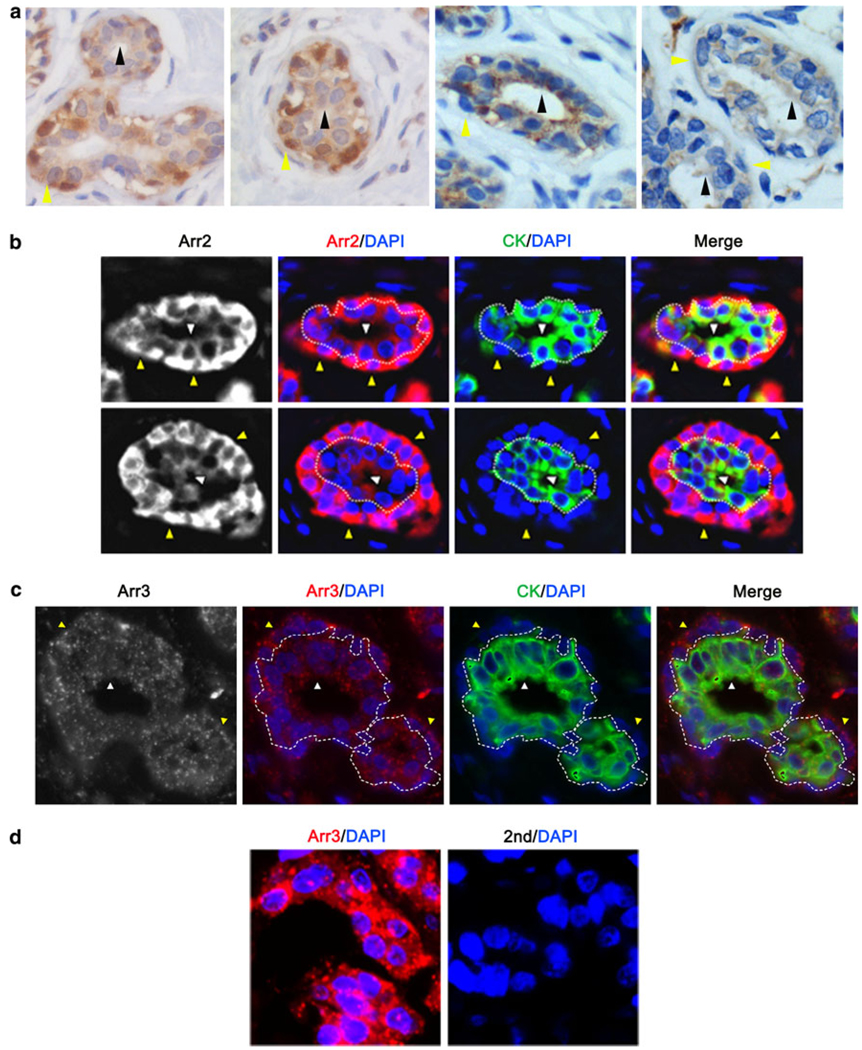
Normal mammary epithelia are composed of two cell types, luminal cells and myoepithelial cells. Luminal cells form the polarized inner layer of the mammary ducts and acini that function in lactating females to produce milk [31]. Myoepithelial cells border the polarized luminal compartment and form contacts with the surrounding stroma where they function as a contractile layer to facilitate movement of milk during lactation and as a mediator of communication between the stroma and luminal cells [31]. Myoepithelial cells are also believed to establish and maintain breast tissue polarity, potentially through the deposition of laminin, and loss of laminin deposition may contribute to de-differentiation and de-polarization of luminal cells during cancer progression [32]. Using the DAB IHC method, we found that arrestin2 was expressed in both cell types of the normal mammary duct (luminal-black arrow and myoepithelial-yellow arrow) with expression being highest in the myoepithelial cells (Fig. 3a, left two panels). Arrestin3 was also expressed in both cell types, albeit at a comparable level (Fig. 3a, right two panels). To confirm these staining patterns, we utilized an immunofluorescent IHC method. Cells positive for cytokeratin (green staining) were identified as luminal cells (Fig. 3b and c, white arrow), whereas cytokeratin-negative cells surrounding the duct were identified as myoepithelial cells (Fig. 3b/c, yellow arrow). A dotted border is also utilized to distinguish myoepithelial and luminal compartments. Similar to the DAB method, arrestin2 (Fig. 3b, white or red) was expressed in both mammary epithelial cell types, with highest expression in myoepithelial cells. In contrast, arrestin3 (Fig. 3c, white or red) had comparable expression in both cell types and also appeared to have a punctate pattern. Staining of breast tumor tissue further demonstrated the specificity of the antibody since the antiarrestin3 produced a punctate pattern of staining, whereas no staining was observed with secondary antibody alone (Fig. 3d).
Fig. 3.
Arrestin is expressed in luminal and myoepithelial cells of the normal mammary duct. a Normal tissue was stained for arrestin2 (left 2 panels) or arrestin3 (right 2 panels) and counterstained with hematoxylin. Arrestin2 is expressed in both cell types, with greatest expression in myoepithelial cells, while arrestin3 is expressed at comparable levels in both cell types (myoepithelial: yellow arrow, luminal: black arrow). b and c Normal tissue was stained through immunofluorescent immunohistochemistry for arrestin2 (Arr2; b) or arrestin3 (Arr3; c) (red), cytokeratin (CK) (green) to identify luminal cells, and DAPI (blue) to identify nuclei. Arrestin2 and arrestin3 are expressed in both myoepithelial (yellow arrow) and luminal cells (white arrow). Arrestin2 displayed greatest expression in myoepithelial cells, whereas arrestin3 was expressed equally in both compartments. d Breast cancer tissue was stained for arrestin3 (red) and DAPI (blue) in the presence (left panel) or absence (right panel) of antiarrestin3
Arrestin expression changes with breast cancer invasiveness
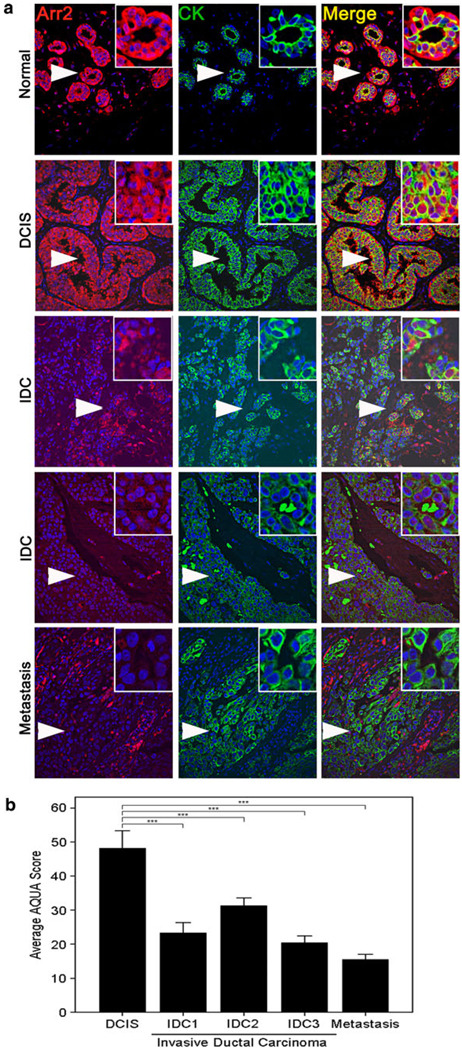
Although a number of studies have alluded to the potential involvement of arrestin in cancer progression, it has yet to be examined whether this could involve changes in arrestin expression. Because the majority of breast cancers are derived from normal luminal epithelial cells, we analyzed the expression of arrestin in luminally derived cancers using automated quantitative analysis. Luminal cancer cells were identified in the tissue sections by staining for cytokeratin (green) and arrestin2 or 3 expression (red), were then analyzed and quantitated in cytokeratin-positive cells. Unfortunately, we were unable to compare arrestin2 expression in normal breast tissue versus breast cancer tissue because we could not accurately quantify arrestin2 expression in normal mammary luminal cells because of its high expression in the bordering myoepithelial cells (Fig. 3a, b). However, we were able to examine the expression of arrestin2 in a progression array of different breast cancer stages and information on arrestin2 expression was obtained for 19 samples of ductal carcinoma in situ (DCIS), 97 invasive (infiltrating) ductal carcinomas (IDC), which included 19 IDC grade 1, 40 IDC grade 2, and 38 IDC grade 3, and 20 lymph node metastases. Interestingly, we found that arrestin2 expression significantly decreased from DCIS to IDC to metastatic lymph node samples (Fig. 4a, b) (ANOVA, P < 0.001). The average expression of arrestin2 was approximately 1.5–2.4-fold higher in DCIS as compared to IDC stages 1–3 (Tukey’s post hoc test, P < 0.001), whereas expression of arrestin2 was 3.1-fold higher in DCIS as compared to metastatic lymph node samples (Tukey’s post hoc test, P < 0.001). These studies reveal that arrestin2 expression decreases with the severity, invasiveness, and progression of breast cancer.
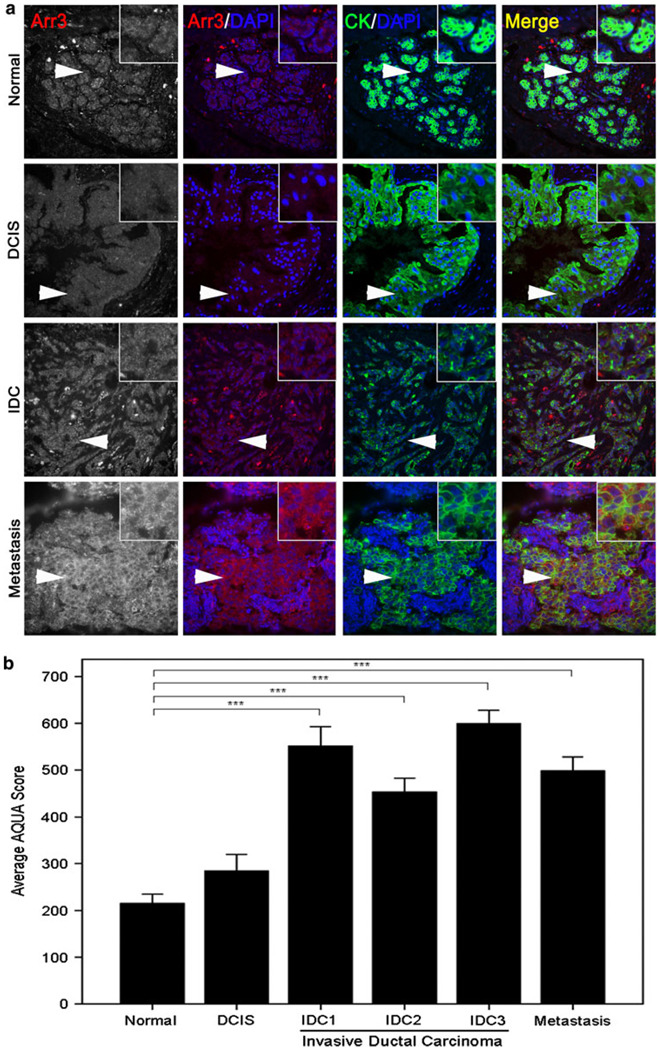
Fig. 4.
Arrestin2 expression decreases with breast cancer progression. a A CEMA breast tumor progression array was stained for arrestin2. About 19 samples of DCIS, 97 samples of IDC, which included 19 cases of IDC grade 1 (IDC1), 40 cases of IDC grade 2 (IDC2), and 38 cases of IDC grade 3 (IDC3), and 20 samples of metastatic lymph nodes were evaluated for arrestin2 expression by AQUA analysis. Representative images of each stage are shown. Arrows denote inset regions. b. Quantitation of arrestin2 intensity in DCIS, IDC, and metastatic lymph nodes. An ANOVA test was utilized to determine statistical significance overall and Tukey’s post hoc test to determine differences between stages, and P < 0.001 (***) were considered statistically significant
Because arrestin3 has comparable expression in myoepithelial and luminal cells, we were able to compare arrestin3 expression in normal and cancer tissue. We obtained information on arrestin3 expression in 35 normal, 16 DCIS, 75 IDC, which included 16 IDC grade 1, 30 IDC grade 2, and 29 IDC grade 3, and 19 metastatic lymph node samples. Interestingly, we found that arrestin3 expression increased with development of invasive breast cancer, specifically with IDC and metastastic lymph nodes (Fig. 5a/b; ANOVA, P < 0.001). Although the 1.3-fold increase in arrestin3 expression was not statistically different between normal and DCIS, statistically significant increases in arrestin3 expression were observed between normal tissue and either IDC or metastatic lymph nodes (Tukey’s post hoc test P < 0.001). A 2.1–2.8-fold increase in arrestin3 expression was seen in IDC grades 1–3 in comparison to normal tissue (Tukey’s post hoc test P < 0.001), whereas an ~2.3-fold increase in arrestin3 expression was observed in mestastatic lymph node samples in comparison to normal tissue (Tukey’s post hoc test P < 0.001). Thus, arrestin3 expression increases with development of invasive breast cancer.
Fig. 5.
Arrestin3 expression increases with breast cancer progression. a A CEMA breast tumor progression array was stained for arrestin3. About 35 samples of normal tissue, 16 samples of DCIS, 75 samples of IDC, which included 16 cases of IDC grade 1 (IDC1), 30 cases of IDC grade 2 (IDC2), and 29 cases of IDC grade 3 (IDC3), and 19 samples of metastatic lymph nodes were evaluated for arrestin3 expression by AQUA analysis. Representative images of each stage are shown. Arrows denote inset regions. b. Quantitation of arrestin3 intensity in normal tissue, DCIS, IDC, and metastatic lymph nodes. An ANOVA test was utilized to determine statistical significance overall and Tukey’s post hoc test to determine differences between stages, and P < 0.001 (***) were considered statistically significant
Arrestin expression is associated with several clinical parameters
We next analyzed a clinical array using quantitative immunofluorescence to determine whether arrestin expression is associated with clinical parameters such as hormone receptor status, tumor size, nuclear grade, and lymph node status. We obtained information on arrestin2 expression in 482 patients. A statistically significant decrease in arrestin2 expression was found to associate with clinical markers, such as ER and triple negative status (Table 1). ER status was available for 473 patients, of which 55% were found to be ER positive. A statistically significant decrease in arrestin2 expression was seen for ER negative patients (P = 0.00045) while no association was seen with PR status. Information on HER2 expression was available for 467 patients, in which 7% were HER2+, and a decrease in arrestin2 expression was seen for the HER2+ subtype, although it was not statistically significant (P = 0.061). Triple negative status was evaluated in 467 patients and 24% were found to be triple negative with arrestin2 expression being reduced in these patients (P = 0.048).
Table 1.
Arrestin2 expression and clinical markers
| Variable | N (%) | Mean intensity Arr2 | Standard error | P value |
|---|---|---|---|---|
| ER status | ||||
| Negative | 214 (45) | 217 | ±3 | 0.00045*** |
| Positive | 259 (55) | 235 | ±4 | |
| PR Status | ||||
| Negative | 223 (48) | 223 | ±4 | 0.178 |
| Positive | 239 (52) | 230 | ±4 | |
| Her2 status | ||||
| Other | 435 (93) | 228 | ±3 | 0.061 |
| HER2 subtype | 32 (7) | 208 | ±10 | |
| Triple negative status | ||||
| ER-/PR-/HER2- | 111 (24) | 217 | ±4 | 0.048* |
| Other | 356 (76) | 230 | ±3 | |
| Lymph node status | ||||
| Negative | 218 (46) | 253 | ±4 | <0.001*** |
| Positive | 258 (54) | 207 | ±3 | |
| Nuclear grade | ||||
| 1 | 69 (16) | 241 | ±7 | 0.004** |
| 2 | 244 (55) | 229 | ±4 | |
| 3 | 130 (29) | 214 | ±4 | |
| Tumor size | ||||
| x<2 cm | 151 (34) | 236 | ±5 | 0.013* |
| 2 cm ≤ x <5 cm | 210 (48) | 230 | ±4 | |
| 5 cm ≤ x | 77 (18) | 212 | ±6 |
P values <0.05
P values <0.01
P values <0.001
Decreased arrestin2 expression also associated with clinical features of increased tumor aggressiveness, such as higher grade (nuclear grade), higher T stage (tumor size), and node-positive status (Table 1). Nuclear grade was used as a read out of tumor grade in this study and 16% of the patients were grade 1, 55% were grade 2, and 29% were grade 3. Tumors of increasing nuclear grade had a statistically significant reduction in arrestin2 between grades 1 and 3 (P = 0.005) and between grades 2 and 3 (P = 0.040). Similar to nuclear grade, tumors of increasing size displayed reduced arrestin2 expression (P = 0.013). Tumor size was grouped according to T staging with <2 cm (T1), 2 to <5 cm (T2), and ≥5 cm (T3) [33]. About 34% of patients were at the T1 stage, 48% were T2, and 18% were T3. T3-stage tumors expressed less arrestin2 than T1 (P = 0.010) while no significant difference in expression was observed between T1 and T2 or T2 and T3. The greatest change in arrestin2 expression was observed in patients who were node negative (218 patients) versus node positive (258 patients) with a decrease in the node-positive patients (P < 0.001).
Data for arrestin3 expression were obtained for 458 patients. We saw no significant association between arrestin3 expression and ER, PR, or triple negative status, although HER2+ patients had a statistically significant decrease in arrestin3 expression (P = 0.014, Table 2). No significant association in arrestin3 expression was observed with tumor size, nuclear grade, or node status (Table 2).
Table 2.
Arrestin3 expression and clinical markers
| Variable | N (%) | Mean intensity Arr3 |
Standard error |
P value |
|---|---|---|---|---|
| ER status | ||||
| Negative | 203 (45) | 1151 | ±25 | 0.092 |
| Positive | 246 (55) | 1207 | ±22 | |
| PR status | ||||
| Negative | 206 (47) | 1203 | ±26 | 0.171 |
| Positive | 234 (53) | 1157 | ±22 | |
| Her2 subtype | ||||
| Other | 409 (93) | 1190 | ±18 | 0.014* |
| HER2 Subtype | 33 (7) | 1033 | ±56 | |
| Triple negative status | ||||
| ER-/PR-/HER2- | 101 (23) | 1227 | ±39 | 0.123 |
| Other | 341 (77) | 1165 | ±19 | |
| Lymph node status | ||||
| Negative | 212 (47) | 1191 | ±23 | 0.771 |
| Positive | 238 (53) | 1181 | ±24 | |
| Nuclear grade | ||||
| 1 | 66 (16) | 1210 | ±45 | 0.493 |
| 2 | 230 (55) | 1170 | ±23 | |
| 3 | 123 (29) | 1212 | ±34 | |
| Tumor size | ||||
| x<2 cm | 141 (34) | 1177 | ±30 | 0.927 |
| 2 cm≤x <5 cm | 195 (47) | 1192 | ±25 | |
| 5 cm ≤ x | 78 (19) | 1192 | ±43 |
P values <0.05
P values <0.01
P values <0.001
Arrestin expression is correlated with survival
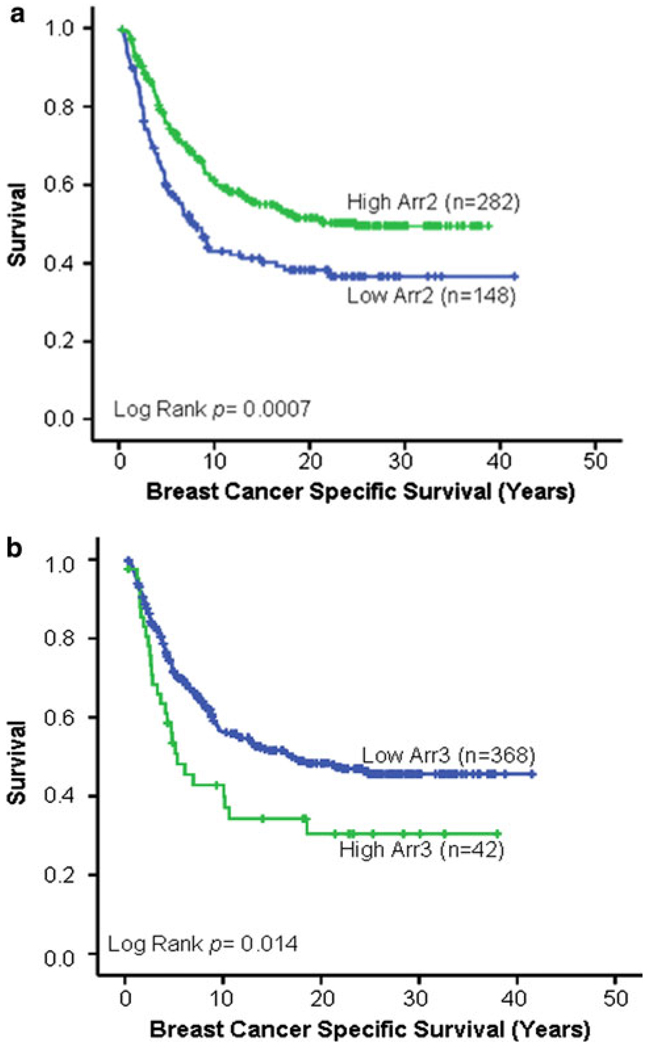
Interestingly, we found that both arrestin2 and arrestin3 expression significantly correlated with patient survival. Table 3 summarizes the number of patients, events, and censored cases evaluated in the univariate and multivariate survival analyses. For arrestin2, survival information was available for 430 patients. Average follow-up time was 12.65 years and median follow-up time was 8.87 years. A cut-point was obtained and validated using X-tile in which patients were grouped into two subcategories: low-arrestin2 expression (148 or 34% of the patients) and high-arrestin2 expression (282 or 66% of the patients; Table 4 and Fig. 6a). Individuals within the low-arrestin2 group expressed an average intensity of 178, whereas individuals within the high-group expressed an average intensity of 254 (Table 4), an ~43% average increase over the low group. Individuals within the low-arrestin2 group survived for the shortest period of time (Fig. 6a; P = 0.0007) with 50% survival of 7.7 years (Table 4). In striking contrast, 50% of individuals within the high-arrestin2 group did not die from the disease until follow-up year 24.8. This finding was supported by univariate analysis, in which a 1.6-fold greater risk of breast cancer-related death was seen for patients expressing low arrestin2 as compared to high arrestin2 (P = 0.001, univariate Cox proportional hazards regression, Table 6).
Table 3.
EMARK diagram summarizing the flow of patients through clinical studies
| Group | Statistical test | N | Death of disease |
Censored |
|---|---|---|---|---|
| Arrestin2 | ||||
| Overall | 430 | 213 | 217 | |
| Arr2 low | Univariate analysis | 148 | 86 | 62 |
| Arr2 high | 282 | 127 | 155 | |
| Overall | 360 | 179 | 181 | |
| Arr2 low | Multivariate analysis | 130 | 76 | 54 |
| Arr2 high | 230 | 103 | 127 | |
| Arrestin3 | ||||
| Overall | 410 | 204 | 206 | |
| Arr3 low | Univariate analysis | 368 | 177 | 191 |
| Arr3 high | 42 | 27 | 15 | |
| Overall | 343 | 173 | 170 | |
| Arr3 low | Multivariate analysis | 306 | 148 | 158 |
| Arr3 high | 37 | 25 | 12 |
Table 4.
Arrestin2 expression in low and high groups
| Arr2 expression | N | Death of disease |
Average intensity of Arr2 |
Intensity of Arr2 range |
50% Survival (Years) |
P value |
|---|---|---|---|---|---|---|
| Low | 148 | 86 | 178 | 119–200 | 7.7 | 0.0007*** |
| High | 282 | 127 | 254 | 200–578 | 24.8 |
P values <0.05
P values <0.01
P values <0.001
Fig. 6.
Arrestin expression is correlated with survival. a Arrestin2 expression information was obtained for 430 patients with survival data by AQUA. Using X-tile analysis, individuals were grouped into either low- (n = 148) or high-arrestin2 (n = 282) expressing groups. Kaplan–Meier survival curves were generated for survival analysis. It was observed that individuals within the low-arrestin2 group experienced poor survival (P = 0.0007). b Arrestin3 expression information was determined for 410 patients with survival data by AQUA. Individuals were grouped into either low-arrestin3 expression (n = 368) or high-arrestin3 expression (n = 42) groups using X-tile analysis. Kaplan–Meier survival curves were generated for survival analysis. It was observed that individuals within the high-arrestin3 groups experienced poor survival (P = 0.014)
Table 6.
Univariate analysis of arrestin2 and arrestin3
| Clinical array |
Univariate analysis |
||||
|---|---|---|---|---|---|
| Variable | n | HR | P | 95% CI | |
| Arr2 | Low | 148 | 1.602 | 0.001*** | 1.218–2.108 |
| High | 282 | – | – | – | |
| Arr3 | Low | 368 | – | – | – |
| High | 42 | 1.650 | 0.016* | 0.404–0.909 | |
P values <0.05
P values <0.01
P values <0.001
For the arrestin3 survival analysis, information was available for 411 patients. Average follow-up time was 12.57 years and median follow-up time was 8.87 years. A cut-point was generated using X-tile and individuals were grouped into either a low-arrestin3 (368 or 90% of the patients) or a high-arrestin3 (42 or 10% of the patients) expressing group (Table 5 and Fig. 6b). Individuals within the low group expressed an average intensity of 1,110 and individuals within the high group expressed an average intensity of 1,873, an ~69% average increase in expression (Table 5). In contrast to arrestin2, increased arrestin3 expression was associated with decreased survival (Fig. 6b) (P = 0.014) with 50% of the individuals in the high group dying by follow-up year 5.3 in comparison to 17.4 years for the low group (Table 5). Univariate analysis supported this observation in which individuals with high-arrestin3 expression had a 1.65-fold greater risk of breast cancer-related death than those in the low-arrestin3 group (P = 0.016, univariate Cox proportional hazards regression, Table 6). These data demonstrate that low-arrestin2 expression or high-arrestin3 expression correlates with breast cancer-specific death.
Table 5.
Arrestin3 expression in low and high groups
| Arr3 expression |
N | Death of disease |
Average intensity of Arr3 |
Intensity of Arr3 range |
50% Survival (Years) |
P value |
|---|---|---|---|---|---|---|
| Low | 368 | 177 | 1110 | 440–1,649 | 17.4 | |
| High | 42 | 27 | 1873 | 1,655–2,429 | 5.3 | 0.014* |
P values <0.05
P values <0.01
P values <0.001
Arrestin3 is an independent breast cancer prognostic marker
To determine whether arrestin2 or arrestin3 is independent prognostic marker of breast cancer, a multivariate analysis was performed. When adjusting for parameters such as age at diagnosis, race, tumor size, tumor grade, ER status, PR status, Her2 subtype, and lymph node status, arrestin2 was not found to be an independent marker (P = 0.520, HR = 1.116, multivariate Cox proportional hazards regression, Table 7). However, when lymph node status was eliminated from the multivariate analysis, arrestin2 was an independent prognostic breast cancer marker (P = 0.039, HR = 1.386, multivariate Cox proportional hazards regression—data not shown). Arrestin3 was found to be an independent prognostic marker in multivariate analysis when adjusting for all parameters (P = 0.027, HR = 1.65, multivariate Cox proportional hazards regression, Table 8). Individuals within the high-arrestin3 group experienced a 1.65-fold greater risk of breast cancer-related death. This is the first demonstration that arrestin3 expression can serve as an independent marker of outcome in individuals with breast cancer.
Table 7.
Arrestin2 multivariate analysis
| Clinical array: Arr2 |
Multivariate analysis |
|||
|---|---|---|---|---|
| Variable | n | HR | P | 95% CI |
| Age at diagnosis | ||||
| <50 | 89 | 1 | – | – |
| ≥50 | 271 | 1.461 | 0.049* | 1.002–2.131 |
| Race | ||||
| Caucasian | 345 | 1 | – | – |
| Other | 15 | 1.400 | 0.309 | 0.732–2.678 |
| Tumor size | ||||
| x<2 cm | 119 | 1 | – | – |
| 2 cm ≤ x < 5 cm | 175 | 1.586 | 0.014* | 1.096–2.294 |
| 5 cm ≤ x | 66 | 2.937 | <0.001*** | 1.927–4.476 |
| Nuclear grade | ||||
| 1 | 54 | 1 | – | – |
| 2 | 201 | 0.928 | 0.741 | 0.598–1.442 |
| 3 | 105 | 1.138 | 0.593 | 0.708–1.828 |
| ER status | ||||
| Negative | 160 | 1 | – | – |
| Positive | 200 | 0.774 | 0.147 | 0.547–1.094 |
| PR status | ||||
| Negative | 176 | 1 | – | – |
| Positive | 184 | 0.985 | 0.926 | 0.713–1.359 |
| Her2 subtype | ||||
| Negative | 334 | 1 | – | – |
| Positive | 26 | 1.457 | 0.230 | 0.788–2.694 |
| Lymph node status | ||||
| Negative | 157 | 1 | – | – |
| Positive | 203 | 1.778 | 0.002** | 1.237–2.554 |
| Arr2 | ||||
| Low | 130 | 1.116 | 0.520 | 0.799–1.559 |
| High | 230 | 1 | – | – |
P values <0.05
P values <0.01
P values <0.001
Table 8.
Arrestin3 multivariate analysis
| Clinical array: Arr3 |
Multivariate adjusted |
|||
|---|---|---|---|---|
| Variable | n | HR | P | 95% CI |
| Age at diagnosis | ||||
| <50 | 88 | 1 | – | – |
| ≥50 | 255 | 1.775 | 0.003** | 1.214–2.595 |
| Race | ||||
| Caucasian | 331 | 1 | – | – |
| Other | 12 | 1.368 | 0.365 | 0.694–2.695 |
| Tumor size | ||||
| X<2 cm | 111 | 1 | – | – |
| 2 cm ≤ x <5 cm | 165 | 1.656 | 0.009** | 1.132–2.422 |
| 5 cm ≤ x | 67 | 3.054 | <0.001*** | 1.989–4.690 |
| Nuclear grade | ||||
| 1 | 50 | 1 | – | – |
| 2 | 191 | 0.866 | 0.541 | 0.547–1.373 |
| 3 | 102 | 1.161 | 0.550 | 0.711–1.898 |
| ER status | ||||
| Negative | 154 | 1 | – | – |
| Positive | 189 | 0.732 | 0.087 | 0.512–1.046 |
| PR status | ||||
| Negative | 163 | 1 | – | – |
| Positive | 180 | 0.932 | 0.680 | 0.665–1.305 |
| Her2 subtype | ||||
| Negative | 317 | 1 | – | – |
| Positive | 26 | 1.816 | 0.049* | 1.004–3.286 |
| Lymph node status | ||||
| Negative | 152 | 1 | – | – |
| Positive | 191 | 1.888 | <0.001*** | 1.343–2.656 |
| Arr3 | ||||
| Low | 306 | 1 | – | – |
| High | 37 | 1.650 | 0.027* | 0.389–0.945 |
P values <0.05
P values <0.01
P values <0.001
Discussion
Because changes in GPCR expression appear to play an important role in the development of some breast cancers, we studied whether proteins that regulate the function of GPCRs, such as arrestins, are also altered in breast cancer. We initially analyzed several breast cancer cell lines of varying aggressiveness and found that arrestin2 expression was significantly reduced in more aggressive basal-like lines as compared to luminal lines, whereas arrestin3 expression did not change significantly between the two subtypes, although its expression was somewhat variable in the basal-like cell lines (Fig. 1). To determine whether arrestin expression changes in vivo, we evaluated primary human breast samples. In normal breast tissue, both arrestins were expressed in the luminal and myoepithelial cells (Fig. 3). Arrestin2 expression was much higher in myoepithelial cells versus luminal cells and may serve as a marker for well-differentiated myoepithelial cells that comprise the normal mammary duct. The high expression of arrestin2 may have relevance to regulation of the oxytocin receptor, a GPCR that is expressed in the brain, ovary, uterus, and testis as well as in mammary myoepithelial cells [34]. Signaling through the oxytocin receptor regulates many critical physiological pathways involved in reproduction and pregnancy, such as differentiation and possibly myoepithelial cell proliferation [35], milk ejection during lactation [36], and development of the postpartum mammary gland [37].
Through the use of two separate breast tumor tissue arrays, a progression CEMA array and a tissue microarray with clinical outcome data, we found that arrestin2 expression was reduced with tumor progression from DCIS to IDC to lymph node metastasis (Fig. 4), whereas arrestin3 expression was increased with the development of invasive breast cancer and remained elevated during metastasis (Fig. 5). Although decreased arrestin2 expression was associated with clinical parameters, such as ER-and triple negative tumors and tumors of increased T stage, increased grade, and positive lymph node status, arrestin3 expression displayed no correlation with clinical parameters or receptor status. Importantly, it was found that the expression pattern of either arrestin correlated with patient survival. Those individuals with reduced arrestin2 expression had a significantly shorter survival compared to individuals within the high-arrestin2 group (Fig. 6a), although arrestin2 expression was not an independent prognostic factor in multivariate analysis, unless node status was eliminated. In contrast, elevated arrestin3 levels were correlated with unfavorable survival in breast cancer patients (Fig. 6b) and arrestin3 expression was found to be an independent prognostic marker by multivariate analysis. If confirmed in independent follow-up studies, more quantitative clinical tests based on arrestin3 levels could be developed with appropriate standardization to help inform physicians and patients about expected outcome.
How might changes in arrestin expression contribute to tumor progression? Based on known arrestin functions, altered arrestin-mediated regulation of cell migration, centrosome function, or MDM2 activity might contribute to the development of cancer. Moreover, although structurally similar, arrestin2 and 3 have a number of distinct functions that could potentially explain their apparent opposite roles in breast cancer progression. For example, arrestin2 is a primary mediator of desensitization for some GPCRs, whereas arrestin3 plays a broader role in MAP kinase signaling. Good examples of this include the angiotensin II type 1A and V2 vasopressin receptors where arrestin3 plays a positive role in ERK1/2 activation while arrestin2 attenuates activation of this pathway [38, 39]. Thus, loss of arrestin2 or overexpression of arrestin3 could ultimately have similar effects on ERK1/2 activation and effects on cell growth.
Arrestin2 and 3 have both been implicated as regulators of cell migration and potentially of metastasis [14, 15, 19, 40]. However, differences exist in how arrestin2 and 3 may regulate migratory and metastatic events, such as through a differential ability to desensitize key receptors involved in migration or the ability to scaffold and regulate proteins involved in cell migration [41]. One example is the CXCL12–CXCR4 signaling axis. CXCL12 activates the chemokine receptor CXCR4, which is overexpressed in 23 different cancers including breast cancer, and plays a role in cancer development through its promotion of cell growth, angiogenesis, and metastasis [42]. While arrestin2 plays a role in endosomal sorting and degradation of CXCR4 [43], arrestin3 plays the predominant role in CXCL12-mediated cell migration. Arrestin3-knockout mice are defective in CXCL12-mediated lymphocyte chemotaxis [44] and knockdown of arrestin3 attenuates CXCL12-dependent activation of p38 and cell migration in HEK293 cells [15]. Similarly, there is also evidence that supports the ability of arrestins to spatially control actin assembly. The PAR2 receptor, which has also been implicated in breast cancer, has served as a particularly useful model for gaining mechanistic insight into arrestin-mediated cell migration. While arrestins have been implicated in regulating PAR2 desensitization, trafficking, and signaling, arrestin2 appears to have a larger role in desensitizing PAR2 signaling and enhancing internalization and degradation, while arrestin3 has a prominent role in cell migration [17, 40, 45]. For example, recent studies suggest that while both arrestin2 and 3 can scaffold cofilin and chronophin and regulate PAR2-mediated actin filament severing and membrane protrusions, arrestin3 plays a larger role in this process [17]. Therefore, both arrestins may promote cell migration and ultimately metastasis through differential mechanisms. Specifically, a loss of arrestin2 expression may lead to increased CXCR4 and PAR2 expression in breast cancer and promote increased signaling and metastatic potential while overexpression of arrestin3 in breast cancer may enhance CXCR4- and/or PAR2-mediated chemotaxis.
Another possible link between arrestins and breast cancer involves their role in regulating centrosome function and genome stability. Chromosomal instability is a driving force for many human cancers and often results from errors that occur during mitosis [46]. The centrosome plays a major role in ensuring mitotic division occurs normally through its role in the nucleation of the bipolar mitotic spindle, cell cycle regulation, and cytokinesis [47]. Alterations in centrosome number and function have been shown to be prevalent in cancer, often resulting in chromosomal instability [48, 49], and a number of studies have demonstrated that centrosomal abnormalities are highly correlated with breast cancer progression [50, 51]. Interestingly, the nonvisual arrestins localize to the centrosome and positively regulate centrosome function [21, 52]. Since loss of arrestin leads to centrosomal abnormalities similar to those observed in breast cancer [21], a decrease in arrestin2 expression in breast cancer may promote the accumulation of centrosomal abnormalities that drive tumor aggression. In support of this hypothesis, overexpression of arrestin2 in the breast cancer cell line MDA-MB-231, which displays high centrosomal abnormalities [50] and express low levels of arrestin2 (Fig. 1), partially rescued the centrosomal defect of multi-nucleation [21].
Nonvisual arrestins might also influence breast cancer progression through an ability to interact with the ring finger E3 ubiquitin ligase MDM2 [53]. MDM2 ubiquitinates and promotes proteasomal degradation of p53 and inhibits p53’s tumor suppressor function [54]. In vivo studies have shown that overexpression of MDM2 can drive tumor development possibly through p53 dependent and independent mechanisms [55, 56]. Because the interaction of arrestins with MDM2 potentially inhibits p53 degradation [53], a loss of arrestin2 in cancer might lead to enhanced p53 degradation and loss of its tumor suppressor function. Interaction of arrestin2 with MDM2 also mediates the ubiquitination and degradation of the insulin-like growth factor-1 receptor (IGF-1R) [57]. The IGF-1R plays an important role in cancer development through its ability to promote proliferation, transformation, invasive/metastatic potential, and protection from apoptosis [58] and in vitro studies have shown that IGF-1R signaling drives oncogenic activity in breast cancer [59]. It has also been demonstrated that IGF-1R is overexpressed in human breast tumors [60, 61] and may exhibit enhanced activity [62]. A decrease in arrestin2 expression in breast tumors might attenuate IGF-1R ubiquitination and degradation leading to enhanced mitogenic and antiapoptotic activity and invasive/metastatic potential.
Overall, we conclude that a loss in arrestin2 or increase in arrestin3 correlates with a more aggressive and advanced form of breast cancer. Further studies will be needed to determine the mechanisms involved in mediating changes in arrestin expression and to determine whether differential changes in arrestin directly affect breast tumor invasiveness and aggressiveness.
Aknowledgements
We would like to thank Dr. Catherine Moore for helping to initiate some of the studies in the breast cancer cell lines and Shashi Rattan for excellent technical assistance. This work was partially supported by National Institutes of Health grants R01 CA129626 and R01 GM047417.
Abbreviations
- AQUA
Automated quantitative analysis
- BP
Blocking peptide
- CEMA
Cutting edge matrix assembly
- DAB
3,3′-Diaminobenzidine
- DAPI
4′,6-Diamidino-2-phenylindole
- DCIS
Ductal carcinoma in situ
- ER
Estrogen receptor
- ERK2
Extracellular signal-regulated kinase 2
- FBS
Fetal bovine serum
- GPCR
G protein-coupled receptor
- GST
Glutathione S-Transferase
- HER2+
Human epidermal growth factor receptor 2
- HR
Hazard ratio
- HRP
Horseradish peroxidase
- IDC
Invasive ductal carcinoma
- IGF-1R
Insulin-like growth factor 1 receptor
- IHC
Immunohistochemistry
- PR
Progesterone receptor
- TBS
Tris buffered saline
Footnotes
The authors declare that there are no conflicts-of-interest.
Contributor Information
Allison M. Michal, Department of Biochemistry and Molecular Biology, Thomas Jefferson University, 233 South 10th Street, Philadelphia, PA 19107, USA
Amy R. Peck, Department of Cancer Biology, Kimmel Cancer Center, Thomas Jefferson University, Philadelphia, PA 19107, USA
Thai H. Tran, Department of Cancer Biology, Kimmel Cancer Center, Thomas Jefferson University, Philadelphia, PA 19107, USA
Chengbao Liu, Department of Cancer Biology, Kimmel Cancer Center, Thomas Jefferson University, Philadelphia, PA 19107, USA.
David L. Rimm, Department of Pathology, Yale University School of Medicine, New Haven, CT 06520, USA
Hallgeir Rui, Department of Cancer Biology, Kimmel Cancer Center, Thomas Jefferson University, Philadelphia, PA 19107, USA.
Jeffrey L. Benovic, Email: benovic@mail.jci.tju.edu, Department of Biochemistry and Molecular Biology, Thomas Jefferson University, 233 South 10th Street, Philadelphia, PA 19107, USA.
References
- 1.American Cancer Society. Breast cancer facts & figures 2009–2010. Atlanta, GA: American Cancer Society Inc.; 2009. [Google Scholar]
- 2.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, Deming SL, Geradts J, Cheang MC, Nielsen TO, Moorman PG, Earp HS, Millikan RC. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 4.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 5.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luttrell LM. Transmembrane signaling by G protein-coupled receptors. Methods Mol Biol. 2006;332:3–49. doi: 10.1385/1-59745-048-0:1. [DOI] [PubMed] [Google Scholar]
- 8.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 9.Lefkowitz RJ. Historical review: a brief history and personal retrospective of seven-transmembrane receptors. Trends Pharmacol Sci. 2004;25:413–422. doi: 10.1016/j.tips.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Krupnick JG, Benovic JL. The role of receptor kinases and arrestins in G protein-coupled receptor regulation. Annu Rev Pharmacol Toxicol. 1998;38:289–319. doi: 10.1146/annurev.pharmtox.38.1.289. [DOI] [PubMed] [Google Scholar]
- 11.Moore CA, Milano SK, Benovic JL. Regulation of receptor trafficking by GRKs and arrestins. Annu Rev Physiol. 2007;69:451–482. doi: 10.1146/annurev.physiol.69.022405.154712. [DOI] [PubMed] [Google Scholar]
- 12.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Beta-arrestins and cell signaling. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 13.Kang J, Shi Y, Xiang B, Qu B, Su W, Zhu M, Zhang M, Bao G, Wang F, Zhang X, Yang R, Fan F, Chen X, Pei G, Ma L. A nuclear function of beta-arrestin1 in GPCR signaling: regulation of histone acetylation and gene transcription. Cell. 2005;123:833–847. doi: 10.1016/j.cell.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 14.Ge L, Ly Y, Hollenberg M, DeFea K. A beta-arrestin-dependent scaffold is associated with prolonged MAPK activation in pseudopodia during protease-activated receptor-2-induced chemotaxis. J Biol Chem. 2003;278:34418–34426. doi: 10.1074/jbc.M300573200. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y, Cheng Z, Ma L, Pei G. Beta-arrestin2 is critically involved in CXCR4-mediated chemotaxis, and this is mediated by its enhancement of p38 MAPK activation. J Biol Chem. 2002;277:49212–49219. doi: 10.1074/jbc.M207294200. [DOI] [PubMed] [Google Scholar]
- 16.Zoudilova M, Kumar P, Ge L, Wang P, Bokoch GM, DeFea KA. Beta-arrestin-dependent regulation of the cofilin pathway downstream of protease-activated receptor-2. J Biol Chem. 2007;282:20634–20646. doi: 10.1074/jbc.M701391200. [DOI] [PubMed] [Google Scholar]
- 17.Zoudilova M, Min J, Richards HL, Carter D, Huang T, DeFea KA. beta-Arrestins scaffold cofilin with chronophin to direct localized actin filament severing and membrane protrusions downstream of protease-activated receptor-2. J Biol Chem. 2010;285:14318–14329. doi: 10.1074/jbc.M109.055806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P, DeFea KA. Protease-activated receptor-2 simultaneously directs beta-arrestin-1-dependent inhibition and Galphaq-dependent activation of phosphatidylinositol 3-kinase. Biochemistry. 2006;45:9374–9385. doi: 10.1021/bi0602617. [DOI] [PubMed] [Google Scholar]
- 19.Buchanan FG, Gorden DL, Matta P, Shi Q, Matrisian LM, DuBois RN. Role of beta-arrestin 1 in the metastatic progression of colorectal cancer. Proc Natl Acad Sci USA. 2006;103:1492–1497. doi: 10.1073/pnas.0510562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakshmikanthan V, Zou L, Kim JI, Michal A, Nie Z, Messias NC, Benovic JL, Daaka Y. Identification of betaArrestin2 as a corepressor of androgen receptor signaling in prostate cancer. Proc Natl Acad Sci USA. 2009;106:9379–9384. doi: 10.1073/pnas.0900258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shankar H, Michal A, Kern RC, Kang DS, Gurevich VV, Benovic JL. Non-visual arrestins are constitutively associated with the centrosome and regulate centrosome function. J Biol Chem. 2010;285:8316–8329. doi: 10.1074/jbc.M109.062521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raghuwanshi SK, Nasser MW, Chen X, Strieter RM, Richardson RM. Depletion of beta-arrestin-2 promotes tumor growth and angiogenesis in a murine model of lung cancer. J Immunol. 2008;180:5699–5706. doi: 10.4049/jimmunol.180.8.5699. [DOI] [PubMed] [Google Scholar]
- 23.Zou L, Yang R, Chai J, Pei G. Rapid xenograft tumor progression in beta-arrestin1 transgenic mice due to enhanced tumor angiogenesis. FASEB J. 2008;22:355–364. doi: 10.1096/fj.07-9046com. [DOI] [PubMed] [Google Scholar]
- 24.LeBaron MJ, Crismon HR, Utama FE, Neilson LM, Sultan AS, Johnson KJ, Andersson EC, Rui H. Ultrahigh density microarrays of solid samples. Nat Methods. 2005;2:511–513. doi: 10.1038/nmeth772. [DOI] [PubMed] [Google Scholar]
- 25.Tran TH, Utama FE, Lin J, Yang N, Sjolund AB, Ryder A, Johnson KJ, Neilson LM, Liu C, Brill KL, Rosenberg AL, Witkiewicz AK, Rui H. Prolactin inhibits BCL6 expression in breast cancer through a Stat5a-dependent mechanism. Cancer Res. 2010;15:1711–1721. doi: 10.1158/0008-5472.CAN-09-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dolled-Filhart M, Ryden L, Cregger M, Jirstrom K, Harigopal M, Camp RL, Rimm DL. Classification of breast cancer using genetic algorithms and tissue microarrays. Clin Cancer Res. 2006;12:6459–6468. doi: 10.1158/1078-0432.CCR-06-1383. [DOI] [PubMed] [Google Scholar]
- 27.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 28.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bioinformatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 29.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23:9067–9072. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- 30.Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, Gray JW, Bissell MJ. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gudjonsson T, Adriance MC, Sternlicht MD, Petersen OW, Bissell MJ. Myoepithelial cells: their origin and function in breast morphogenesis and neoplasia. J Mammary Gland Biol Neoplasia. 2005;10:261–272. doi: 10.1007/s10911-005-9586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gudjonsson T, Ronnov-Jessen L, Villadsen R, Rank F, Bissell MJ, Petersen OW. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J Cell Sci. 2002;115:39–50. doi: 10.1242/jcs.115.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sainsbury JR, Anderson TJ, Morgan DA. ABC of breast diseases: breast cancer. BMJ. 2000;321:745–750. doi: 10.1136/bmj.321.7263.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gimpl G, Fahrenholz F. The oxytocin receptor system: structure, function, and regulation. Physiol Rev. 2001;81:629–683. doi: 10.1152/physrev.2001.81.2.629. [DOI] [PubMed] [Google Scholar]
- 35.Sapino A, Macri L, Tonda L, Bussolati G. Oxytocin enhances myoepithelial cell differentiation and proliferation in the mouse mammary gland. Endocrinology. 1993;133:838–842. doi: 10.1210/endo.133.2.8344220. [DOI] [PubMed] [Google Scholar]
- 36.Nishimori K, Young LJ, Guo Q, Wang Z, Insel TR, Matzuk MM. Oxytocin is required for nursing but is not essential for parturition or reproductive behavior. Proc Natl Acad Sci USA. 1996;93:11699–11704. doi: 10.1073/pnas.93.21.11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner KU, Young WS, 3rd, Liu X, Ginns EI, Li M, Furth PA, Hennighausen L. Oxytocin and milk removal are required for post-partum mammary-gland development. Genes Funct. 1997;1:233–244. doi: 10.1046/j.1365-4624.1997.00024.x. [DOI] [PubMed] [Google Scholar]
- 38.Ahn S, Wei H, Garrison TR, Lefkowitz RJ. Reciprocal regulation of angiotensin receptor-activated extracellular signal-regulated kinases by beta-arrestins 1 and 2. J Biol Chem. 2004;279:7807–7811. doi: 10.1074/jbc.C300443200. [DOI] [PubMed] [Google Scholar]
- 39.Ren XR, Reiter E, Ahn S, Kim J, Chen W, Lefkowitz RJ. Different G protein-coupled receptor kinases govern G protein and beta-arrestin-mediated signaling of V2 vasopressin receptor. Proc Natl Acad Sci USA. 2005;102:1448–1453. doi: 10.1073/pnas.0409534102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ge L, Shenoy SK, Lefkowitz RJ, DeFea K. Constitutive protease-activated receptor-2-mediated migration of MDA MB-231 breast cancer cells requires both beta-arrestin-1 and -2. J Biol Chem. 2004;279:55419–55424. doi: 10.1074/jbc.M410312200. [DOI] [PubMed] [Google Scholar]
- 41.Defea KA. Stop that cell! beta-arrestin-dependent chemotaxis: a tale of localized actin assembly and receptor desensitization. Annu Rev Physiol. 2007;69:535–560. doi: 10.1146/annurev.physiol.69.022405.154804. [DOI] [PubMed] [Google Scholar]
- 42.Balkwill F. The significance of cancer cell expression of the chemokine receptor CXCR4. Semin Cancer Biol. 2004;14:171–179. doi: 10.1016/j.semcancer.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Bhandari D, Trejo J, Benovic JL, Marchese A. Arrestin-2 interacts with the ubiquitin-protein isopeptide ligase atrophininteracting protein 4 and mediates endosomal sorting of the chemokine receptor CXCR4. J Biol Chem. 2007;282:36971–36979. doi: 10.1074/jbc.M705085200. [DOI] [PubMed] [Google Scholar]
- 44.Fong AM, Premont RT, Richardson RM, Yu YR, Lefkowitz RJ, Patel DD. Defective lymphocyte chemotaxis in beta-arrestin2-and GRK6-deficient mice. Proc Natl Acad Sci USA. 2002;99:7478–7483. doi: 10.1073/pnas.112198299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar P, Lau CS, Mathur M, Wang P, DeFea KA. Differential effects of beta-arrestins on the internalization, desensitization and ERK1/2 activation downstream of protease activated receptor-2. Am J Physiol Cell Physiol. 2007;293:C346–C357. doi: 10.1152/ajpcell.00010.2007. [DOI] [PubMed] [Google Scholar]
- 46.Saunders W. Centrosomal amplification and spindle multipolarity in cancer cells. Semin Cancer Biol. 2005;15:25–32. doi: 10.1016/j.semcancer.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15:303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 48.Salisbury JL, D’Assoro AB, Lingle WL. Centrosome amplification and the origin of chromosomal instability in breast cancer. J Mammary Gland Biol Neoplasia. 2004;9:275–283. doi: 10.1023/B:JOMG.0000048774.27697.30. [DOI] [PubMed] [Google Scholar]
- 49.Lingle WL, Lukasiewicz K, Salisbury JL. Deregulation of the centrosome cycle and the origin of chromosomal instability in cancer. Adv Exp Med Biol. 2005;570:393–421. doi: 10.1007/1-4020-3764-3_14. [DOI] [PubMed] [Google Scholar]
- 50.D’Assoro AB, Barrett SL, Folk C, Negron VC, Boeneman K, Busby R, Whitehead C, Stivala F, Lingle WL, Salisbury JL. Amplified centrosomes in breast cancer: a potential indicator of tumor aggressiveness. Breast Cancer Res Treat. 2002;75:25–34. doi: 10.1023/a:1016550619925. [DOI] [PubMed] [Google Scholar]
- 51.Lingle WL, Barrett SL, Negron VC, D’Assoro AB, Boeneman K, Liu W, Whitehead CM, Reynolds C, Salisbury JL. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci USA. 2002;99:1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Molla-Herman A, Boularan C, Ghossoub R, Scott MG, Burtey A, Zarka M, Saunier S, Concordet JP, Marullo S, Benmerah A. Targeting of beta-arrestin2 to the centrosome and primary cilium: role in cell proliferation control. PLoS ONE. 2008;3:e3728. doi: 10.1371/journal.pone.0003728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strous GJ, Schantl JA. Beta-arrestin and Mdm2, unsuspected partners in signaling from the cell surface. Sci STKE. 2001;2001(110):pe41. doi: 10.1126/stke.2001.110.pe41. [DOI] [PubMed] [Google Scholar]
- 54.Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harbor Perspect Biol. 2009;1:a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones SN, Hancock AR, Vogel H, Donehower LA, Bradley A. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci USA. 1998;95:15608–15612. doi: 10.1073/pnas.95.26.15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turbin DA, Cheang MC, Bajdik CD, Gelmon KA, Yorida E, De Luca A, Nielsen TO, Huntsman DG, Gilks CB. MDM2 protein expression is a negative prognostic marker in breast carcinoma. Mod Pathol. 2006;19:69–74. doi: 10.1038/modpathol.3800484. [DOI] [PubMed] [Google Scholar]
- 57.Girnita L, Shenoy SK, Sehat B, Vasilcanu R, Girnita A, Lefkowitz RJ, Larsson O. β-Arrestin is crucial for ubiquitination and down-regulation of the insulin-like growth factor-1 receptor by acting as adaptor for the MDM2 E3 ligase. J Biol Chem. 2005;280:24412–24419. doi: 10.1074/jbc.M501129200. [DOI] [PubMed] [Google Scholar]
- 58.Werner H, Bruchim I. The insulin-like growth factor-I receptor as an oncogene. Arch Physiol Biochem. 2009;115:58–71. doi: 10.1080/13813450902783106. [DOI] [PubMed] [Google Scholar]
- 59.Dunn SE, Ehrlich M, Sharp NJ, Reiss K, Solomon G, Hawkins R, Baserga R, Barrett JC. A dominant negative mutant of the insulin-like growth factor-I receptor inhibits the adhesion, invasion, and metastasis of breast cancer. Cancer Res. 1998;58:3353–3361. [PubMed] [Google Scholar]
- 60.Bonneterre J, Peyrat JP, Beuscart R, Demaille A. Prognostic significance of insulin-like growth factor 1 receptors in human breast cancer. Cancer Res. 1990;50:6931–6935. [PubMed] [Google Scholar]
- 61.Ellis MJ, Jenkins S, Hanfelt J, Redington ME, Taylor M, Leek R, Siddle K, Harris A. Insulin-like growth factors in human breast cancer. Breast Cancer Res Treat. 1998;52:175–184. doi: 10.1023/a:1006127621512. [DOI] [PubMed] [Google Scholar]
- 62.Resnik JL, Reichart DB, Huey K, Webster NJ, Seely BL. Elevated insulin-like growth factor I receptor autophosphorylation and kinase activity in human breast cancer. Cancer Res. 1998;58:1159–1164. [PubMed] [Google Scholar]