Abstract
The glmS riboswitch regulates gene expression through a self-cleavage activity. The reaction is catalyzed with the assistance of the metabolite cofactor glucosamine-6-phosphate (GlcN6P), whose amino group is proposed to serve as the general acid during the reaction. This reaction is pH-dependent with a pKa that is lower than the observed pKa for the amine of GlcN6P in solution. GlcN6P, like other pyranose sugars, undergoes spontaneous and rapid interconversion between the α and β anomers at the C1 position. Here we demonstrate by NMR that the Bacillus anthracis glmS riboswitch selectively binds the α-anomer of GlcN6P with a maximum binding affinity of 0.36 mM, and that binding is pH-dependent. We also report that the anomeric ratio between α and β is pH-dependent and the pKas of the two amines differ by 0.5 pH units, α being the higher of the two (pKa = 8.3). The pH dependence of binding reveals a pKa of 6.7, suggesting that the glmS RNA reduces the pKa of the GlcN6P amine by 1.6 units in the ground state. We reevaluated previously obtained kinetic data and found the reaction pKa is 6.9, within error of the binding data. The data support a model where the reaction pKa corresponds to that of the GlcN6P amine. This observation has broader relevance for considering how the microenvironment of an RNA, despite its anionic character, can reduce the pKas of functional groups for use in catalysis.
The glmS gene codes for the enzyme glutamine-fructose-6-phosphate amidotransferase, which produces glucosamine-6-phosphate (GlcN6P) from fructose-6-phosphate and glutamine (1). GlcN6P is an essential metabolite that is processed to the activated form N-acetylglucosamine and used to make peptidoglycans, lipopolysaccharides and teichoic acids in bacteria; chitin in fungi, insects and crustaceans; and glycoproteins, glycosaminoglycans and mucopolysacharides in mammals (1). Given the importance of GlcN6P, it is not surprising that deletion of the glmS gene is lethal in bacteria, making this gene an attractive antibiotic target (1–4).
In many Gram-positive bacteria, the glmS gene is regulated by a riboswitch that is found in the 5' untranslated region of the gene (5). This glmS riboswitch senses the concentration of GlcN6P and regulates the expression of the amidotransferase (5, 6). However, unlike the majority of riboswitches described to date, the glmS riboswitch does not function by stabilizing alternative structural conformations upon ligand binding. Instead, GlcN6P functions as a metabolite cofactor that promotes self-cleavage of the glmS RNA (5, 7). Cleavage destabilizes the downstream mRNA by generating a 5'-terminal OH that is recognized as an mRNA degradation signal by ribonuclease J1 (6).
Biochemical and structural data have identified the overall mechanism of RNA strand scission in this ribozyme. Upon GlcN6P binding, the glmS riboswitch is proposed to react via a general acid-base mechanism with the amine of GlcN6P functioning as the general acid (7–10). Crystal structures of the Bacillus anthracis and the Thermoanaerobacter tengcongensis glmS ribozymes bound to GlcN6P have been reported and in each case the amino group is in close proximity to the O5' leaving group (8, 10). The reaction is pH-dependent with a pKa between 7.6 and 7.8 (7, 9, 11). Because this pKa is near the pKa of the amine of GlcN6P in solution (8.2), it has been attributed to the pKa of the amine in the transition state (7, 9). However, this would require a shift of ~0.5 pH units in the acidic direction in the transition state. The observation that other ligands, including serinol and glucosamine, with a primary amine can catalyze the reaction provided additional evidence that the amine acts as the general acid (7). Furthermore it has been observed that the reaction pKa for serinol and glucosamine match the solution pKa of the amine groups of these ligands (7). Thus, it seems likely that the pKa of the GlcN6P amine is being lowered, at least in the transition state.
Like all pyranose sugars, GlcN6P undergoes spontaneous isomerization at the anomeric carbon (C1 position) to produce an α and a β-isomer (Figure 1) (12). Previous efforts to interpret the glmS ribozyme reaction did not consider the role this isomerization might play, or that the riboswitch may selectively bind and react with only one of the two anomers. A previous attempt to determine if the reaction is preferentially catalyzed by one of the anomers was inconclusive because neither of the GlcN6P analogs used to mimic the α or β conformations was reactive (13). All structures of the glmS riboswitch have modeled GlcN6P using the α-anomer. The high resolution T. tengcongensis structure (1.7Å resolution) is at a sufficient resolution to be reasonably confident about such an assignment, but even then, it would be difficult to identify fractional binding of the β-anomer (10).
Figure 1.
Isomerization of GlcN6P between the two pyranose anomers. The position for the C1, or anomeric, carbon is numbered.
Surprisingly, several aspects of GlcN6P isomerization chemistry are unknown. These include the α/βisomeric ratio at equilibrium, the pH dependence of that ratio, the pKas of the amines for the individual isomers, and the rate of GlcN6P interconversion. Furthermore, it is unclear if the glmS RNA has a preference for one anomer over the other. It is possible that the discrepancy between the solution pKa of GlcN6P and the reaction pKa is due to anomeric specificity of the glmS riboswitch. Characterization of these parameters is necessary to properly interpret the pH-dependent reaction kinetics of GlcN6P-induced glmS RNA cleavage and to understand how the metabolite is used as a catalytic cofactor.
In the present study we have used NMR to determine the pKa of the amine of GlcN6P bound to the glmS riboswitch. We also determined the anomer specificity of GlcN6P and the nature of GlcN6P isomerization in the free form. We report that the glmS riboswitch specifically binds to the α-anomer in a pH-dependent fashion, with a preference for the deprotonated form and a pKa of 6.7. This is consistent with the active site of the glmS riboswitch shifting the pKa of the amine in the acidic direction, which has implications for other RNA active sites where general acid-base chemistry has been proposed (14–19).
Materials and Methods
Substrate preparation
The inactive glmS RNA with a 2' O-methyl substitution at the cleavage site was prepared as a two-piece construct as previously described (8). The transcribed portion of the RNA was purified from the HDV ribozyme and abortive products by size exclusion chromatography using a Hi Load 26/60 Superdex™ 75 prep-grade column (GE Healthcare). The oligonucleotide portion of the riboswitch was purchased from Thermo Scientific. Both RNAs were purified using a G-25 Sephadex column to remove salts. The RNAs were diluted to between 50 and 75 µM in 50 mM NaCl, 10 mM sodium acetate (pKa 4.8), 10 mM triethanolamine (pKa 7.8), and 10 mM MgCl2 at the pH needed for the experiment. The D-glucosamine-6-phosphate sodium salt (GlcN6P) and D-glucose-6-phosphate sodium salt (Glc6P) was purchased from Sigma-Aldrich®. A 1 M solution of both sugars was prepared in 50 mM NaCl, 10 mM sodium acetate (pKa 4.8) and 10 mM triethanolamine (pKa 7.8) at the desired pH. All GlcN6P samples contained 10% D2O. Because the water peak overlapped with the chemical shift of the H1 proton for the α–anomer, the Glc6P sample was dried and resuspended in 100% D2O.
NMR spectroscopy
All NMR experiments were performed using a Varian 500 MHz spectrometer with a HCN triple axis probe at 25° C running VNMR-J. Chemical shifts of the H2 and H1 protons were obtained by the known chemical shifts and J-coupling of the two isomers of other glucose sugars (12), and confirmed using NOESY data. All 1D data were analyzed using MestReNova 6.6.1. 2D data were analyzed using Sparky 3.115.
1H NMR pH titration
NMR samples were titrated to the appropriate pH using NaOH. Ten 3 mM samples of GlcN6P were prepared between pH 5.5 and 10.5 at intervals of 0.5 pH units. The pH was tested before and after the NMR experiment. No significant change in pH was observed over the course of the experiment. 1D 1H spectra with water suppression were collected at each pH. The percent α-anomer was calculated using the area under the H2 peak of the two anomers. The H2 chemical shift for each anomer was used to determine the pKa. The Ka was calculated using the equation:
| (1) |
where ω is the H2 chemical shift.
2D 1H-1H NOESY at varying mixing times
A NOESY of 5 mM GlcN6P with a 0.5, 1, 1.5, 2, and 5 second mixing time of 5 mM GlcN6P at pH 7.5 was used to determine the isomerization rate. Positive off-diagonal peaks arise from chemical exchange between the anomers. Positive NOEs were observed between the H2 of α-anomer and the H2 of the β-anomer. Peak volumes were determined using Sparky (20). The rates of α to β isomer (kβα) and β to α isomer (kαβ) conversion was calculated using the equation:
| (4) |
were A is matrix that contains peak volumes and R is a matrix that contains the kinetic parameters with EXSY CALC (MESTRALAB RESEARCH) (21, 22). The optimum mixing time (τmopt) was determined using the equation (23):
| (3) |
WaterLOGSY spectra
Two samples of GlcN6P were prepared at pH 7.5, one in the absence of any RNA and one in the presence of 500 µM glmS RNA. A WaterLOGSY was performed using 512 scans of both samples (24). The sign of the H2 proton was used to determine if GlcN6P was binding to the RNA, a positive signal arising from interactions between the RNA and the ligand. Only the sample containing RNA displayed any positive signals.
Binding affinities measured by T1 relaxation rates
For the pH 5.5 GlcN6P and pH 7.5 Glc6P titrations, the ligand was added in nine steps between 0.25 and 9 mM to 75 µM glmS RNA. For the pH titrations of GlcN6P above 5.5, GlcN6P was added in eight steps between 0.25 and 5 mM to 35 µM glmS RNA in 550 µl. The addition of ligand diluted the glmS RNA concentration by less than 2%. The actual concentration of ligand was determined by using sodium acetate as an internal standard. T1 values of GlcN6P and Glc6P in the absence of RNA were measured at each pH tested and at varying concentrations. There was little to no pH or concentration dependence on free T1 values. 512 scans were used to accurately determine T1 at low ligand concentration and only 8 scans were used at the highest concentrations. The Kd values were calculated using the equation:
| (4) |
where [R]o is the concentration of glmS RNA, [L]o is the concentration of ligand, and T1bound is the T1 of the bound ligand, τbound is the lifetime of the bound state, and ΔT1 is [1/T1obs − 1/T1free] (38). The pH binding data were fit to the equation:
| (5) |
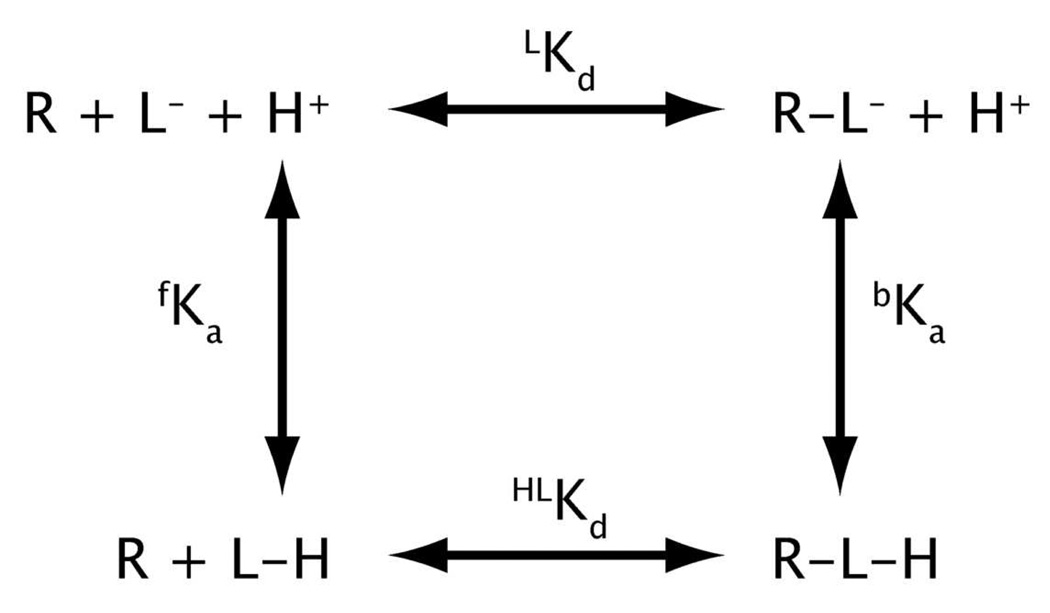
where Kapp is the observed Kd, LHKd is the binding affinity of the protonated ligand, and fKa and bKa are the Ka of the bound and free ligand (25). This equation assumes only one protonation state and is derived from Scheme 1. From Scheme 1 one can also derive:
| (6) |
Thus, if the ligand binds the deprotonated form of the ligand tighter than the protonated form, then the pKa of the bound ligand will be lower than the pKa of the free ligand.
Scheme 1.
Binding of protonated and deprotonated ligand (L) to RNA (R), where LKd and LHKd is the binding affinity of the deprotonated and protonated ligand respectively, and fKa and bKa are the binding affinity of the bound and free ligand respectively.
Reevaluation of kmax as a function of pH
The previously published glmS ribozyme kinetic data were fit to the equation:
| (7) |
were fHA+ is the fraction of the general acid that is protonated and fB− is the fraction of deprotonated base and k1 is the intrinsic rate of the reaction, which is dependent on the Brϕnsted equation (16). The data fit equally well to the more simple equation:
| (8) |
Results
We determined the GlcN6P α/β isomer ratio in solution as a function of pH. If the glmS riboswitch binds only one isomer and the isomeric ratio is pH-dependent, then the concentration of the relevant ligand will be different at each pH. We collected 1D NMR data of GlcN6P between pH 5.5 and 10.5 and determined the ratio of the isomers by integrating the area under the peak for the H-2 proton (Figure 2a). The ratio of anomers, α:β, changes from 37:63 at pH 10.5 to 64:36 at pH 5.5 (Figure 2b). The ratio is 1:1 near pH 8.0. The largest change in isomeric ratio occurs near the pKa of the amine (~8) while there are smaller changes in isomeric ratio near the pKa of the phosphate (6.1). Thus, the solution pH has complex effects on the relative concentrations of the GlcN6P isomers and their relative protonation states.
Figure 2.
pKa and isomer ratios of GlcN6P. (a) the region of the 1H spectrum of 3 mM GlcN6P at 25° C with the chemical shift of the H2 proton for both isomers is shown as a function of pH. The chemical shift assignments for the isomers were made by comparison to known J-coupling and chemical shift data for α- and β- anomers of glucosamine. The area under the peaks for both anomers was used to determine the fraction of α-anomer as a function of pH (b). The pKas of the two anomers were determined by fitting the chemical shift change as a function of pH (c). The two vertical lines indicate the pKas of the isomers.
It is possible that if the amine pKa of the two isomers in solution were different, the observed shift in pKa could be due to isomeric preference of the glmS riboswitch. In fact, the NMR data demonstrated that the amino pKas of the two GlcN6P isomers were not equal. These equilibrium constants were determined by fitting the change in chemical shift of both the 15N labeled amine nitrogen and the H-2 proton of the α- and β-isomer versus pH (data not shown and Figure 2c). We found that the amino group pKa of the α-anomer is 8.3 and the β-anomer is 7.8, a difference of almost half a pH unit. The pKa values of the two anomers are just above and below the bulk solution measurement for GlcN6P amine pKa (8.2) (7). These measurements show that the pKa of the β-anomer is closest to the observed pKa of the cleavage reaction (7.6) (9).
GlcN6P is synthesized as the α-anomer in vivo (26). Unless the isomerization is rapid, it is possible that GlcN6P is enzymatically converted to GlcN1P before it undergoes significant anomerization to the β-form (1). The rates of exchange for glucose are known and are relatively slow (t1/2 = 7 min) (27). Amino substitution (glucosamine) increases the rate by 5–10 fold and a phosphate substitution (glucose-6-phosphate or Glc6P) increases the rate by 200-fold (27, 28), but the rate of the doubly substituted GlcN6P has not been measured. We used NOE chemical exchange to determine the rate of exchange at pH 7.5. The rate of conversion from the α- to the β-anomer kαβ and conversion back to the α-anomer k were first estimated at various mixing times from 0.5 to 5 seconds. These rates were used to determine the optimum mixing time (approximately 1 second). The rates of conversion using the optimum mixing time are kαβ = 0.06 s−1 and k = 0.08 s−1 giving an equilibration half-life of 5 sec. These fast exchange rates indicate that both anomers of GlcN6P are likely to be present in vivo and both would be available for binding to the glmS riboswitch. The question then becomes; does the riboswitch differentiate between them?
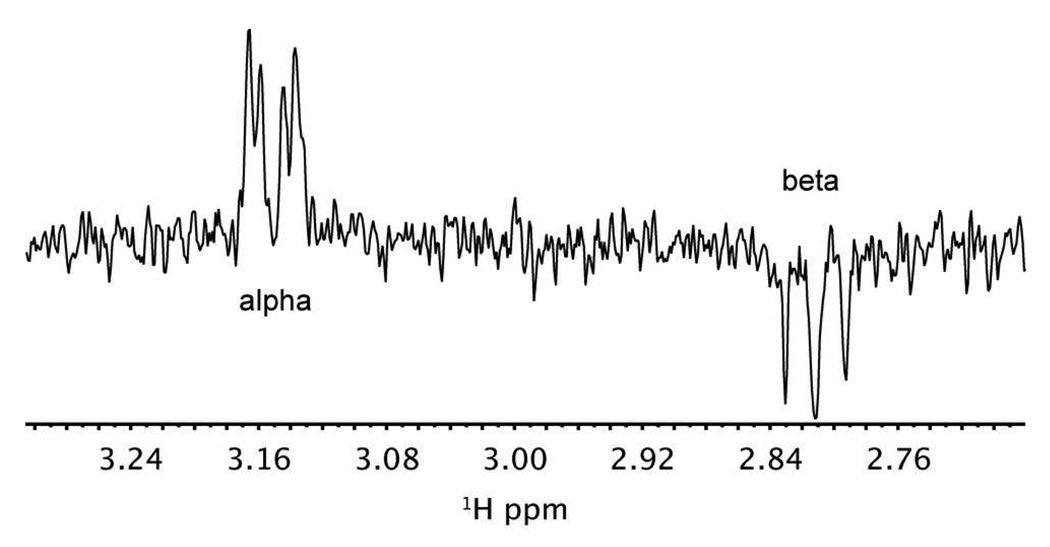
We performed a WaterLOGSY NMR experiment to test if GlcN6P binding to the Bacillus anthracis glmS riboswitch is isomer specific (Figure 3) (24). In this experiment, bulk magnetization is transferred to the ligand though the ligand-RNA complex. If the ligand binds to the RNA and is in fast exchange, then magnetization transfer from RNA-bound water to RNA-bound ligand results in a positive signal. In contrast, if the ligand does not bind the RNA, then magnetization transfer from the bulk solvent will result in a negative ligand signal. When 500 µM glmS RNA was added to 3 mM GlcN6P at pH 7.5, 1H signals from the α-isomer in the WaterLOGSY experiment were positive (same sign as the RNA) while signals from the β-isomer were negative (Figure 3). Therefore, at this concentration there was no detectable binding from the β-isomer, which suggests that the glmS riboswitch selectively binds the α-isomer.
Figure 3.
WaterLOGSY of the H2 protons of GlcN6P in the presence of glmS riboswitch. GlcN6P concentration is 3 mM and RNA concentration was 500 uM. The glmS RNA construct used was inactivated as previously described by a 2´-O-methyl substitution.
To further determine if GlcN6P binding is isomer specific, dissociation constants (Kd) were determined for the α- and the β-isomer interaction with the substrate form of the glmS riboswitch inactivated with a 2'-OMe substitution at the cleavage site. Several NMR methods could be used to determine ligand-binding affinities; however, the large size of the RNA and fast exchange of the ligand precluded direct observation of the RNA bound ligand. Therefore, efforts were focused on ligand-observed NMR methods. Of the parameters tested, the change in the 1/T1 relaxation rate of the ligand (1 to 2 seconds) in the presence of the RNA showed the largest change. We measured the change in relaxation of the α-isomer H1 proton and the β-isomer H2 proton as a function of GlcN6P concentration at 45 µM glmS riboswitch (Figure 4a). The H2 proton signal was used for the β-isomer because the water peak interfered with the H1 proton signal. The H2 proton signal gave the same value, but had a lower signal to noise ratio. The affinity of glmS RNA for the α-isomer was 0.4 mM at pH 7.5. After adjusting for the isomeric partitioning at pH 7.5, the apparent binding affinity of the anomeric mixture is 0.8 mM, which is within two-fold of the kinetic measurements of the apparent Kd for the Bacillus subtilis (1.5 mM) and the Bacillus anthracis glmS RNA (1.4 mM) (9, 11). Unlike the α-isomer, there was little change in signal intensity of the WaterLOGSY at any concentration of GlcN6P and no detectable change in T1 relaxation rates for the β-isomer (Figure 4a). Based on the detection limit for the T1 data of the H2 proton, the tightest binding affinity of the β-isomer is >7 mM, which is at least 15-fold weaker than the affinity for the α-isomer. Because the β-isomer does not bind the glmS riboswitch to a significant degree, only the solution pKa of the α-isomer (8.3) is relevant. Thus, the different pKa values for the two isomers does not explain the observed reaction pKa (9).
Figure 4.
The binding affinity of GlcN6P for the glmS riboswitch. (a) The T1 relaxation rates for the H1 proton of the α-isomer (red) and H2 proton of the β-isomer (blue) as a function of GlcN6P concentration in the presence of 45 µM glmS RNA at pH 7.5. (b) Binding affinities of GlcN6P at varying pH fit to equation 4. Error bars arise from triplicate measurements at each pH.
To explore the extent of amine recognition by the riboswitch, we tested binding of Glc6P, which has a hydroxyl in place of the amine group. The amine donates hydrogen bonds to both the 5′-O leaving group and the O4 of U43 (9). These hydrogen bonds would likely be altered when the amine is protonated. There was no observable change in T1 when Glc6P was added to the glmS RNA. Based on the minimum observable change in T1, the binding affinity of Glc6P is >10 mM or at least 20-fold less than GlcN6P. Thus amine recognition has a significant effect on cofactor binding affinity.
The glmS riboswitch reaction rate displays a strong pH dependence (9), including effects on both the kmax and the K1/2 in a system were the cofactor (GlcN6P) is in excess over the riboswitch. Prior to the experiments reported here, it had not been possible to directly measure the GlcN6P binding affinity. We explored the pH dependence of cofactor binding by determining the Kd as a function of pH from 5.5 to 8.5 using the T1 relaxation approach outlined above. There was little change in the Kd between pH 8.5 and 7.5, (0.43 ± 0.06 and 0.36 ± 0.06 mM respectively), but there was a five-fold loss of affinity between pH 7.5 and 5.5 (Kd = 1.8 ± 0.2) (Figure 4b). The binding data fit to equation 4 with a pKa of 6.7 ± 0.2, which is intermediate between the pKa of the phosphate and the amine of GlcN6P. The loss of affinity observed at lower pH is consistent with the K1/2 effects observed for glmS ribozyme reactivity.
Discussion
The glmS ribozyme binds to GlcN6P, resulting in cleavage of the RNA. This reaction is thought to proceed though a general acid-base mechanism with the amine of GlcN6P acting as the general acid (9, 29). Both the kmax and K1/2 of the reaction display a pH dependence with pKa values of 7.5 and 6.3 respectively (9). If these pKa values reflect the pKa of the amine of GlcN6P bound to the RNA, then binding of the RNA must shift the pKa of the amine. Can RNA (an anionic molecule) shift the pKa of a functional group in the acidic direction? If so, what is the magnitude of this equilibrium shift? While pKa values of RNA active site residues have been shown to be shifted in the basic direction, measurement of a pKa shift in the acid direction has not been observed. We have used NMR to begin to answer these questions as well as determine the nature of GlcN6P recognition.
Increased affinity at high pH indicates that the riboswitch preferentially binds the deprotonated form of GlcN6P (Figure 4b). Near physiological pH there are two titratable groups on GlcN6P: the amine and the phosphate with pKas of 8.3 and 6.1, respectively. These pKa values flank the observed binding pKa, which raises the question as to which functional group has a pKa of 6.7. From scheme 1 and equation 6 any change in binding affinity of a ligand as a function of pH will have a corresponding shift in pKa of a functional group on the ligand. Tighter binding at high pH indicates stabilization of the deprotonated form of the relevant group, which will reduce the pKa of that group (25). Thus, the binding data support a model in which the pKa of the amine is reduced and disfavors a model in which the pKa of the phosphate is elevated. This represents a pKa shift of 1.6 units from the unperturbed value of the amine in solution. A pKa shift of this magnitude is also consistent with a modeling study that compared the binding of the protonated and deprotonated form of GlcN6P and predicted a pKa shift of approximately 2 units for the amino group (30). Our data are not consistent with a second MD simulation study, which predicted that the amine is more likely to be protonated upon RNA binding (29). It is possible that phosphate protonation also reduces binding affinity, but we could not monitor the pKa of this group. If the pKa of the phosphate were shifted, it would be to a value less than 6.0, which places it into the range where adenine and cytosine protonation on the RNA interfere with the analysis (31).
Previous kinetic data displayed a complex pH dependence that could not be readily modeled assuming a fixed cofactor concentration as a function of pH (9). The current work establishes that only the α-anomer of GlcN6P binds the glmS riboswitch and the concentration of the α-anomer changes with pH. Using this information we reevaluated the kinetic data using the actual concentration of the α-anomer at each pH (Figure 5). The result was an excellent fit of the reaction data to a standard pH profile equation. The concentration of GlcN6P required to react at half the maximal rate (K1/2) matches the Kd in both magnitude and its dependence on pH. The pKa of 6.5 +/− 0.4 for the K1/2 is within error of the pKa measured for binding (6.7+/− 0.2). In addition to revisiting the K1/2 data we reevaluated the kmax data and determined that the pKa of the reaction is 6.9 +/− 0.1 (Figure 5). This value is within the error of the pKa we measured for binding, suggesting that the pKa of the group we are measuring, which is likely to be the amine, is acting as either the general acid, in a general acid-base mechanism or alone as a general base.
Figure 5.
Reevaluation of previously acquired kinetic data. (a) K1/2 as a function of pH. K1/2 values were determined by the amount of GlcN6P required to reach half-maximal rate. The data were fit to equation 5 to determine the pKa. (b) kmax (maximal rate) as a function of pH. The data were fit to equation 7 to determine the reaction pKa.
The kinetic data are not sufficient to determine if the reaction pKa is from a general acid, in a general acid-base mechanism with a general base pKa above 9.5, or acting as alone as a general base (the pH rate profile would look the same). However, structural data show the amine of GlcN6P in position to protonate the 5′-O leaving-group as a general acid and the N1 of G33 (G40 in T. tengcongensis) in position to act as the general base by deprotonating the 2′-OH nucleophile (Figure 6). Biochemical data indicate that both the amine of GlcN6P and the nucleobase G33 are required for catalysis (7, 8). The kinetic data fit well with an unperturbed pKa of approximately 10 for guanine and a pKa of 6.9 for the amine general acid. This leads us to propose that upon binding to the glmS riboswitch the pKa of the amine of GlcN6P is shifted toward neutrality and acts as the general acid in a general acid-base mechanism with G33 acting as the general base (Figure 6).
Figure 6.
Proposed mechanism of the glmS riboswitch cleavage. In this mechanism, upon binding to the glmS riboswitch the pKa of the amine of GlcN6P is shifted to 6.9 and acts as the general acid donating a proton to the 5′-O leaving-group and G33 acts as the general base by deprotonating the 2′-OH nucleophile.
It has been proposed that RNA can alter the pKa of active site groups (14–16, 32); however, prior to this study, pKa perturbation had only been reported for cases where RNA stabilized the protonated form of adenosine or cytosine, thus increasing the pKa (32–37). One commonly observed equilibrium shift involves the protonation of the adenine N1 (solution pKa near 4) in the context of an A+-C pair near neutral pH (6.8) (31, 33, 35, 36). Elevated pKa values between 4.5 and 6.4 have also been observed for the catalytic cytidine in the HDV ribozyme active site using both NMR and Raman spectroscopy (32, 34). In all these studies the pKa shift is in the basic direction. Given the anionic character of RNA it is not entirely unexpected that an RNA active site can increase the pKa of functional groups. A pKa shift in the acidic direction is less anticipated, yet this is what is observed for GlcN6P binding within the glmS ribozyme. While the pKa of the β-anomer is 7.7, which is close to the pKa of the reaction, the relevant pKa is that of the α-anomer, with a solution pKa of 8.3. Yet when bound in the glmS riboswitch active site, the pKa of the amine appears to be reduced to 6.7, corresponding to a shift of 1.6 units.
Two previous studies provide some insight into which functional groups that may be responsible for this pKa shift within the glmS ribozyme (7, 30). Molecular dynamics simulations predicted that the shift in pKa between the bound and the free form was dependent upon G33 (30). Mutation of G33 to an A results in an inactive riboswitch (8). They hypothesized that this observation is at least partially due to the increased pKa of the amine (30). It has also been observed that the shift in pKa is ligand-dependent (7). When glucosamine or serinol is used in place of GlcN6P the reaction pKa is equal to the solution pKa of glucosamine or serinol, respectively (7). Of the substrates tested, only GlcN6P has a reaction pKa that does not match the pKa in solution. Since the only difference between glucosamine and GlcN6P is the phosphate, and the phosphate is coordinated to metal ions, one hypothesis is that these metals may be at least partially responsible for the observed equilibrium shift. Further study is required to determine if these or other functional groups are responsible for the observed effect. The NMR approach described here provides a means to readily test these possibilities in future studies.
The observation that the glmS riboswitch active site shifts the amine pKa in the acidic direction gives rise to the possibility that other RNA active sites can achieve the same result. A general acid-base model has been proposed for several RNA active sites (14–19). However, unlike proteins, RNA does not contain functional groups with pKa values near neutrality that would dramatically enhance rates in such a mechanism (31). Guanine is repeatedly observed in ribozyme active sites and these bases have been proposed to play a catalytic role as general acid or general base catalysts, yet the pKa of the N1 of G is approximately 10, well removed from neutrality (8, 14, 16–18, 38, 39). If the pKa of guanine could be decreased, then it would enhance its ability to play this catalytic role. However, there is still no direct evidence of an acidic pKa shift of an active site guanine. In fact the only direct pKa measurement of an active site guanine failed to show a pKa shift toward neutrality (40). Because of the lack of a pKa shift for the guanine, it is debated whether the ribozymes employ a general acid-base mechanism (14, 40). Our measurement of a 1.6 unit pKa shift for a functional group in an RNA active site shows that the microenvironment of an RNA active site can reduce a pKa. This gives rise to the possibility that other ribozyme active sites can perform the same function allowing them to catalyze cleavage using general acid-base chemistry with pKas that are closer to neutrality.
Acknowledgments
We thank the staff at the Yale chemical instrument center, Dr. Xiaoling Wu and Dr. Eric Paulson, for their support in obtaining NMR data. We also thank David Hiller for helpful discussions. This work was supported by NIH grant 54839 to SAS, NIH Postdoctoral Fellowship 087070 to JHD and NIH training grant 007223 to BFD.
Abbreviations
- GlcN6P
glucosamine-6-phosphate
- Glc6P
glucose-6-phosphate
- GlcN1P
glucosamine-1-phosphate
- NMR
nuclear magnetic resonance
References
- 1.Milewski S. Glucosamine-6-phosphate synthase--the multi-facets enzyme. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 2002;1597:173–192. doi: 10.1016/s0167-4838(02)00318-7. [DOI] [PubMed] [Google Scholar]
- 2.Wu HC, Wu TC. Isolation and Characterization of a Glucosamine-Requiring Mutant of Escherichia coli K-12 Defective in Glucosamine-6-Phosphate Synthetase. J. Bacteriol. 1971;105:455–466. doi: 10.1128/jb.105.2.455-466.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarvas M. Mutant of Escherichia coli K-12 Defective in D-Glucosamine Biosynthesis. J. Bacteriol. 1971;105:467–471. doi: 10.1128/jb.105.2.467-471.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freese EB, Cole RM, Klofat W, Freese E. Growth, Sporulation, and Enzyme Defects of Glucosamine Mutants of Bacillus subtilis. J. Bacteriol. 1970;101:1046–1062. doi: 10.1128/jb.101.3.1046-1062.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 6.Collins JA, Irnov I, Baker S, Winkler WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 2007;21:3356–3368. doi: 10.1101/gad.1605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarthy TJ, Plog MA, Floy SA, Jansen JA, Soukup JK, Soukup GA. Ligand requirements for glmS ribozyme self-cleavage. Chem. Biol. 2005;12:1221–1226. doi: 10.1016/j.chembiol.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem. Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cochrane JC, Lipchock SV, Smith KD, Strobel SA. Structural and Chemical Basis for Glucosamine 6-Phosphate Binding and Activation of the glmS Ribozyme. Biochemistry. 2009;48:3239–3246. doi: 10.1021/bi802069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein DJ, Wilkinson SR, Been MD, Ferré-D’Amaré AR. Requirement of helix P2.2 and nucleotide G1 for positioning the cleavage site and cofactor of the glmS ribozyme. J. Mol. Biol. 2007;373:178–189. doi: 10.1016/j.jmb.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hampel KJ, Tinsley MM. Evidence for Preorganization of the glmS Ribozyme Ligand Binding Pocket. Biochemistry. 2006;45:7861–7871. doi: 10.1021/bi060337z. [DOI] [PubMed] [Google Scholar]
- 12.Horton D, Jewell JS, Philips KD. Anomeric Equilibria in Derivatives of Amino Sugars. Some 2-Amino-2-deoxy-D-hexose Derivatives1–3. J. Org. Chem. 1966;31:4022–4025. doi: 10.1021/jo01279a048. [DOI] [PubMed] [Google Scholar]
- 13.Lim J, Grove BC, Roth A, Breaker RR. Characteristics of Ligand Recognition by aglmS Self-Cleaving Ribozyme. Angew. Chem. Int. Ed. 2006;45:6689–6693. doi: 10.1002/anie.200602534. [DOI] [PubMed] [Google Scholar]
- 14.Wilson TJ, Lilley DMJ. Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid–base catalysis? A critical assessment of available experimental data. RNA. 2011;17:213–221. doi: 10.1261/rna.2473711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson TJ, McLeod AC, Lilley DMJ. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J. 2007;26:2489–2500. doi: 10.1038/sj.emboj.7601698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bevilacqua PC. Mechanistic Considerations for General Acid–Base Catalysis by RNA: Revisiting the Mechanism of the Hairpin Ribozyme. Biochemistry. 2003;42:2259–2265. doi: 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- 17.Cochrane JC, Strobel SA. Catalytic Strategies of Self-Cleaving Ribozymes. Acc. Chem. Res. 2008;41:1027–1035. doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]
- 18.Martick M, Scott WG. Tertiary Contacts Distant from the Active Site Prime a Ribozyme for Catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Burke JM. Model for General Acid–Base Catalysis by the Hammerhead Ribozyme: pH–Activity Relationships of G8 and G12 Variants at the Putative Active Site. Biochemistry. 2005;44:7864–7870. doi: 10.1021/bi047941z. [DOI] [PubMed] [Google Scholar]
- 20.Goddard TD, Kneller DG. Sparky 3. San Francisco: University of California; [Google Scholar]
- 21.Zolnai, Juranic, Vikic-Topic, Macura Quantitative determination of magnetization exchange rate constants from a series of two-dimensional exchange NMR spectra. J. Chem. Inf. Comput. Sci. 2000;40:611–621. doi: 10.1021/ci990107+. [DOI] [PubMed] [Google Scholar]
- 22.Lu J, Ma D, Hu J, Tang W, Zhu D. Nuclear magnetic resonance spectroscopic studies of pyridine methyl derivatives binding to cytochrome c. J. Chem. Soc., Dalton Trans. 1998:2267–2274. [Google Scholar]
- 23.Perrin CL, Dwyer TJ. Application of two-dimensional NMR to kinetics of chemical exchange. Chem. Rev. 1990;90:935–967. [Google Scholar]
- 24.Dalvit C, Pevarello P, Tatò M, Veronesi M, Vulpetti A, Sundström M. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J. Biomol. NMR. 2000;18:65–68. doi: 10.1023/a:1008354229396. [DOI] [PubMed] [Google Scholar]
- 25.Bradshaw JM, Waksman G. Calorimetric Investigation of Proton Linkage by Monitoring both the Enthalpy and Association Constant of Binding: Application to the Interaction of the Src SH2 Domain with a High-Affinity Tyrosyl Phosphopeptide. Biochemistry. 1998;37:15400–15407. doi: 10.1021/bi9814991. [DOI] [PubMed] [Google Scholar]
- 26.Teplyakov A, Obmolova G, Badet-Denisot M-A, Badet B. The mechanism of sugar phosphate isomerization by glucosamine 6-phosphate synthase. Protein Sci. 2008;8:596–602. doi: 10.1110/ps.8.3.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey JM, Fishman PH, Pentchev PG. Anomalous mutarotation of glucose 6-phosphate. An example of intramolecular catalysis. Biochemistry. 1970;9:1189–1194. doi: 10.1021/bi00807a020. [DOI] [PubMed] [Google Scholar]
- 28.Neuberger A, Fletcher AP. Dissociation constants of 2-amino-2-deoxy-D-glucopyranose. J. Chem. Soc. B. 1969:178–181. [Google Scholar]
- 29.Banás P, Walter NG, Sponer J, Otyepka M. Protonation States of the Key Active Site Residues and Structural Dynamics of the glmS Riboswitch As Revealed by Molecular Dynamics. J. Phys. Chem. B. 2010;114:8701–8712. doi: 10.1021/jp9109699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xin Y, Hamelberg D. Deciphering the role of glucosamine-6-phosphate in the riboswitch action of glmS ribozyme. RNA. 2010;16:2455–2463. doi: 10.1261/rna.2334110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blackburn GM, Gait MJ, Loakes D, Williams DM. Nucleic acids in chemistry and biology. 3rd ed. Cambridge, UK: Royal Society of Chemistry; 2006. [Google Scholar]
- 32.Gong B, Chen J-H, Chase E, Chadalavada DM, Yajima R, Golden BL, Bevilacqua PC, Carey PR. Direct Measurement of a pKa near Neutrality for the Catalytic Cytosine in the Genomic HDV Ribozyme Using Raman Crystallography. J. Am. Chem. Soc. 2007;129:13335–13342. doi: 10.1021/ja0743893. [DOI] [PubMed] [Google Scholar]
- 33.Puglisi JD, Wyatt JR, Tinoco I. Solution conformation of an RNA hairpin loop. Biochemistry. 1990;29:4215–4226. doi: 10.1021/bi00469a026. [DOI] [PubMed] [Google Scholar]
- 34.Luptá kA, Ferré-D’Amaré AR, Zhou K, Zilm KW, Doudna JA. Direct pKa Measurement of the Active-Site Cytosine in a Genomic Hepatitis Delta Virus Ribozyme. J. Am. Chem. Soc. 2001;123:8447–8452. doi: 10.1021/ja016091x. [DOI] [PubMed] [Google Scholar]
- 35.Cai Z, Tinoco I. Solution Structure of Loop A from the Hairpin Ribozyme from Tobacco Ringspot Virus Satellite. Biochemistry. 1996;35:6026–6036. doi: 10.1021/bi952985g. [DOI] [PubMed] [Google Scholar]
- 36.Ravindranathan S, Butcher SE, Feigon J. Adenine Protonation in Domain B of the Hairpin Ribozyme. Biochemistry. 2000;39:16026–16032. doi: 10.1021/bi001976r. [DOI] [PubMed] [Google Scholar]
- 37.Guo M, Spitale RC, Volpini R, Krucinska J, Cristalli G, Carey PR, Wedekind JE. Direct Raman Measurement of an Elevated Base pKa in the Active Site of a Small Ribozyme in a Precatalytic Conformation. J. Am. Chem. Soc. 2009;131:12908–12909. doi: 10.1021/ja9060883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rupert PB, Ferre-D’Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 39.Knobloch B, Sigel H, Okruszek A, Sigel RKO. Acid–base properties of the nucleic-acid model 2′-deoxyguanylyl(5′→3′)-2′-deoxy-5′-guanylate, d(pGpG)3–, and of related guanine derivatives. Org. Biomol. Chem. 2006;4:1085. doi: 10.1039/b517904a. [DOI] [PubMed] [Google Scholar]
- 40.Liu L, Cottrell JW, Scott LG, Fedor MJ. Direct measurement of the ionization state of an essential guanine in the hairpin ribozyme. Nat. Chem. Biol. 2009;5:351–357. doi: 10.1038/nchembio.156. [DOI] [PMC free article] [PubMed] [Google Scholar]